Introduction
Epithelial-mesenchymal interactions are important
for tooth development and various molecules, including bone
morphogenetic protein 4 (Bmp4), lymphoid enhancer binding factor 1,
Wnt family member 10A, distal-less homeobox 2 (Dlx2), Dlx5 and msh
homeobox 1 (Msx1) are expressed during these processes and have
been demonstrated to be important components of the tooth
initiation and odontogenic patterning signaling pathways (1–6).
Dlx2 is a member of the vertebrate Dlx gene family, which is
composed of six members organized into three convergent pairs of
genes on chromosome 2 in mice (7,8).
Dlx1 and Dlx2 are expressed in the epithelium and the cranial
neural crest cell (CNCC)-derived mesenchyme of the mandibular and
maxillary processes, and these genes are important for the
development of craniofacial skeleton tissues (9–11).
Previous studies revealed that Dlx2-null and overexpressing mutants
exhibited malformation of craniofacial tissues (11–16).
Thomas et al (17) reported
that the development of maxillary molars required regional
specification of a population of CNCCs by the Dlx1 and Dlx2
homeobox genes, and newborn mice with null mutations in the Dlx1
and Dlx2 genes had no maxillary molars; however, all other teeth
were present (17). This phenotype
may be due to a defect in the mesenchyme whereby odontogenic cells
are reprogrammed to become chondrogenic, resulting in the
replacement of maxillary molar teeth with ectopic cartilage. An
‘odontogenic homeobox code’ for dentition patterning based on the
spatially restricted expression of homeobox genes in the first
branchial arch mesenchyme was previously proposed. It was also
proposed that the Dlx1 and Dlx2 genes were specifically involved in
the pattern of molar tooth development (17). Lézot et al (18,19)
also reported that Dlx2 expression was evident in the molar and
incisor root epithelia during initial root formation and may
constitute a landmark for cementoblast subpopulations of epithelial
origin involved in root morphogenesis and cementogenesis (18,19).
A previous study revealed that the deletion or mutation of Dlx3 may
lead to major dentin defects through changes in the regulation of
dentin sialophosphoprotein (20).
However, it is clear from previous Dlx2-knockout
studies that Dlx2 may contribute to tooth development and whether
Dlx2 overexpression may influence the in vivo phenotypes of
dental structures in mammals remains to be elucidated. The present
study used a transgenic mouse overexpressing Dlx2 in neural crest
cells (NCCs) to determine how Dlx2 overexpression influences dental
and periodontal tissues in mice. This analysis revealed that the
mice exhibited tooth abnormalities, including incisor cross-bite,
shortened tooth roots, increased cementum deposition, periodontal
ligament (PDL) disorganization and osteoporotic alveolar bone.
Materials and methods
Mouse strains
Wnt1-Cre transgenic mice were obtained from the
Jackson Laboratory for Genomic Medicine (Farmington, CT, USA).
Previous studies used Wnt1-Cre mice crossed with Rosa R26R reporter
mice to indicate precisely where and when the Cre recombinase was
active during tooth development, including condensed dental
mesenchyme, dental papilla, dental pulp, odontoblasts, dentine
matrix, cementum and PDL, and used these mice to investigate the
functions of genes in CNCCs during tooth development (2,21).
Transgenic mice conditionally overexpressing Dlx2 (iZEG-Dlx2) were
constructed as described in our previous study (13). Wnt1-Cre transgenic mice were mated
with iZEG-Dlx2 transgenic mice to obtain double transgenic
offspring (Wnt1Cre::iZEG-Dlx2) that specifically overexpressed Dlx2
in tissues derived from NCCs and the mice were genotyped with
polymerase chain reaction (PCR) using primers to detect Cre
recombinase (Cre) and enhanced green fluorescent protein (EGFP), as
described in our previous study (13). Mice (including male and female
mice) from a C57BL/6J genetic background were used in the current
study and non-recombined littermates were used as controls. All
mice were housed in a specific pathogen-free laboratory animal room
at a temperature of 22°C. The light cycle consisted of 12 h light
and 12 h dark. The animal experimental procedures were performed in
compliance with the guidelines of the Institutional Animal Care and
Use Committees of the Shanghai Ninth People's Hospital (Shanghai,
China) and were approved by the Institutional Animal Care and Use
Committees of the Shanghai Ninth People's Hospital (Shanghai,
China).
Tooth preparation and
measurements
Adult (P90) control (n=6) and Wnt1Cre::iZEG-Dlx2
(n=6) mice (body weight, 25.2–28.3 g) were sacrificed using 0.8%
pentobarbital sodium via intraperitoneal injection (100 ml/10 g
body weight), skinned and eviscerated, then transferred to 95%
ethanol for 2 days. The skulls of mice were then stained with
Alcian blue and Alizarin red as previously described (13). The stained teeth were then
separated from the alveolar bone under an integrated microscope and
were transferred to a solution of 50% glycerol and 50% water for
imaging.
The root lengths and the ratios of crown/root length
in both the maxillary and mandibular first molars in six control
and Wnt1Cre::iZEG-Dlx2 mice (twelve teeth respectively) were
quantified using digital hand calipers, and all were performed in
triplicate.
Micro-computed tomography (CT)
scans
The skulls of P90 iZEG-Dlx2 and Wnt1Cre::iZEG-Dlx2
mice were collected and fixed with 4% paraformaldehyde (PFA).
Subsequently, micro-CT data were collected using an eXplore Locus
MicroCT scanner (GE Healthcare Life Sciences, Milwaukee, WI, USA),
using 0.01-mm-thick slices. The 3D reconstructions of the skulls
and bone mineral density calculations were completed using GE
MicroView software version 2.2 (GE Healthcare Life Sciences).
Histological analysis
Whole heads of embryonic day 13.5 (E13.5) iZEG-Dlx2
and Wnt1Cre::iZEG-Dlx2 mice (n=6) obtained from pregnant mice
following anesthetization with 0.8% pentobarbital sodium via
intraperitoneal injection (100 ml/10 g body weight). Jaws of P90
iZEG-Dlx2 and Wnt1Cre::iZEG-Dlx2 mice (n=6) were obtained following
anesthetization with 0.8% pentobarbital sodium via intraperitoneal
injection (100 ml/10 g body weight) and were dissected and fixed in
4% PFA. P90 jaws were subsequently demineralized in 0.5 M EDTA. The
tissues were embedded in paraffin, and 5-µm tissue sections were
cut, stained with hematoxylin and eosin (H&E) and mounted with
resinous mounting medium.
