Introduction
Hepatocellular carcinoma (HCC) is the fifth most
common cancer and the most frequent liver cancer globally (1). The majority of HCC cases occur in
cirrhotic liver, and the primary risk factors are chronic hepatitis
B virus or chronic hepatitis C virus (HCV) infection, which account
for almost all HCC cases (2). The
incidence of HCC varies between different geographical areas;
however, it is increasing globally, particularly in Asia, with 6–11
per 100,000 people with the disease (3,4). A
study of HCC epidemiology in Germany indicated that, despite the
availability of various advanced chemotherapies and radiotherapies,
including chemoembolization with drug-eluting beads, sorafenib and
selective internal radiotherapy, the overall survival rate has not
improved (5). Therefore, the
development of more effective therapeutic methods, including
molecular targeting therapy, is necessary.
Multiple studies have been conducted to investigate
the molecular mechanisms underlying HCC pathogenesis and numerous
gene markers have been identified in HCC, including
alpha-fetoprotein, glypican-3 (a serum and histochemical marker)
and transforming growth factor-β (6–8). As
small, non-coding RNAs, microRNAs (miRNAs) are important regulators
of cellular function and physiology (9). Controlling miRNA expression is
essential for the maintenance of the steady state of cellular
machinery (10). Various microRNAs
(miRNAs) have been proposed as novel biomarkers of HCC prognosis,
including the chromosome 19 miRNA cluster, which is overexpressed
in HCC (11). HCV-induced
alteration of miRNA expression regulates inflammation, leading to
liver fibrosis. In addition, miRNA (miR)-449a has been reported to
serve as an inhibitor in HCV patients, acting via the
downregulation of chitinase-3-like protein 1 expression, which is
an inflammatory marker for chronic liver diseases with fibrosis
(12). Dysregulation of other
miRNAs has been detected in HCC, including upregulated miR-23a,
−146a and −181a, and downregulated miR-17, −338-3p and −378
(13). Notably, miR-122 is
involved in HCC pathogenesis. It is commonly downregulated in HCC
and the loss of miR-122 contributes to hepatocarcinogenesis in mice
(14). miR-122 has additionally
been reported to induce apoptosis in human HCC cell lines via
targeting the anti-apoptosis gene B-cell lymphoma-2-like 2
(15). Furthermore, miR-122
inhibits cell proliferation in HCC by targeting the Wnt/β-catenin
signaling pathway (16). To
further understand the modulation of miR-122 in HCC development,
previous studies have investigated the consequences of miR-122
deletion. Hsu et al (17)
revealed that deletion of mouse mir-122 resulted in
hepatocarcinogenesis and HCC-like tumor development. Although
increased expression of multiple targets of miR-122 has been
detected in miR-122 knockout (KO) mice, including aldolase,
fructose bisphosphate A (ALDOA), solute carrier family 7
member 1, citrate synthase and cyclin G1, the functions and
pathways of the targets, and the potential associations between
them at the protein level, remain to be elucidated.
In the present study, the expression profile dataset
generated by Hsu et al (17), GSE31731, was reanalyzed and
differentially expressed genes (DEGs) were identified between
miR-122 KO mice and age-matched wild-type (WT) mice. Enrichment
analysis of the identified DEGs was subsequently conducted,
followed by protein-protein interaction (PPI) and module analysis.
Furthermore, targets of miR-122 of these DEGs were selected by
combining the results with relevant databases and literature
mining, followed by transcription factor (TF) analysis. By means of
these comprehensive bioinformatics analyses, the present study
aimed to further elucidate the involvement of miR-122 in HCC, and
identify potential regulators among its targets.
Materials and methods
Data resources
Gene expression of GSE31731 of HCCs, which was
deposited by Hsu et al (17) in the public Gene Expression Omnibus
(GEO; www.ncbi.nlm.nih.gov/geo) database,
was utilized in the present study. The miR-122 KO mice (liver tumor
samples) and age-matched WT mice (healthy liver samples) were
contained in this dataset. There were 8 biological replicates.
Two-channel microarray experiments were conducted for generation of
the dataset, based on the platform GPL13912 (accession no.
Agilent-028005 SurePrint G3 Mouse GE 8×60 K Microarray; Agilent
Technologies, Inc., Santa Clara, CA, USA).
Data pretreatment and differential
expression analysis
Raw data were preprocessed using R package in
Bioconductor (version 3.4; www.bioconductor.org/packages/3.0/bioc/). Following
background correction and normalization, the expression value was
converted from the probe level to the gene level. Subsequently,
DEGs between liver tumor samples and healthy liver samples were
screened out, based on the Student's t-test in Linear Models
for Microarray Analysis package (version 3.30.3; www.bioconductor.org/packages/release/bioc/html/limma.html)
(18). The Benjamini-Hochberg
method (19) was used to adjust
the P-value. The selection criteria for significant DEGs were a
false discovery rate <0.01 and a log2 fold change
>1.5.
Enrichment analysis for DEGs
The Database for Annotation, Visualization and
Integrated Discovery (DAVID; david.ncifcrf.gov/home.jsp) is a common tool for gene
function and pathway annotation (20–22).
To examine biological functions and pathways of the identified
DEGs, Gene Ontology (GO; www.geneontology.org/) (23) and Kyoto Encyclopedia of Genes and
Genomes (KEGG; www.genome.jp/kegg/pathway.html) (24) pathway enrichment analyses were
performed using DAVID (version 6.8). Cut-off values for significant
GO and KEGG pathway terms were P<0.05 and enriched gene number
≥2.
PPI network construction
To predict potential interactions between DEGs at
the protein level, the DEGs were entered into the Search Tool for
the Retrieval of Interacting Genes (string-db.org/) database (25). The parameter of interplayed PPIs
was set as 0.7, and a prerequisite for the network construction was
that all the PPI nodes were DEGs. Finally, the PPI network was
visualized by Cytoscape (version 3.4.0; cytoscape.org/) software (26).
Furthermore, module analysis was performed for the
PPI network using ClusterONE (version 1.0; www.paccanarolab.org/clusterone/) (27), followed by KEGG pathway enrichment
analysis. The threshold for significant module selection was
P<1.0×10−6.
Analysis of miR-122 targets
Initially, targets of miR-122 in mouse were
downloaded from three databases: miRecords (c1.accurascience.com/miRecords/) (28), TargetScan (www.targetscan.org/) (29) and microrna.org
(www.microrna.org) (30), and only genes that appeared in at
least two databases were deemed to be targets of miR-122. These
predicted targets were compared with the DEGs, and the overlapping
genes were screened out. Following this, TFs of the overlapped
targets were predicted by the iRegulon plugin of Cytoscape
(iregulon.aertslab.org), which integrates
a set of TF databases including Transfac, Jaspar, Encode,
Swissregulon and Homer to detect enriched TF motifs and their
optimal sets of direct targets (31). Normalized Enrichment Score (NES)
was the measurement index for TFs of the targets and the threshold
used was NES >3. Furthermore, the Agilent Literature Search
plugin (Agilent Technologies, Inc.) (32), which is complementary for protein
interaction data, was used to analyze the literature mining
association network. In the present study, the search terms were
set as ‘targets of miR-122’, context ‘liver cancer’ and species
‘Mus’, to select the reported HCC-associated literature involving
miR-122.
Results
DEGs between miR-122 KO and WT
groups
Using predefined criteria, DEGs between miR-122 KO
and WT groups were screened out, including 713 upregulated and 395
downregulated genes.
Altered functions and pathways of
DEGs
Based on GO and KEGG enrichment analyses,
upregulated DEGs were identified to be significantly enriched in
cell cycle associated biological processes (BPs), including the
cell cycle, M phase and mitotic cell cycle [for example nucleolar
and spindle associated protein 1 (NUSAP1); Table I], the cytokine-cytokine receptor
interaction pathway [for example C-X-C motif chemokine receptor 4
(CXCR4) and C-C motif chemokine receptor 2 (CCR2);
Table II], and various
cancer-associated pathways, including small cell lung cancer and
pathways in cancer [for example integrin subunit alpha V
(ITGAV); Table II]. The
downregulated DEGs were associated with oxidation-reduction
(Table I) and
metabolism-associated pathways, including drug metabolism, linoleic
acid metabolism and retinol metabolism (Table II).
 | Table I.Functions altered by differentially
expressed genes. |
Table I.
Functions altered by differentially
expressed genes.
| Category | Term | Count | P-value |
|---|
| Upregulated |
|
|
|
| BP | GO:0007049~cell
cycle | 57 |
4.96×10−11 |
|
| GO:0000279~M
phase | 34 |
1.64×10−09 |
|
| GO:0000278~mitotic
cell cycle | 31 |
2.64×10−09 |
|
|
GO:0007067~mitosis | 27 |
3.21×10−09 |
|
| GO:0000280~nuclear
division | 27 |
3.21×10−09 |
| CC |
GO:0005576~extracellular region | 133 |
9.92×10−18 |
|
|
GO:0044421~extracellular region part | 74 |
2.00×10−13 |
|
|
GO:0005578~proteinaceous extracellular
matrix | 35 |
7.33×10−09 |
|
|
GO:0031012~extracellular matrix | 35 |
2.01×10−08 |
|
|
GO:0044420~extracellular matrix part | 18 |
4.96×10−08 |
| MF | GO:0005509~calcium
ion binding | 60 |
1.62×10−07 |
|
|
GO:0008009~chemokine activity | 10 |
4.56×10−06 |
|
| GO:0008201~heparin
binding | 14 |
5.26×10−06 |
|
|
GO:0042379~chemokine receptor binding | 10 |
5.74×10−06 |
|
| GO:0001871~pattern
binding | 17 |
9.14×10−06 |
| Downregulated |
|
|
|
| BP |
GO:0055114~oxidation-reduction | 64 |
9.82×10−28 |
|
| GO:0006631~fatty
acid metabolic process | 24 |
4.36×10−13 |
|
| GO:0008202~steroid
metabolic process | 21 |
1.75×10−11 |
|
| GO:0006694~steroid
biosynthetic process | 15 |
3.73×10−11 |
|
|
GO:0006956~complement activation | 10 |
1.37×10−08 |
| CC |
GO:0005777~peroxisome | 22 |
4.28×10−15 |
|
|
GO:0042579~microbody | 22 |
4.28×10−15 |
|
|
GO:0005792~microsome | 26 |
5.15×10−14 |
|
|
GO:0042598~vesicular fraction | 26 |
1.13×10−13 |
|
|
GO:0005739~mitochondrion | 66 |
7.86×10−11 |
| MF | GO:0009055~electron
carrier activity | 34 |
4.36×10−21 |
|
| GO:0020037~heme
binding | 23 |
9.42×10−14 |
|
|
GO:0046906~tetrapyrrole binding | 23 |
2.57×10−13 |
|
| GO:0005506~iron ion
binding | 33 |
3.02×10−13 |
 | Table II.Pathways altered by differentially
expressed genes. |
Table II.
Pathways altered by differentially
expressed genes.