Analysis of cell proliferation and
apoptosis
A total of 10 mg/ml 5-bromo-20-deoxyuridine (BrdU;
Sigma-Aldrich; Merck Millipore, Darmstadt, Germany) solution was
injected intraperitoneally at a dose of 100 µg/g body weight into
pregnant mice (n=3) at E13.5. The mice were sacrificed by
anesthetization with 0.8% pentobarbital sodium via intraperitoneal
injecion (100 ml/10 g body weight) 2 h after BrdU injection and the
embryos were fixed using 4% PFA, embedded in paraffin and cut into
5-µm-thick sections. Subsequently, BrdU-labeled cells were detected
using immunofluorescence staining. Terminal
deoxynucleotidyl-transferase-mediated dUTP nick end labeling
(TUNEL) assays were used to determine apoptosis in the E13.5 mice
and the FITC In Situ Cell Death Detection kit (KeyGen
Biotech Co., Ltd, Nanjing, China) was used according to the
manufacturer's protocol.
Immunohistochemistry
Heads (E10.5) of iZEG-Dlx2 and Wnt1Cre::iZEG-Dlx2
mice (n=6) obtained from pregnant mice, and jaws of P90 iZEG-Dlx2
and Wnt1Cre::iZEG-Dlx2 mice (n=6) were obtained after sacrifice
with 0.8% pentobarbital sodium via intraperitoneal injection (100
ml/10 g body weight), following sacrifice with 0.8% pentobarbital
sodium via intraperitoneal injection (100 ml/10 g body weight and
were embedded in paraffin and sectioned at a thickness of 5 µm. A
bone antigen restoration liquid kit (Sunteam Biotech, Shanghai,
China) was used for antigen retrieval. Slides were then washed with
PBS and blocked for 60 min with 3% bovine serum albumin (Gibco;
Thermo Fisher Scientific, Inc., Waltham, MA, USA) in PBS containing
0.2% Triton X-100. The sections were subsequently incubated with
primary antibodies for anti-Dlx2 (1:100; cat. no. ab18188; Abcam,
Cambridge, UK), anti-osteopontin (1:100; cat. no. ab8448; Abcam),
anti-BrdU (1:200; cat. no. ab6326; Abcam), anti-Msx2 (1:150; cat.
no. ab69058; Abcam), anti-SMAD family member 4 (Smad4; 1:100; cat.
no. BS2050; Bioworld Technology, Inc., St. Louis Park, MN, USA),
sex determining region Y-box 9 (Sox9; 1:100; cat. no. BS1597;
Bioworld Technology, Inc.), anti-transforming growth factor β
receptor 1 (TGFβR1)/TGFβR2 (1:100; cat. nos. BS3257 and BS1696,
respectively; Bioworld Technology, Inc.) or anti-runt related
transcription factor 2 (Runx2; 1:100; cat. no. MAB2006; R&D
Systems, Inc., Minneapolis, MN, USA) overnight at 4°C. The donkey
anti-rabbit AlexaFluor 488 and donkey anti-rat AlexaFluor 568 (cat.
nos. A21206 and A11077, respectively; Jackson ImmunoResearch
Laboratories, Inc., West Grove, PA, USA) secondary antibodies were
diluted at 1:300 and incubated with sections for 60 min at room
temperature. Finally, slides were mounted with Vectashield mounting
medium containing DAPI (Invitrogen; Thermo Fisher Scientific, Inc.)
and images were captured using a fluorescence microscope.
Statistical analysis
Data are presented as the mean ± standard deviation.
The differences between experimental and control groups were
compared with an independent Student's t-test using SPSS version
18.0 (SPSS, Inc., Chicago, IL, USA). P<0.05 was considered to
indicate a statistically significant difference.
Results
Overexpression of Dlx2 in the teeth of
Wnt1-Cre::iZEG-Dlx2 double transgenic mice
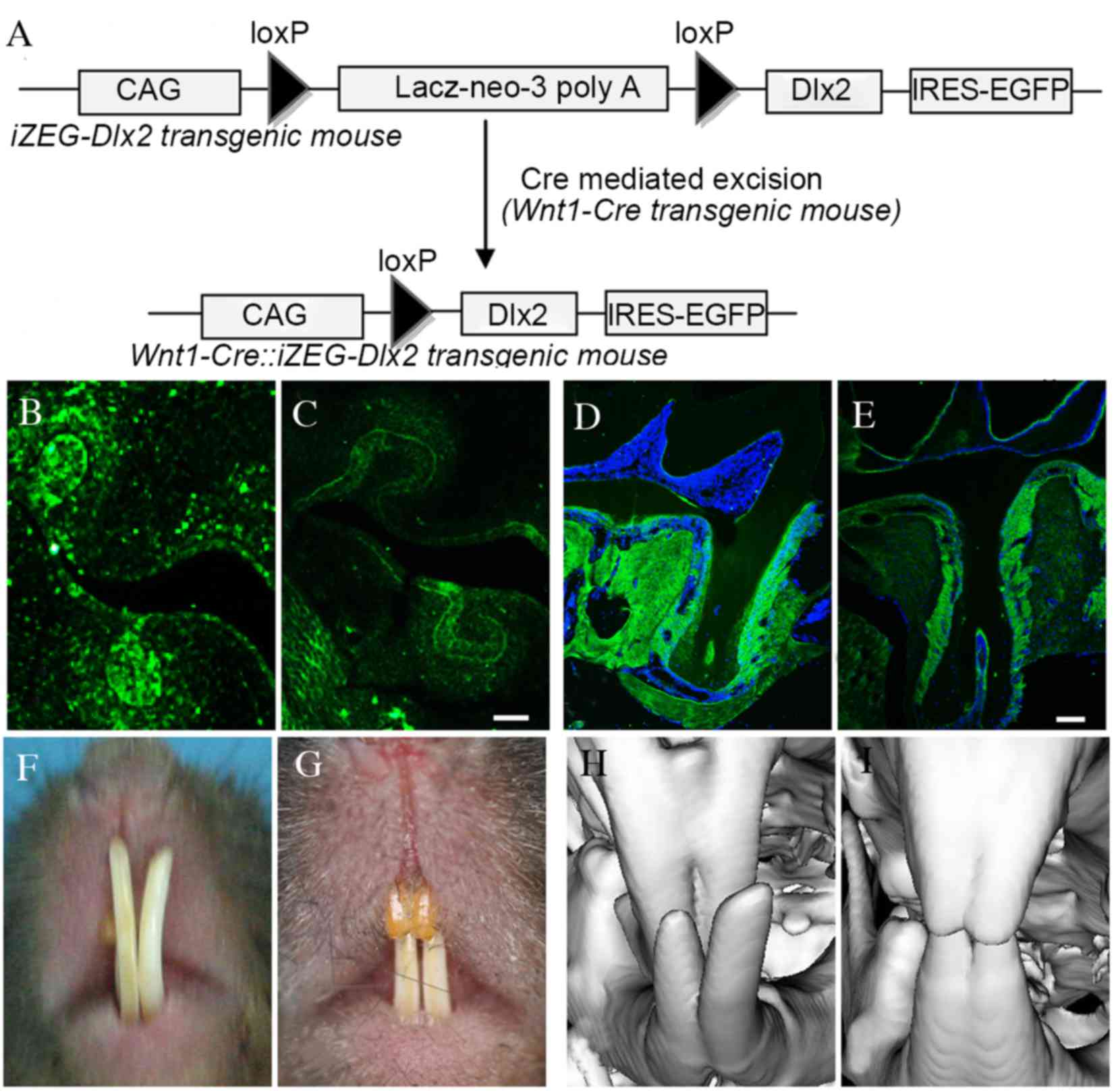
A schematic diagram of the Wnt1-Cre::iZEG-Dlx2
transgene is presented in Fig. 1A,
and the mice were genotyped via PCR using primers specific for EGFP
and Cre. Cre-mediated NCC-specific recombination was also detected
by PCR analysis using a pair of primers spanning the CAG promoter
and the Dlx2 coding sequence (CAG-Dlx2 primers), as previously
described (13).