| Term | Count | Genes | P-value |
|---|
| Upregulated |
|
|
|
|
mmu04060:Cytokine-cytokine
receptor interaction | 27 | CCL2, CXCL5, CXCR4,
CXCL14, CCR2 |
4.82×10−06 |
|
mmu04512:ECM-receptor
interaction | 15 | COL3A1, LAMA2,
ITGAV, COL1A2, LAMC1 |
4.87×10−06 |
|
mmu04510:Focal adhesion | 22 | COL3A1, LAMA2,
ITGAV, LAMC1 |
4.43×10−05 |
|
mmu00480:Glutathione
metabolism | 10 | GPX2, GSTA1, GSTA2,
GSTM3, G6PDX |
2.05×10−04 |
|
mmu04110:Cell cycle | 14 | CCNB2, KMYT1,
BUB1B, ESPL1, CDC20 | 0.001940583 |
|
mmu05222:Small cell lung
cancer | 11 | LAMA2, COL4A1,
ITGAV, LAMC2, LAMC1 | 0.002161153 |
|
mmu04810:Regulation of actin
cytoskeleton | 19 | ITGAX, ITGAV,
PDGFRB, PAK1, DIAP3 | 0.002891419 |
|
mmu04062:Chemokine signaling
pathway | 16 | CCL2, CXCR4,
CXCL16, CCR2, CX3CR1 | 0.006803694 |
|
mmu05200:Pathways in
cancer | 23 | COL4A1, LAMA2,
ITGAV, LAMC2, LAMC1 | 0.011257034 |
|
mmu00590:Arachidonic acid
metabolism | 9 | GPX2, CBR1,
CYP4F18, GPX3, GGT1 et al | 0.018763259 |
| Downregulated |
|
|
|
|
mmu00982:Drug metabolism | 18 | CYP2C37, CYP3A16,
CYP2C54, CYP2C44, ADH4 |
1.83×10−12 |
|
mmu00980:Metabolism of
xenobiotics by cytochrome P450 | 17 | CYP2C37, CYP3A16,
CYP2C54, CYP2C44, CYP2C68 |
2.81×10−12 |
|
mmu00591:Linoleic acid
metabolism | 14 | CYP2J5, CYP2C37,
CYP3A16, CYP2C54, CYP2C44 |
4.16×10−11 |
|
mmu00830:Retinol
metabolism | 16 | CYP2C37, CYP3A16,
CYP2C54, CYP2C44, CYP2C68 |
6.09×10−11 |
|
mmu03320:PPAR signaling