Immunofluorescence confirmed that Dlx2 expression was higher in the
NCCs derivative tissues, including periodontal tissue, cementum and
alveolar bone in embryonic and adult Wnt1Cre::iZEG-Dlx2 mice
(Fig. 1B-E) when compared with
iZEG-Dlx2 control mice, whereas Dlx2 expression exhibited similar
level in others tissues, including long bone and liver, between the
Wnt1Cre::iZEG-Dlx2 mice and iZEG-Dlx2 control mice.
Overexpression of Dlx2 leads to tooth
dysmorphia
P90 Wnt1Cre::iZEG-Dlx2 mice exhibited cross-bite
malocclusion (Fig. 1F-I). General
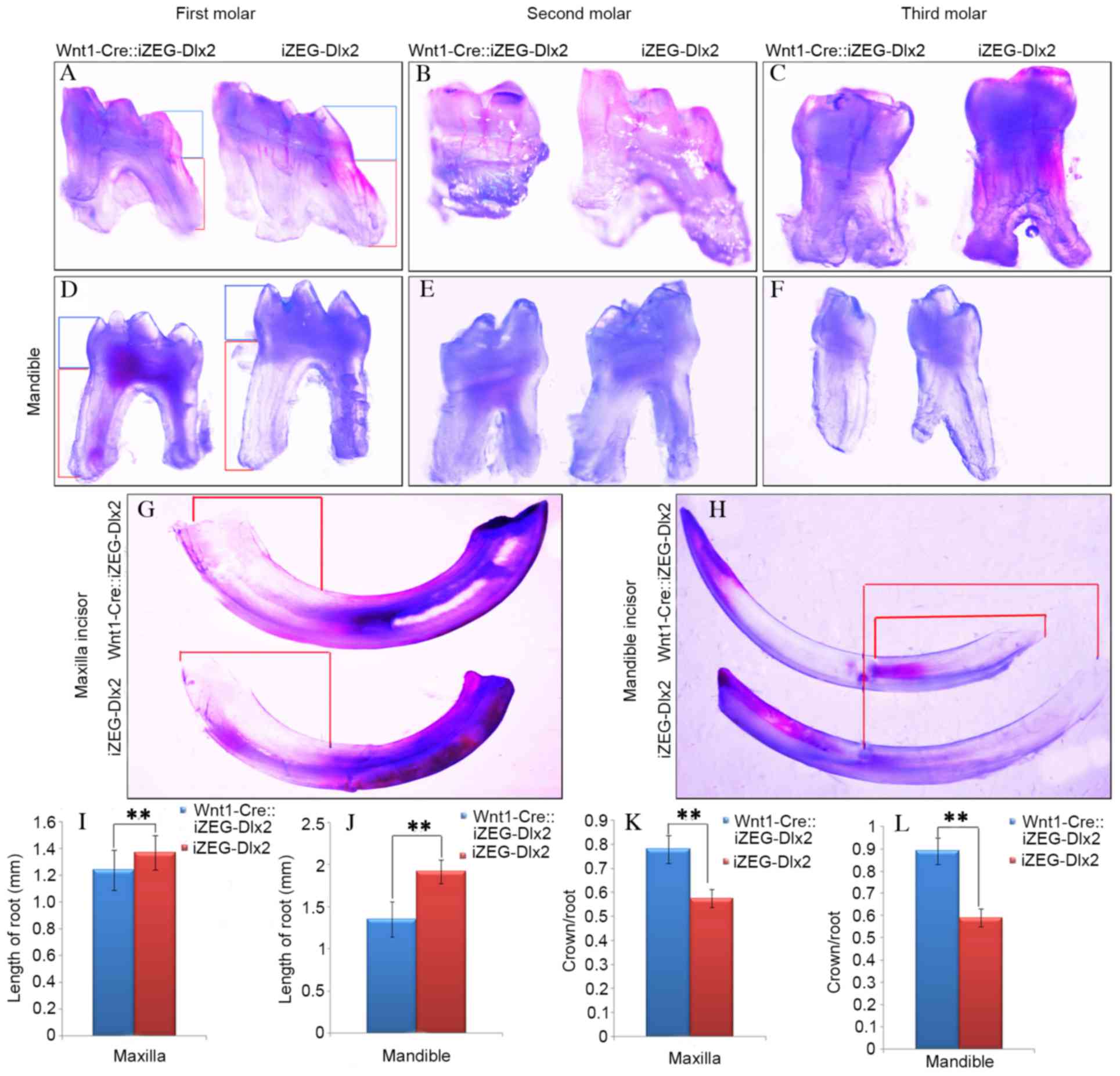
observation under an integrated microscope demonstrated that the
mandibular and maxillary teeth exhibited shortened root lengths and
root morphology dysmorphia (Fig.
2A-H). Measurements of teeth from Wnt1Cre::iZEG-Dlx2 mice and
iZEG-Dlx2 mice revealed that root length was significantly reduced
(P<0.01; Fig. 2I and J) and the
ratios of crown/root length were significantly increased
(P<0.01; Fig. 2K and L) in both
the maxillary and mandibular first molars in Wnt1Cre::iZEG-Dlx2
mice compared with iZEG-Dlx2 mice.
Overexpression of Dlx2 leads to
increased cementum deposition and osteoporotic alveolar bone
The present study performed detailed analyses of the
maxillary first molar of Wnt1Cre::iZEG-Dlx2 mice at P90. H&E
staining of sagittal sections revealed bone loss and bone defects
in the interdental septum and root furcation region in alveolar
bone compared with control iZEG-Dlx2 mice (Fig. 3A-H). Notably, increased acellular
cementum deposition in Wnt1Cre::iZEG-Dlx2 mice was observed and the
molars had sparse and disorganized PDL compared with iZEG-Dlx2
mice. Although the alveolar bone-PDL attachments were defective in
Wnt1Cre::iZEG-Dlx2 molars, the cementum-PDL attachments had a
normal appearance (Fig. 3E and F).