pathway | 16 | ACOX1, ACSL1,
CYP4A12A, HMGCS2, SCP2 |
5.86×10−10 |
|
mmu00590:Arachidonic acid
metabolism | 15 | CYP2J5, CYP2C37,
CYP2C54, CYP2C44, CYP2J8 |
1.14×10−08 |
|
mmu00120:Primary bile acid
biosynthesis | 8 | CYP7B1, HSD3B7,
CYP7A1, CYP8B1, SCP2 |
2.81×10−08 |
|
mmu00071:Fatty acid
metabolism | 9 | CYP4A12B, GCDH,
ACOX1, ACSL1, ADH4 |
1.21×10−05 |
|
mmu00140:Steroid hormone
biosynthesis | 9 | CYP7B1, CYP3A16,
HSD3B6, HSD17B2, CYP7A1 |
1.21×10−05 |
|
mmu04610:Complement and
coagulation cascades | 11 | MBL1, C8A, MBL2,
C8B, CD55 |
1.40×10−05 |
PPI network and functional module
analysis
A PPI network was established, involving 549 nodes
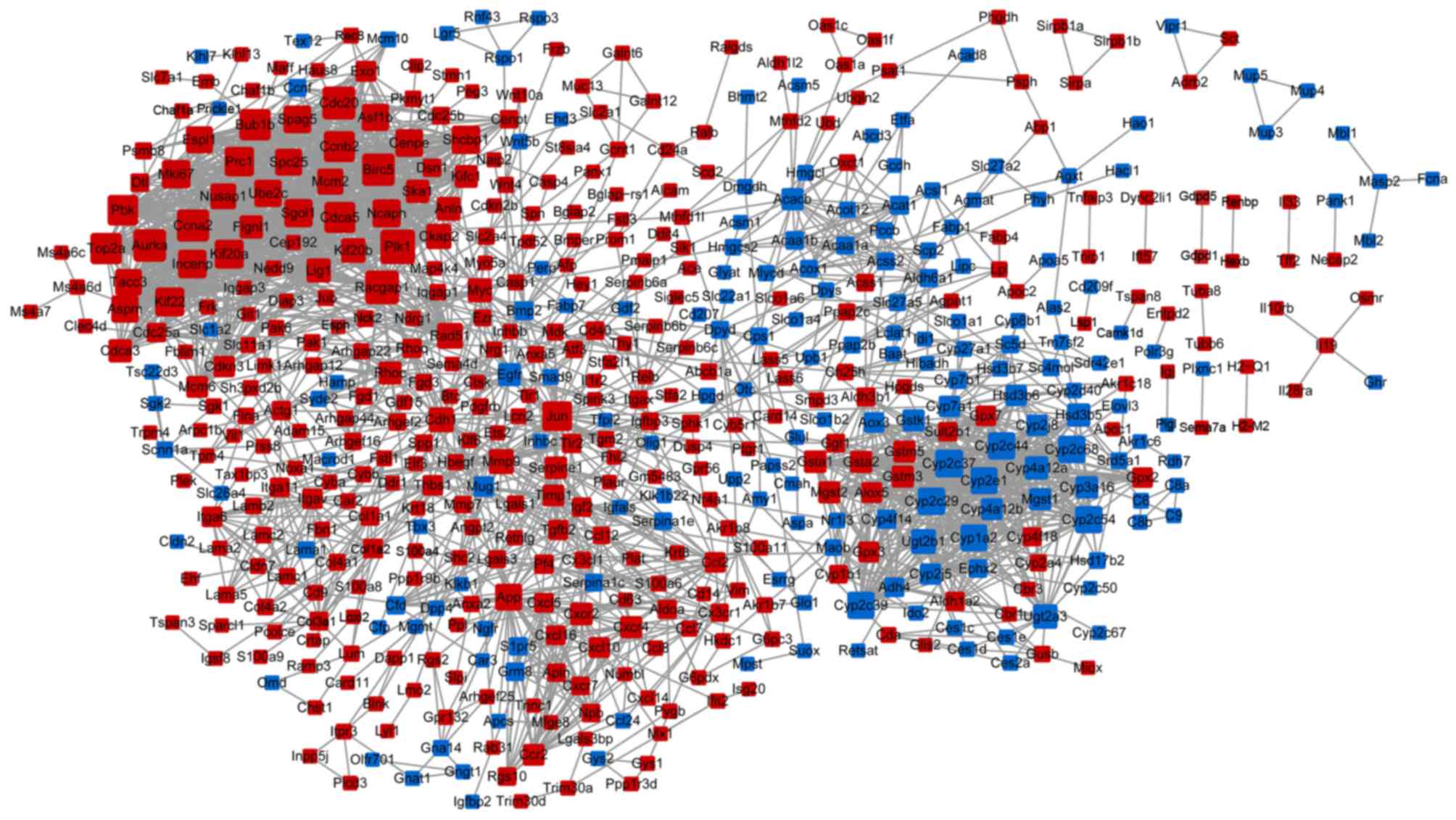
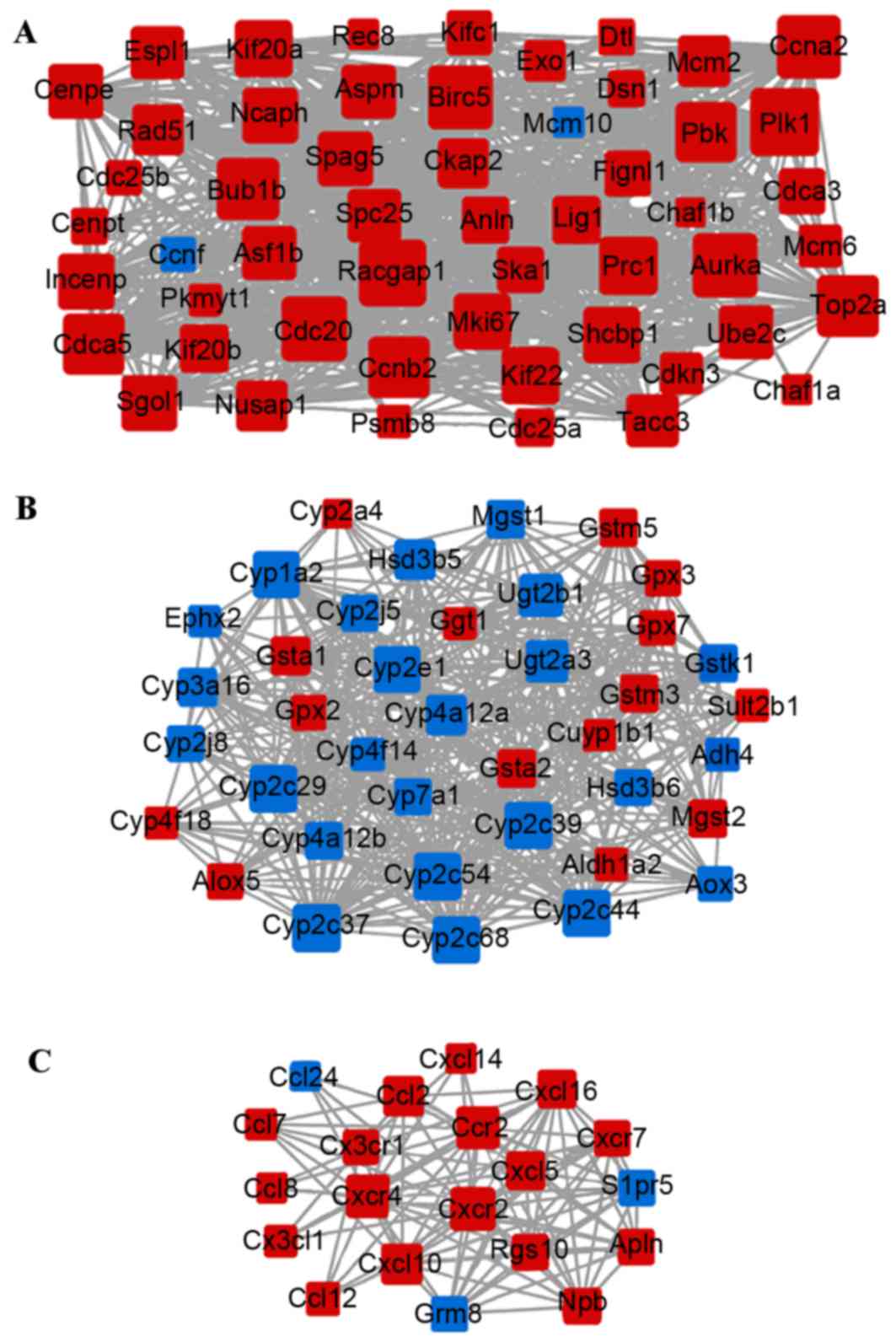
and 2,243 interplayed protein interactions (Fig. 1). Three sub-networks [modules A
(Fig. 2A), B (Fig. 2B) and C (Fig. 2C)] were extracted from the PPI
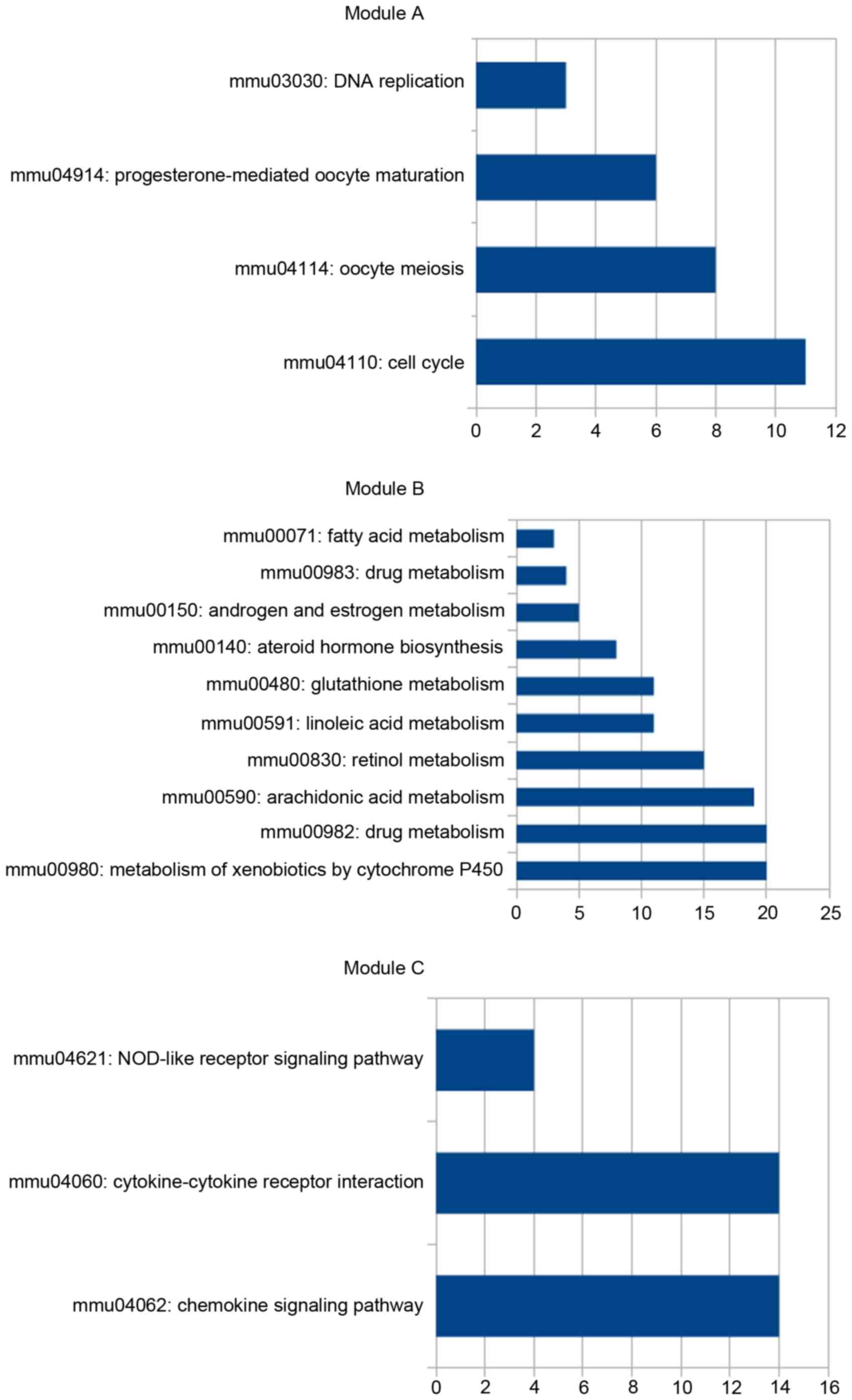
network. Enrichment analysis revealed that the majority of the
genes in module A were upregulated and enriched in DNA replication
and cell cycle-associated pathways, whereas the majority of genes
in module B were downregulated and enriched in metabolism of
xenobiotics by cytochrome P450 pathway (Fig. 3). In module C, the majority of the
genes were upregulated and involved with the chemokine signaling
pathway and the cytokine-cytokine receptor interaction pathway
(Fig. 3).
Targets of miR-122