Immunofluorescence results confirmed that the deposition of
osteopontin, an osteogenic marker, on the alveolar bone and
periodontal tissues was reduced in Wnt1Cre::iZEG-Dlx2 mice compared
with iZEG-Dlx2 mice (Fig. 3I and
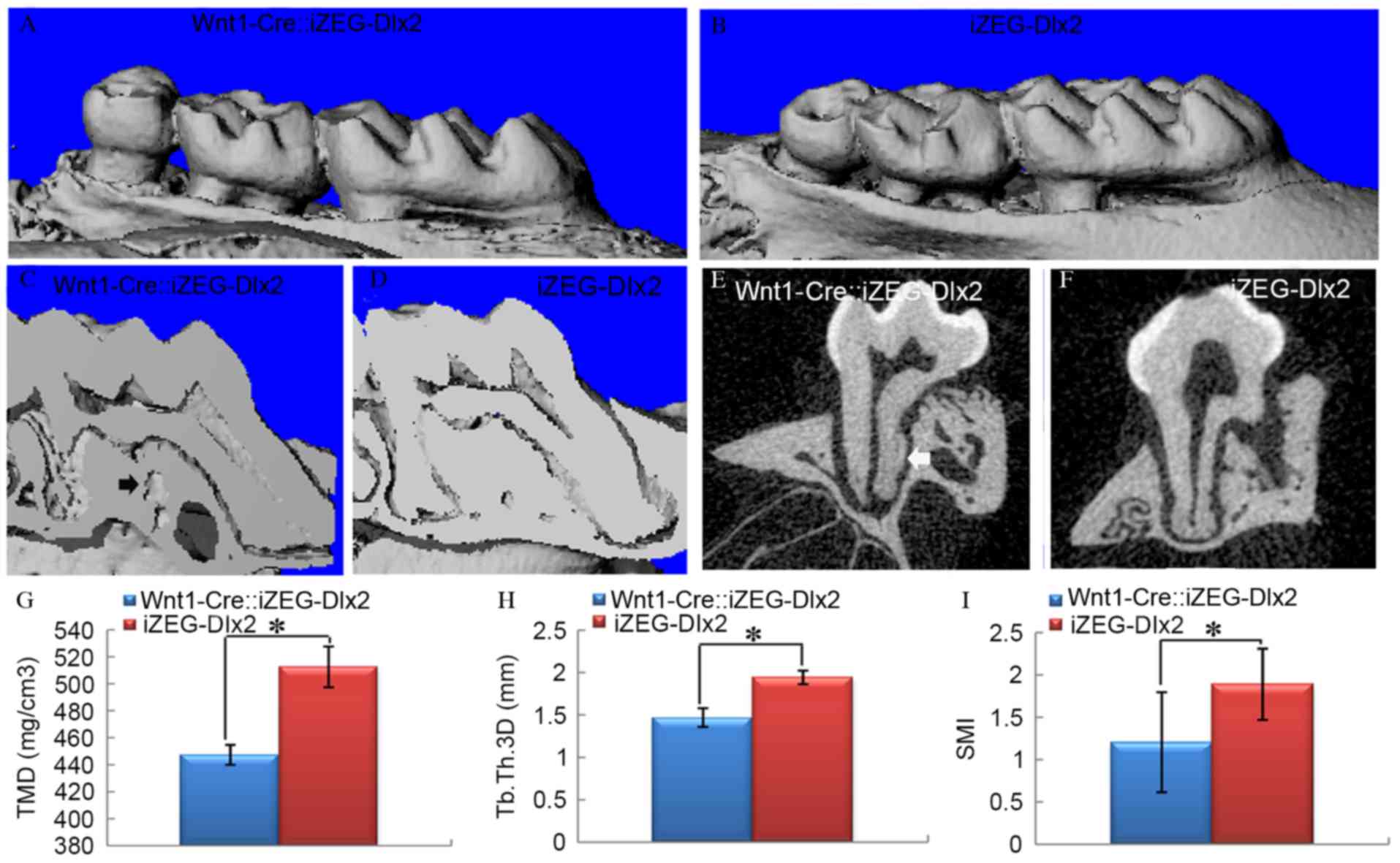
J). The 3D reconstructed images from micro-CT analysis at P90
also confirmed these results (Fig.
4A-F). At P90, tissue mineral density (TMD), trabecular
thickness 3D (Tb.Th 3D) and structure model index (SMI) were
quantified using micro-CT examination, which indicated a
significant reduction in TMD and Tb.Th 3D (P<0.05; Fig. 4G and H), and a significant increase
in SMI (P<0.05; Fig. 4I) in the
surrounding alveolar bone of Wnt1Cre::iZEG-Dlx2 mice compared with
iZEG-Dlx2 mice (Fig. 4), which
clearly demonstrated the osteoporosis of the alveolar bone
tissue.
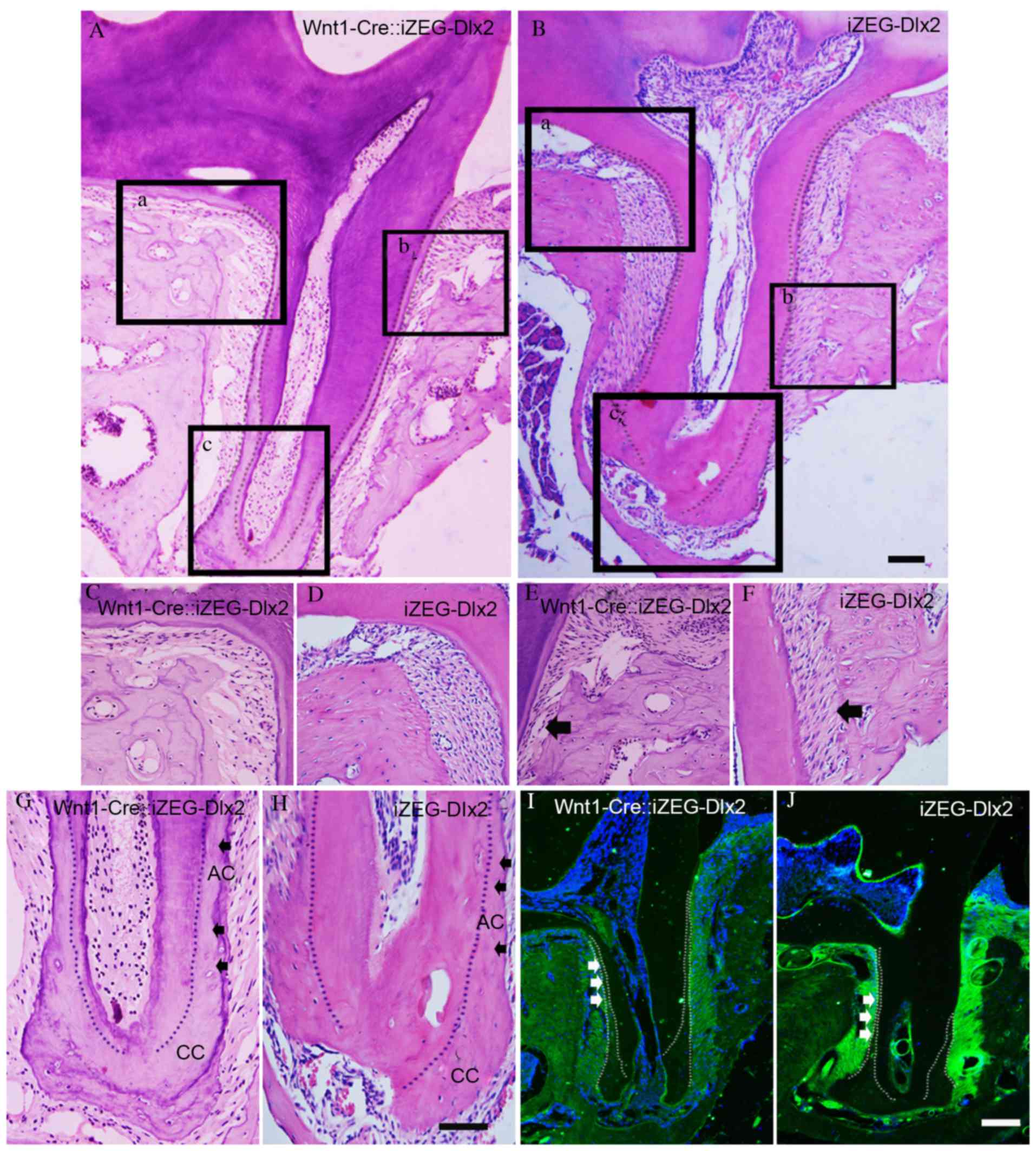 | Figure 3.Overexpression of Dlx2 led to
increased deposition of cementum and osteoporotic alveolar bone.