Integrating the information from miRNA databases
with identified DEGs, a total of 76 overlapping genes were selected
as the targets of miR-122. Enrichment analysis indicated that these
genes were significantly involved in pathways in cancer (for
example ITGAV; Table II),
regulation of actin cytoskeleton (for example ITGAV;
Table II) and cytokine-cytokine
receptor interaction (for example CXCR4 and CCR2;
Table II).
Notably, 39 genes of the 76 overlapping targets were
additionally the predominant nodes with high degree in the PPI
network, including upregulated NUSAP1 (degree=30),
CXCR4 (degree=21), CCR2 (degree=20), ITGAV
(degree=17) and ALDOA (degree=14); and the downregulated
acyl-CoA synthetase short-chain family member 2 (degree=10).
NUSAP1 was also highlighted in module A, whereas
CXCR4 and CCR2 were prominent in module C.
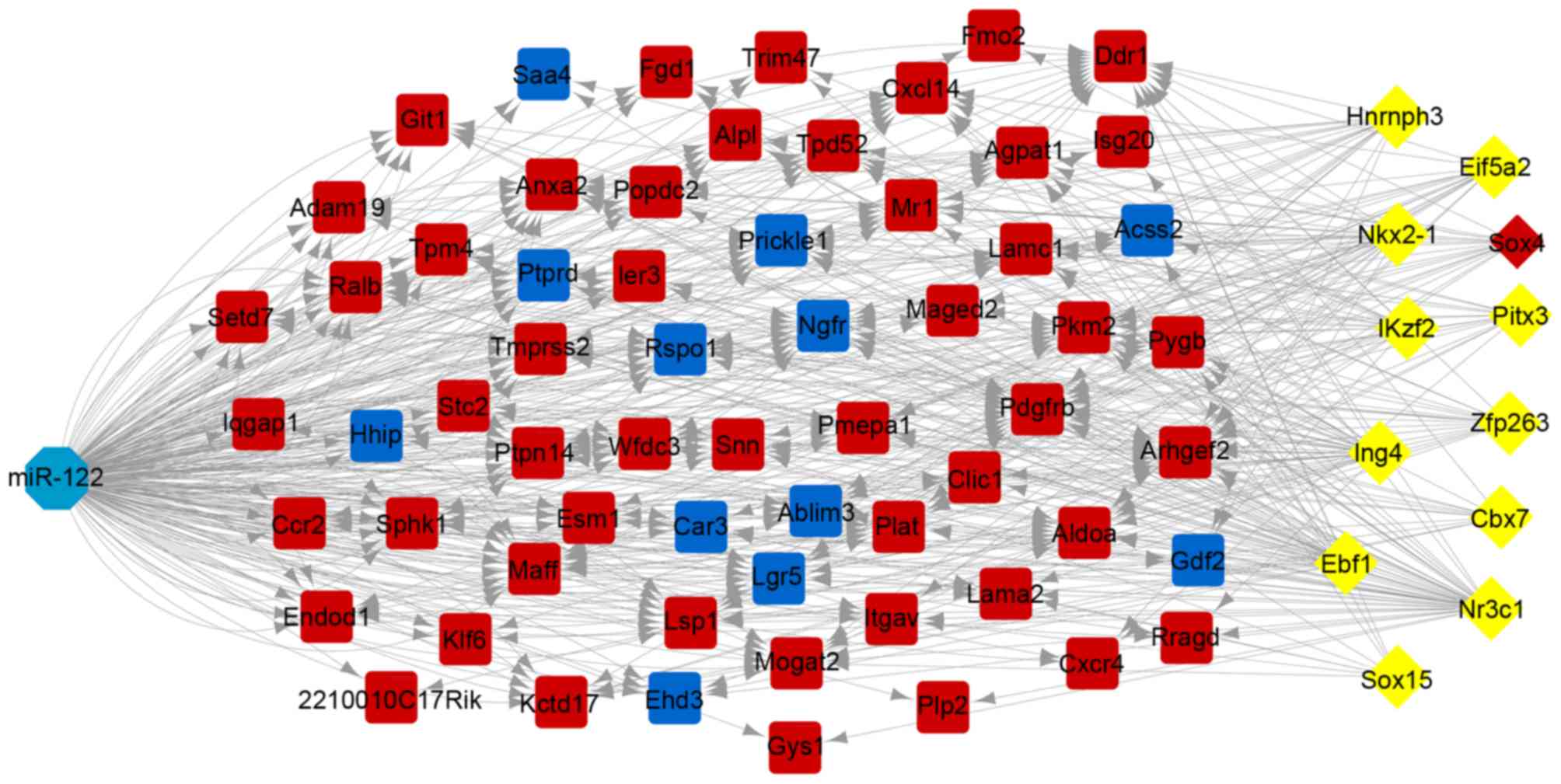
In total, 12 TFs targeting 62 overlapping genes were
predicted, including sex determining region Y-box 4 (SOX4),
heterogeneous nuclear ribonucleoprotein H3, NK2 homeobox 1,
inhibitor of growth family member 4, early B-cell factor 1, sex
determining region Y-box 15, nuclear receptor subfamily 3 group C
member 1, zinc finger protein 263, IKAROS family zinc finger 2,
paired like homeodomain 3, eukaryotic translation initiation factor
5A2 and chromobox 7. The TF-target regulatory network is presented
in Fig. 4. Notably, of the TFs,
SOX4 was additionally an upregulated DEG.
According to literature mining, a total of 47 genes
of the 76 overlapping targets were reported to be associated with
HCC, including ITGAV and CXCR4. Furthermore,
significantly altered expression of these genes was detected
following miR-122 KO, which suggested the involvement of miR-122 in
HCC development.
Discussion
miR-122 deficiency results in chronic
steatohepatitis and spontaneous HCC (14). By re-analyzing the dataset
GSE31731, numerous DEGs were identified between miR-122 KO and WT
groups. Of these, upregulated genes were significantly enriched in
cell cycle-associated processes (for example NUSAP1),
cytokine-cytokine receptor interaction pathways (for example
CXCR4 and CCR2), and extracellular matrix (ECM)
-receptor interactions (for example ITGAV). Various
overlapping targets were highlighted in the PPI network, including
NUSAP1, CXCR4, CCR2 and ITGAV. Notably,
NUSAP1 was predominant in module A, which was associated
with the cell cycle-associated pathway, whereas CXCR4 and
CCR2 were linked in module C, enriched in the
cytokine-cytokine receptor interaction pathway. Furthermore,
upregulated SOX4 was identified as a TF.
Restoration of miR-122 suppressed HCC tumor cell
growth, and the antitumor activity was closely associated with cell
cycle arrest (33). The protein
encoded by NUSAP1 is a nucleolar-spindle-associated protein
that acts as a positive regulator of mitosis (34). It has been identified as a cell
cycle progression gene in numerous cancer types, including prostate
and lung cancer (35,36). In aggressive HCC, expression of
NUSAP1 is affected by other cell cycle-associated genes,
including L2DTL (37). In
the present study, upregulated NUSAP1 in the miR-122 KO
group was significantly enriched in cell cycle-associated BPs, and
additionally served as a node in module A of the PPI network, as
well as one of the overlapped targets of miR-122. However,
NUSAP1 has not been previously reported to be directly
involved in HCC based on the literature mining results, suggesting
that this gene may be a novel target of miR-122 involved in cell
cycle-associated processes in HCC progression.
Dysregulation of the cytokine-cytokine receptor
interaction pathway has been detected in HCC development (38,39).
Activation of various chemokines in this pathway is involved in the
development of numerous cancers, including HCC (40). As a chemokine receptor, the
function of CXCR4 has been extensively investigated.
Increased expression of CXCR4 has previously been reported
to have a close association with the progression of HCC (41). In addition, CXCR4 inhibition
results in antitumor effects, including the inhibition of tumor
growth and the improvement of survival in mice with HCC (42). Elevated expression of CXCR4
in the miR-122 deficiency group, combined with the target
information in miRNA databases, indicated that CXCR4 may be
a potential target of miR-122 in HCC. CCR2 is the chemokine
receptor of C-C motif chemokine ligand 2 (CCL2). Increased
expression of CCR2 has previously been observed in liver
leukocytes from patients with HCC (43). CCL2 is also involved in the
cytokine-cytokine receptor interaction pathway and its expression
is associated with HCC progression (38). Notably, CCL2 has been
identified as a target of miR-122, and restoration of miR-122
results in the suppression of CCL2 in HCC (44). The results of the present study
indicated that the DEG CCR2 was upregulated in the miR-122
KO group and significantly enriched in the cytokine-cytokine
receptor interaction pathway. Notably, it was additionally
identified as an overlapping target based on the miRNA targeting
database, and was linked to CXCR4 in module C of the PPI
network. These data collectively suggested that CCR2 may be
a target of miR-122, and co-regulate the cytokine-cytokine receptor
interaction pathway with CXCR4 in HCC development.
ECM-receptor interaction is a common pathway that is
disturbed by altered gene expressions in various cancers (45,46).
The gene ITGAV encodes a protein that belongs to the
integrin superfamily. It has been reported to be involved in the
ECM-receptor interaction pathway. It is a part of the ECM system
(47), suggesting its involvement
in the ECM-receptor interaction pathway. In other cancer types,
including gastric cancer, ITGAV is enriched in this pathway
(48). Notably, overexpressed
ITGAV is induced by the gene forkhead box Q1 (FOXQ1),
a member of the forkhead TF family that influences HCC metastasis
(49), suggesting the potential
involvement of this gene in HCC. Upregulated ITGAV in the
miR-122 KO group, combined with enrichment analysis and the
overlapped prediction target, indicated that this gene may be a
target of miR-122 that modulates the ECM-receptor interaction
pathway in HCC progression.