Hematoxylin and eosin staining of sagittal sections revealed
osteoporosis in alveolar bone tissue, increased acellular cementum
deposition and defective alveolar bone-PDL attachment in the
maxillary first molars of (A) P90 adult Wnt1-Cre::iZEG-Dlx2 mice
and (B) iZEG-Dlx2 mice (Scale bars, 200 µm). Bone resorption in the
root furcation region in alveolar bone of (C) P90 adult
Wnt1-Cre::iZEG-Dlx2 mice and (D) iZEG-Dlx2 mice. Bone resorption
and defective alveolar bone-PDL attachment (arrow) in the
interdental septum of (E) P90 Wnt1-Cre::iZEG-Dlx2 mice and (F)
iZEG-Dlx2 mice. Increased acellular cementum and normal cellular
cementum deposition (arrow) in the maxillary first molars of (G)
Wnt1-Cre::iZEG-Dlx2 mice and (H) iZEG-Dlx2 mice (Scale bars, 50
µm). The areas presented in (C-H) are higher magnification images
corresponding to the boxed regions in A (a, b, c) and B (a, b, c),
respectively. Immunofluorescence staining revealed the decreased
deposition of osteopontin on the alveolar bone and periodontal
tissues of (I) Wnt1-Cre::iZEG-Dlx2 mice and (J) iZEG-Dlx2 mice
(Scale bars, 100 µm). PDL, periodontal ligament, AC, acellular
cementum; CC, cellular cementum; Dlx2, distal-less homeobox 2;
iZEG-Dlx2, transgenic mice conditionally overexpressing Dlx2;
Wnt1-Cre::iZEG-Dlx2, double transgenic iZEG-Dlx2 mice expressing
Wnt1-Cre. |
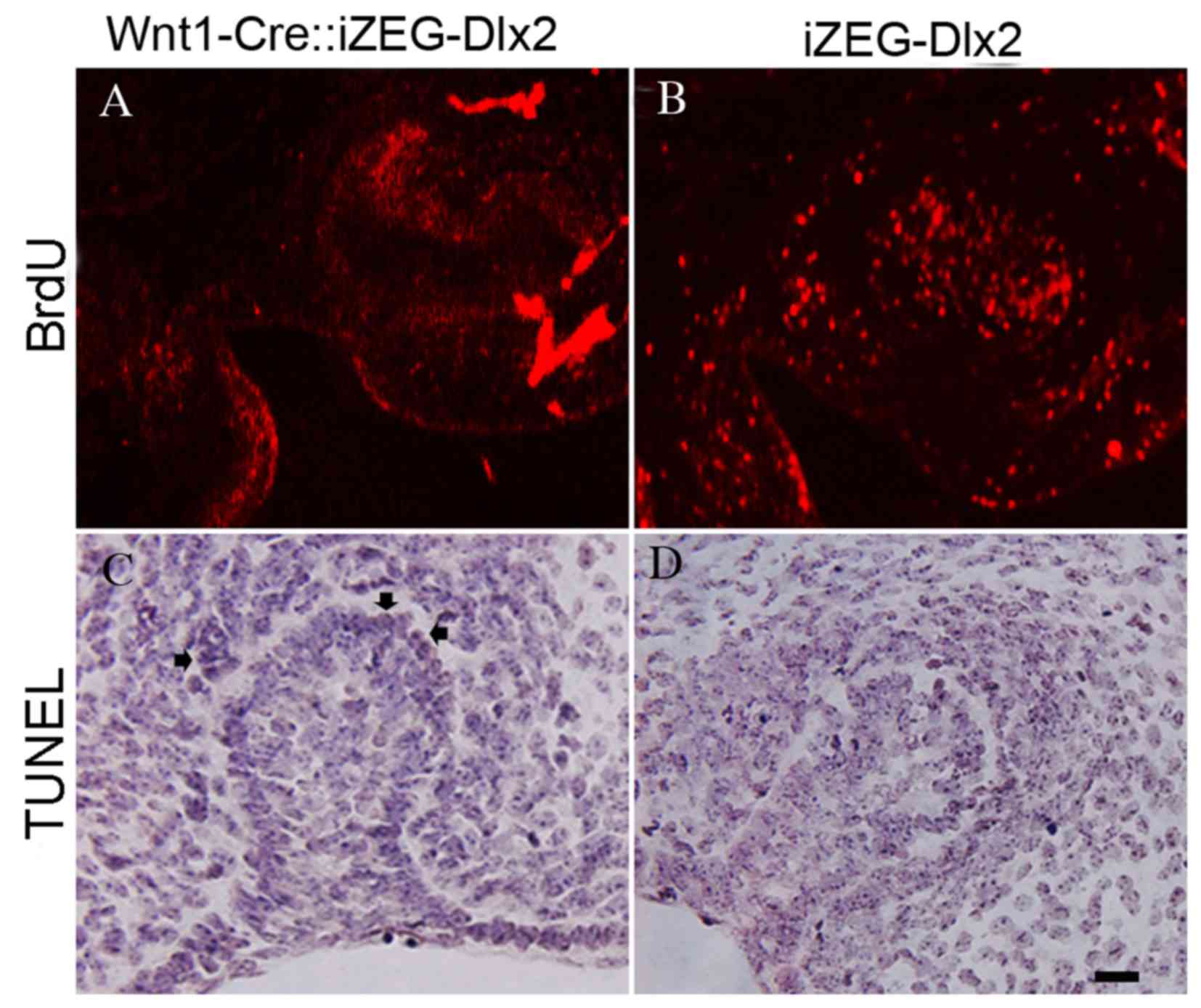
Reduced cellular proliferation and
increased apoptosis within the dental germ and alveolar bone of
E13.5 Wnt1Cre::iZEG-Dlx2 embryos
A marked decrease in cellular proliferation was
detected using BrdU-labeling of the dental germ and alveolar bone
in E13.5 Wnt1Cre::iZEG-Dlx2 embryos (Fig. 5A and B). TUNEL assays revealed
increased apoptosis in the dental germ and alveolar bone in E13.5
Wnt1Cre::iZEG-Dlx2 embryos compared with iZEG-Dlx2 embryos
(Fig. 5C and D).
Expression of genes associated with
tooth development in Wnt1Cre::iZEG-Dlx2 mice
To identify whether the expression of certain genes
that are involved in tooth development was altered by Dlx2
overexpression, the present study used immunofluorescence staining
to detect the expression levels of TGFβR1, TGFβR2, Smad4, Msx2,
Sox9 and Runx2. The expression levels of TGFβR1, TGFβR2, Smad4 and
Msx2, which have previously been demonstrated to contribute to
tooth development, were upregulated in the dental germ of E13.5
Wnt1Cre::iZEG-Dlx2 embryos (Fig.
6A-H). Msx2 was also upregulated in the epithelium (Fig. 6G and H). A previous study revealed
that a null mutation of Dlx2 may lead to the reprogramming of
odontogenic cells to become chondrogenic and express Sox9 (17). The present study revealed that
Dlx2-overexpression may also increase Sox9 expression in the dental
germ of E13.5 Wnt1Cre::iZEG-Dlx2 mice (Fig. 6I and J). By contrast, the
expression of Runx2, an osteogenic and odontogenic marker, was
downregulated in the dental germ and alveolar bone of E13.5
Wnt1Cre::iZEG-Dlx2 mice compared with iZEG-Dlx2 (Fig. 6K and L).
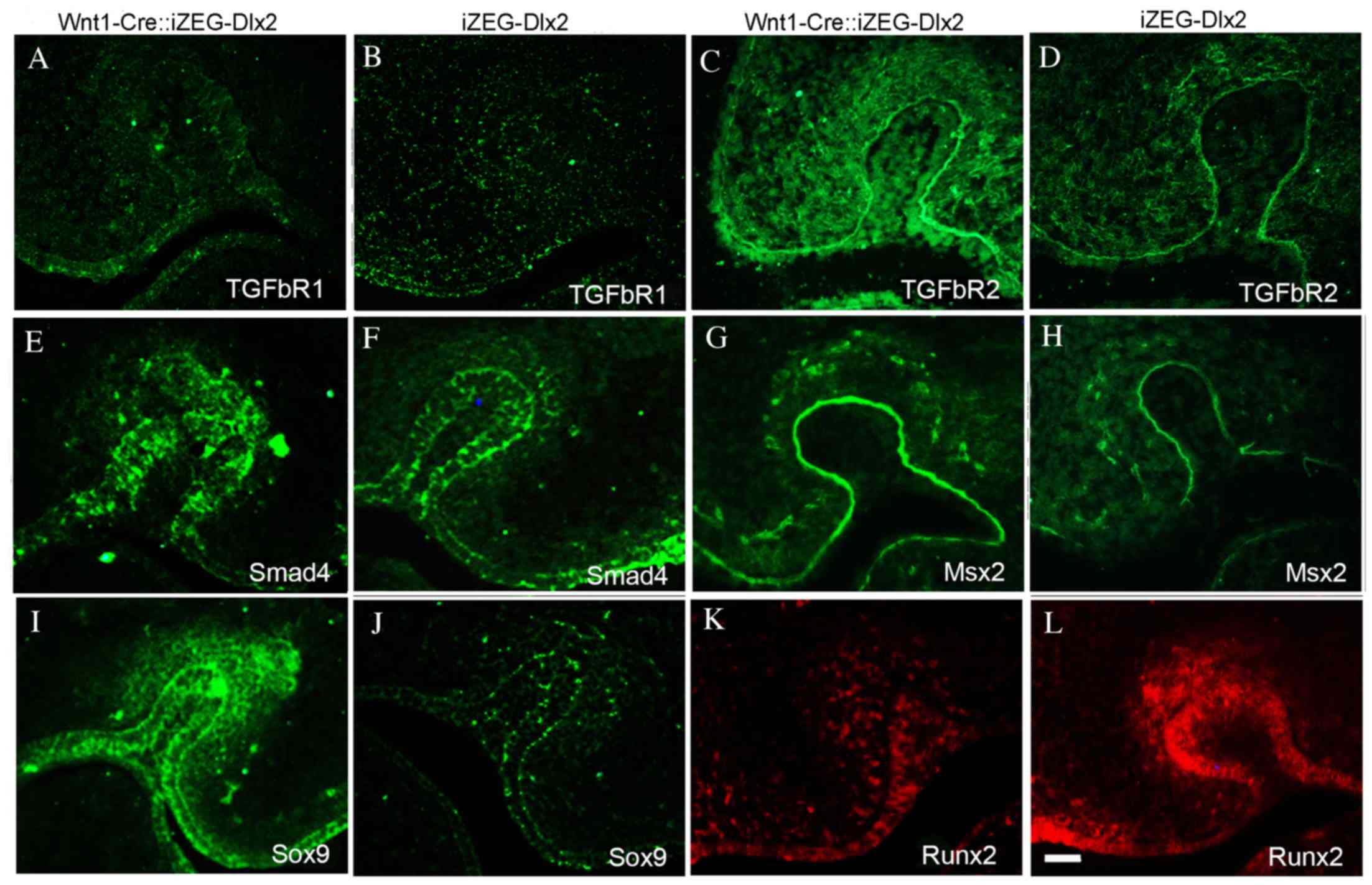 | Figure 6.Expression of genes implicated in
tooth development in Wnt1Cre::iZEG-Dlx2 mice. Immunofluorescence
staining demonstrated enhanced TGFβR1 protein expression in the
tooth region of (A) E13.5 Wnt1Cre::iZEG-Dlx2 compared with (B)
iZEG-Dlx2 embryos. TGFβR2 protein expression is significantly
upregulated in the tooth region of (C) E13.5 Wnt1Cre::iZEG-Dlx2
compared with (D) iZEG-Dlx2 embryos. Smad4 protein expression was
upregulated in the tooth region of (E) E13.5 Wnt1Cre::iZEG-Dlx2 and
(F) iZEG-Dlx2 embryos. Msx2 was upregulated in the dental germ,
especially in the epithelium of (G) E13.5 Wnt1Cre::iZEG-Dlx2
compared with (H) iZEG-Dlx2 embryos. Sox9 expression was
upregulated in the dental germ of (I) E13.5 Wnt1Cre::iZEG-Dlx2
compared with (J) iZEG-Dlx2 embryos. Runx2 expression was
downregulated in the dental germ of (K) E13.5 Wnt1Cre::iZEG-Dlx2
compared with (L) iZEG-Dlx2 embryos. Scale bars, 100 µm. E13.5,
embryonic day 13.5; Dlx2, distal-less homeobox 2;
Wnt1-Cre::iZEG-Dlx2, double transgenic iZEG-Dlx2 mice expressing
Wnt1-Cre; iZEG-Dlx2, transgenic mice conditionally overexpressing
Dlx2; TGFβR1/2, transforming growth factor β receptor 1/2; Smad4,
SMAD family member 4; Msx2, msh homeobox 2; Sox9, SRY (sex
determining region Y)-box 9; Runx2, runt related transcription
factor 2. |
Discussion
To the best of our knowledge the present study is
the first to describe the effects of Dlx2 overexpression on the
formation of dental and periodontal tissues. The current findings
suggest that the Dlx2 may be involved in the control of cementum
formation. Previous studies revealed that during root cementum
formation, cementoblasts exhibited similar characteristics of bone
formation to osteoblasts and contribute to matrix deposition in
cooperation with periodontal ligament cells (22,23).