The intron-lacking gene SOX4 contributes to
hepatocarcinogenesis and its overexpression may be a useful
prognostic marker for survival after surgical resection (50). It has been demonstrated in
vitro that overexpressed SOX4 is involved in
p53-mediated apoptosis in HCC (50). In addition, SOX4
overexpression has previously been reported to control the
metastasis of HCC (51). Thus,
SOX4 has been identified as a marker gene for HCC (52). Furthermore, SOX4 serves as a
TF that regulates cell differentiation. In HCC, various targets
have been experimentally validated using chromatin
immunoprecipitation and small interfering RNA assays, including
aldo-keto reductase family 1 member B10, coiled-coil domain
containing 97, dickkopf Wnt signaling pathway inhibitor 1,
FOXQ1 and microtubule associated protein 4 (51). miR-191 inhibition has previously
been reported to result in the upregulation of SOX4, and
increased SOX4 expression promotes cell apoptosis and
suppresses tumorigenesis of HCC (53). The present study indicated that the
TF SOX4 may additionally be a target of miR-122.
Despite these comprehensive bioinformatics analyses,
the present study has limitations. The data was downloaded from the
GEO database, and the sample size was relatively small. In
addition, experimental validation of the associations between
miR-122 and the predicted targets was lacking, and will be
addressed in follow-up studies. Furthermore, the target expression
levels following miR-122 restoration were not investigated,
although this would have further confirmed the targeting
associations. However, the present study has significant value as
it provides novel insights into the consequences of miR-122 KO in
HCC progression.
In conclusion, various crucial targets of miR-122 in
HCC progression were identified, including NUSAP1,
CXCR4, CCR2 and ITGAV. Cell cycle-associated
processes, the cytokine-cytokine receptor interaction pathway,
which may be co-regulated by CXCR4 and CCR2, and the
ECM-receptor interaction pathway were altered by these targets. In
addition, the target SOX4 may be a TF. The results of the
present study may provide a theoretical basis for further studies
on the mechanisms of miR-122 in the development of HCC.
Acknowledgements
The present study was supported by the Social
Development Fund of Nantong (grant no. HS2014063), the 12th Talent
Summit of Top Six Industries in Jiangsu Province (grant no. WSW080)
and the Project of Enhancing Medicine with Science and Education
under the 13th Five-Year Plan of Nantong [grant no. Nantong MED Sci
& Edu (2016) 23].
References
|
1
|
Yang J, Cai X, Lu W, Hu C, Xu X, Yu Q and
Cao P: Evodiamine inhibits STAT3 signaling by inducing phosphatase
shatterproof 1 in hepatocellular carcinoma cells. Cancer Lett.
328:243–251. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Borel F, Konstantinova P and Jansen PL:
Diagnostic and therapeutic potential of miRNA signatures in
patients with hepatocellular carcinoma. J Hepatol. 56:1371–1383.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Al-Mahtab M, Uddin H and Fazle Akbar SM:
Epidemiology and Risk Factors of Hepatocellular Carcinoma in Asia.
J Gastroenterology and Hepatology Res. 3:2014.
|
|
4
|
Paranaguá-Vezozzo DC, Ono SK,
Alvarado-Mora MV, Farias AQ, Cunha-Silva M, França JI, Alves VA,
Sherman M and Carrilho FJ: Epidemiology of HCC in Brazil: Incidence
and risk factors in a ten-year cohort. Ann Hepatol. 13:386–393.
2014.PubMed/NCBI
|
|
5
|
Weinmann A, Koch S, Niederle IM,
Schulze-Bergkamen H, König J, Hoppe-Lotichius M, Hansen T, Pitton
MB, Düber C, Otto G, et al: Trends in epidemiology, treatment, and
survival of hepatocellular carcinoma patients between 1998 and
2009: An analysis of 1066 cases of a German HCC registry. J Clin
Gastroenterol. 48:279–289. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Soresi M, Magliarisi C, Campagna P, Leto
G, Bonfissuto G, Riili A, Carroccio A, Sesti R, Tripi S and
Montalto G: Usefulness of alpha-fetoprotein in the diagnosis of
hepatocellular carcinoma. Anticancer Res. 23:1747–1753.
2003.PubMed/NCBI
|
|
7
|
Capurro M, Wanless IR, Sherman M, Deboer
G, Shi W, Miyoshi E and Filmus J: Glypican-3: A novel serum and
histochemical marker for hepatocellular carcinoma.
Gastroenterology. 125:89–97. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Murata M, Matsuzaki K, Yoshida K, Sekimoto
G, Tahashi Y, Mori S, Uemura Y, Sakaida N, Fujisawa J, Seki T, et
al: Hepatitis B virus X protein shifts human hepatic transforming
growth factor (TGF)-beta signaling from tumor suppression to
oncogenesis in early chronic hepatitis B. Hepatology. 49:1203–1217.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Beta M, Venkatesan N, Vasudevan M,
Vetrivel U, Khetan V and Krishnakumar S: Identification and
insilico analysis of retinoblastoma serum microRNA profile and gene
targets towards prediction of novel serum biomarkers. Bioinform
Biol Insights. 7:21–34. 2013.PubMed/NCBI
|
|
10
|
Kosaka N, Iguchi H and Ochiya T:
Circulating microRNA in body fluid: A new potential biomarker for
cancer diagnosis and prognosis. Cancer Sci. 101:2087–2092. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Augello C, Vaira V, Caruso L, Destro A,
Maggioni M, Park YN, Montorsi M, Santambrogio R, Roncalli M and
Bosari S: MicroRNA profiling of hepatocarcinogenesis identifies
C19MC cluster as a novel prognostic biomarker in hepatocellular
carcinoma. Liver Int. 32:772–782. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sarma NJ, Tiriveedhi V, Subramanian V,
Shenoy S, Crippin JS, Chapman WC and Mohanakumar T: Hepatitis C
virus mediated changes in miRNA-449a modulates inflammatory
biomarker YKL40 through components of the NOTCH signaling pathway.