Molar roots contain two types of cementum: The acellular cementum
that is initially deposited, and the cellular cementum that
progressively increases in thickness towards the root apex. By
contrast, continuously erupted incisors only have acellular
cementum (22). A previous study
demonstrated that Dlx2 may be continuously present in the
epithelial root sheath, from tooth initiation and morphogenesis to
the last stages of dental tissue formation and implied that Dlx2
may be involved in the control of cementogenesis (19). The present study determined that
Dlx2 overexpression may result in an increased deposition of
acellular cementum, indicating that Dlx2 is crucial for the
regulation of cementum formation.
Dlx1 and Dlx2 were the first genes to be identified
to contribute to odontogenic patterning and their null mutations
solely disrupt the development of the maxillary molar, indicating
that there are distinct genetic pathways directing the development
of different teeth (17,24). However, the present study revealed
that the overexpression of Dlx2 disrupted cementogenesis in
maxillary and mandibular molars and incisors. It is possible that
the expression of other Dlx genes, such as Dlx5 and Dlx1 may
compensate for the absence of Dlx2; however, however that cannot
inhibit the effects of Dlx2 overexpression. Unlike cementum
deposition, Dlx2 overexpression may lead to osteoporotic alveolar
bone and downregulation of Runx2 expression, which is consistent
with our previous study, which determined that Dlx2 overexpression
may impair craniofacial bone development (13). One potential explanation for this
discrepancy is that alveolar bone and cementum develop via
different molecular mechanisms.
Previous studies have revealed that the development
of maxillary molars requires the regional specification of a
population of CNCCs by Dlx1 and Dlx2, indicating that the ectoderm
mesenchyme may influence tooth morphogenesis (17,24).
In the current study, the overexpression of Dlx2 in the neural
crest led to disrupted tooth development, confirming this model.
Notably, the present study identified that the overexpression of
Dlx2 resulted in an ectopic patch of Sox9-expressing cells in the
dental germ, whereas a previous study revealed that a null mutation
of Dlx2 may also reprogram odontogenic cells to express Sox9
(17). The upregulation and
downregulation of Dlx2 expression have similar effects on the
expression of Sox9, and the mechanism that leads to these
expression patterns remains to be elucidated.
Mice overexpressing Dlx2 exhibited short molar tooth
roots, and certain maxillary and mandibular incisors had long
crowns and an over-cross bite or curved shape. Potentially, the
continuously erupting mouse incisors may have acellular cementum on
the lingual root analog, revealing a weaker effect of Dlx2
overexpression when compared with cellular cementum. Additionally,
due to the deviation of the maxilla, no natural tooth wear was
observed through the consumption of hard foodstuffs and gnawing
behavior.
Numerous genes have been reported to be involved in
tooth development, including Bmp4, Msx2 and Dlx5, which have also
been demonstrated to interact with Dlx2 (1,25).
The immunofluorescence results in the present study revealed that
Dlx2 overexpression may alter the expression of TGFβR1, TGFβR2,
Smad4 and Msx2, the four genes that are crucial for tooth
development (1,26,27),
in tooth tissue, indicating that Dlx2 may disrupt tooth development
by interaction with these genes. However, there are various
candidate genes that may interact with Dlx2 to regulate tooth
development, the exact molecular associations among these genes
remain to be elucidated, and require investigation in the future
using bioinformatics analysis and experimental verification of
their functions.
In conclusion the present study demonstrated that
Dlx2 overexpression in mouse CNCCs resulted in incisor cross-bite,
shortened tooth roots, increased deposition of cementum,
periodontal ligament disorganization and osteoporotic alveolar
bone. Additionally, Dlx2 overexpression increased the expression
levels of odontogenesis-associated genes, including TGFβR1, TGFβR2,
Smad4, Msx2 and Sox9 in tooth regions. Therefore, the present study
suggested that Dlx2 overexpression may alter the alveolar bone,
cementum and periodontal ligament phenotypes in mice. It is of note
that that the present study was limited in that the dental and
periodontal phenotypes were assessed in adult mice, whereas the
genetic modification usually occurs during embryogenesis; thus, the
primary effects of Dlx2 overexpression may occur early in
development. Therefore, it is unclear whether the phenotypes
observed were due to developmental defects or postnatal degradation
secondary to craniofacial deformity and malnutrition. The exact
molecular mechanisms underlying the effects of Dlx2 overexpression
on dental and periodontal tissue phenotypes require further
investigated in the future.
Acknowledgements
The authors thank Professor Rulang Jiang (Division
of Developmental Biology and Plastic Surgery, Cincinnati Children's
Hospital Medical Center, Cincinnati, OH, USA) for his advice in
experimental design and biological techniques, and Professor John
L.R. Rubenstein (University of California San Francisco, San
Francisco, CA, USA) for providing the pCAGGS-Dlx2 plasmid. The
current study was supported by National Nature Science Foundation
of China (grant nos. 81300842 and 81271122), Open Foundation of
Beijing Key Laboratory of Tooth Regeneration and Function
Reconstruction and Program for Innovation Research Team of Shanghai
Municipal Education Commission.
References
|
1
|
Saadi I, Das P, Zhao M, Raj L, Ruspita I,
Xia Y, Papaioannou VE and Bei M: Msx1 and Tbx2 antagonistically
regulate Bmp4 expression during the bud-to-cap stage transition in
tooth development. Development. 140:2697–2702. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jia S, Zhou J, Gao Y, Baek JA, Martin JF,
Lan Y and Jiang R: Roles of Bmp4 during tooth morphogenesis and
sequential tooth formation. Development. 140:423–432. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhang Z, Lan Y, Chai Y and Jiang R:
Antagonistic actions of Msx1 and Osr2 pattern mammalian teeth into
a single row. Science. 323:1232–1234. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhao Z, Stock D, Buchanan A and Weiss K:
Expression of Dlx genes during the development of the murine
dentition. Dev Genes Evol. 210:270–275. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fujimori S, Novak H, Weissenböck M,
Jussila M, Gonçalves A, Zeller R, Galloway J, Thesleff I and
Hartmann C: Wnt/β-catenin signaling in the dental mesenchyme
regulates incisor development by regulating Bmp4. Dev Biol.
348:97–106. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sasaki H, Muramatsu T, Kwon HJ, Yamamoto
H, Hashimoto S, Jung HS and Shimono M: Down-regulated genes in
mouse dental papillae and pulp. J Dent Res. 89:679–683. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ozcelik T, Porteus MH, Rubenstein JL and
Francke U: DLX2 (TES1), a homeobox gene of the Distal-less family,
assigned to conserved regions on human and mouse chromosomes 2.
Genomics. 13:1157–1161. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Stock DW, Ellies DL, Zhao Z, Ekker M,
Ruddle FH and Weiss KM: The evolution of the vertebrate Dlx gene
family. Proc Natl Acad Sci USA. 93:10858–10863. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Qiu M, Bulfone A, Ghattas I, Meneses JJ,
Christensen L, Sharpe PT, Presley R, Pedersen RA and Rubenstein JL:
Role of the Dlx homeobox genes in proximodistal patterning of the
branchial arches: Mutations of Dlx-1, Dlx-2, and Dlx-1 and −2 alter
morphogenesis of proximal skeletal and soft tissue structures
derived from the first and second arches. Dev Biol. 185:165–184.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Eisenstat DD, Liu JK, Mione M, Zhong W, Yu
G, Anderson SA, Ghattas I, Puelles L and Rubenstein JL: DLX-1,
DLX-2, and DLX-5 expression define distinct stages of basal
forebrain differentiation. J Comp Neurol. 414:217–237. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Depew MJ, Simpson CA, Morasso M and
Rubenstein JL: Reassessing the Dlx code: The genetic regulation of
branchial arch skeletal pattern and development. J Anat.
207:501–561. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Qiu M, Bulfone A, Martinez S, Meneses JJ,
Shimamura K, Pedersen RA and Rubenstein JL: Null mutation of Dlx-2
results in abnormal morphogenesis of proximal first and second
branchial arch derivatives and abnormal differentiation in the
forebrain. Genes Dev. 9:2523–2538. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Dai J, Kuang Y, Fang B, Gong H, Lu S, Mou
Z, Sun H, Dong Y, Lu J, Zhang W, et al: The effect of
overexpression of Dlx2 on the migration, proliferation and
osteogenic differentiation of cranial neural crest stem cells.
Biomaterials. 34:1898–1910. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
McKeown SJ, Newgreen DF and Farlie PG:
Dlx2 over-expression regulates cell adhesion and mesenchymal
condensation in ectomesenchyme. Dev Biol. 281:22–37. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gordon CT, Brinas IM, Rodda FA, Bendall AJ
and Farlie PG: Role of Dlx genes in craniofacial morphogenesis:
Dlx2 influences skeletal patterning by inducing ectomesenchymal
aggregation in ovo. Evol Dev. 12:459–473. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jeong J, Cesario J, Zhao Y, Burns L,
Westphal H and Rubenstein JL: Cleft palate defect of Dlx1/2-/−
mutant mice is caused by lack of vertical outgrowth in the
posterior palate. Dev Dyn. 241:1757–1769. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Thomas BL, Tucker AS, Qui M, Ferguson CA,
Hardcastle Z, Rubenstein JL and Sharpe PT: Role of Dlx-1 and Dlx-2
genes in patterning of the murine dentition. Development.
124:4811–4818. 1997.PubMed/NCBI
|
|
18
|
Lezot F, Thomas B, Greene SR, Hotton D,
Yuan ZA, Castaneda B, Bolaños A, Depew M, Sharpe P, Gibson CW and
Berdal A: Physiological implications of DLX homeoproteins in enamel
formation. J Cell Physiol. 216:688–697. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lezot F, Davideau JL, Thomas B, Sharpe P,
Forest N and Berdal A: Epithelial Dlx-2 homeogene expression and
cementogenesis. J Histochem Cytochem. 48:277–284. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Duverger O, Zah A, Isaac J, Sun HW,
Bartels AK, Lian JB, Berdal A, Hwang J and Morasso MI: Neural crest
deletion of Dlx3 leads to major dentin defects through
down-regulation of Dspp. J Biol Chem. 287:12230–12240. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chai Y, Jiang X, Ito Y, Bringas P Jr, Han
J, Rowitch DH, Soriano P, McMahon AP and Sucov HM: Fate of the
mammalian cranial neural crest during tooth and mandibular
morphogenesis. Development. 127:1671–1679. 2000.PubMed/NCBI
|
|
22
|
Foster BL, Soenjaya Y, Nociti FH Jr, Holm
E, Zerfas PM, Wimer HF, Holdsworth DW, Aubin JE, Hunter GK,
Goldberg HA and Somerman MJ: Deficiency in acellular cementum and
periodontal attachment in bsp null mice. J Dent Res. 92:166–172.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Fong H, Chu EY, Tompkins KA, Foster BL,
Sitara D, Lanske B and Somerman MJ: Aberrant cementum phenotype
associated with the hypophosphatemic hyp mouse. J Periodontol.
80:1348–1354. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Thomas BL and Sharpe PT: Patterning of the
murine dentition by homeobox genes. Eur J Oral Sci. 106:(Suppl 1).
S48–S54. 1998. View Article : Google Scholar
|
|
25
|
Thomas BL, Liu JK, Rubenstein JL and
Sharpe PT: Independent regulation of Dlx2 expression in the
epithelium and mesenchyme of the first branchial arch. Development.
127:217–224. 2000.PubMed/NCBI
|
|
26
|
Zhao H, Oka K, Bringas P, Kaartinen V and
Chai Y: TGF-beta type I receptor Alk5 regulates tooth initiation
and mandible patterning in a type II receptor-independent manner.
Dev Biol. 320:19–29. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Huang XF and Chai Y: Molecular regulatory
mechanism of tooth root development. Int J Oral Sci. 4:177–181.
2012. View Article : Google Scholar : PubMed/NCBI
|