PLoS One. 7:e508262012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang ZZ, Liu X, Wang DQ, Teng MK, Niu LW,
Huang AL and Liang Z: Hepatitis B virus and hepatocellular
carcinoma at the miRNA level. World J Gastroenterol. 17:3353–3358.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hsu SH, Wang B, Kutay H, Bid H, Shreve J,
Zhang X, Costinean S, Bratasz A, Houghton P and Ghoshal K: Hepatic
loss of miR-122 predisposes mice to hepatobiliary cyst and
hepatocellular carcinoma upon diethylnitrosamine exposure. Am J
Pathol. 183:1719–1730. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lin CJ, Gong HY, Tseng HC, Wang WL and Wu
JL: miR-122 targets an anti-apoptotic gene, Bcl-w, in human
hepatocellular carcinoma cell lines. Biochem Biophys Res Commun.
375:315–320. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xu J, Zhu X, Wu L, Yang R, Yang Z, Wang Q
and Wu F: MicroRNA-122 suppresses cell proliferation and induces
cell apoptosis in hepatocellular carcinoma by directly targeting
Wnt/β-catenin pathway. Liver Int. 32:752–760. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hsu SH, Wang B, Kota J, Yu J, Costinean S,
Kutay H, Yu L, Bai S, La Perle K, Chivukula RR, et al: Essential
metabolic, anti-inflammatory, and anti-tumorigenic functions of
miR-122 in liver. J Clin Invest. 122:2871–2883. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Smyth GK: Limma: Linear models for
microarray dataBioinformatics and computational biology solutions
using R and Bioconductor. Springer; pp. 397–420. 2005, View Article : Google Scholar
|
|
19
|
Haynes W: Benjamini-Hochberg
MethodEncyclopedia of Systems Biology. Springer; pp. 78. 2013,
View Article : Google Scholar
|
|
20
|
Dennis G Jr, Sherman BT, Hosack DA, Yang
J, Gao W, Lane HC and Lempicki RA: DAVID: Database for annotation,
visualization and integrated discovery. Genome Biol. 4:P32003.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
da W Huang, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009.PubMed/NCBI
|
|
22
|
da W Huang, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gene Ontology Consortium, ; Blake JA,
Dolan M, Drabkin H, Hill DP, Li N, Sitnikov D, Bridges S, Burgess
S, Buza T, McCarthy F, et al: Gene Ontology annotations and
resources. Nucleic Acids Res. 41:D530–D535. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kanehisa M, Goto S, Sato Y, Furumichi M
and Tanabe M: KEGG for integration and interpretation of
large-scale molecular data sets. Nucleic Acids Res. 40:D109–D114.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43:D447–D452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Smoot ME, Ono K, Ruscheinski J, Wang PL
and Ideker T: Cytoscape 2.8: New features for data integration and
network visualization. Bioinformatics. 27:431–432. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wan FC, Cui YP, Wu JT, Wang JM-Z, Liu Q
and Gao ZL: The PPI network and cluster ONE analysis to explain the
mechanism of bladder cancer. Eur Rev Med Pharmacol Sci. 17:618–623.
2013.PubMed/NCBI
|
|
28
|
Xiao F, Zuo Z, Cai G, Kang S, Gao X and Li
T: miRecords: An integrated resource for microRNA-target
interactions. Nucleic Acids Res. 37:D105–D110. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lewis BP, Shih IH, Jones-Rhoades MW,
Bartel DP and Burge CB: Prediction of mammalian microRNA targets.
Cell. 115:787–798. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Betel D, Wilson M, Gabow A, Marks DS and
Sander C: The microRNA. org resource: Targets and expression.
Nucleic Acids Res. 36:D149–D153. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Janky R, Verfaillie A, Imrichová H, Van de
Sande B, Standaert L, Christiaens V, Hulselmans G, Herten K,
Sanchez M Naval, Potier D, et al: iRegulon: from a gene list to a
gene regulatory network using large motif and track collections.
PLoS Comput Biol. 10:e10037312014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cline MS, Smoot M, Cerami E, Kuchinsky A,
Landys N, Workman C, Christmas R, Avila-Campilo I, Creech M, Gross
B, et al: Integration of biological networks and gene expression
data using Cytoscape. Nat Protoc. 2:2366–2382. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ma L, Liu J, Shen J, Liu L, Wu J, Li W,
Luo J, Chen Q and Qian C: Expression of miR-122 mediated by
adenoviral vector induces apoptosis and cell cycle arrest of cancer
cells. Cancer Biol Ther. 9:554–561. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Raemaekers T, Ribbeck K, Beaudouin J,
Annaert W, Van Camp M, Stockmans I, Smets N, Bouillon R, Ellenberg
J and Carmeliet G: NuSAP, a novel microtubule-associated protein
involved in mitotic spindle organization. J Cell Biol.
162:1017–1029. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Cuzick J, Swanson GP, Fisher G, Brothman
AR, Berney DM, Reid JE, Mesher D, Speights VO, Stankiewicz E,
Foster CS, et al: Prognostic value of an RNA expression signature
derived from cell cycle proliferation genes in patients with
prostate cancer: A retrospective study. Lancet Oncol. 12:245–255.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhou C, Chen H, Han L, Wang A and Chen LA:
Identification of featured biomarkers in different types of lung
cancer with DNA microarray. Mol Biol Rep. 41:6357–6363. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Pan HW, Chou HY, Liu SH, Peng SY, Liu CL
and Hsu HC: Role of L2DTL, cell cycle-regulated nuclear and
centrosome protein, in aggressive hepatocellular carcinoma. Cell
Cycle. 5:2676–2687. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li T, Wan B, Huang J and Zhang X:
Comparison of gene expression in hepatocellular carcinoma, liver
development, and liver regeneration. Mol Genet Genomics.
283:485–492. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lin ZY, Chuang WL and Chuang YH:
Amphotericin B up-regulates angiogenic genes in hepatocellular
carcinoma cell lines. Eur J Clin Invest. 39:239–245. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Huang F and Geng XP: Chemokines and
hepatocellular carcinoma. World J Gastroenterol. 16:1832–1836.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Schimanski CC, Bahre R, Gockel I, Müller
A, Frerichs K, Hörner V, Teufel A, Simiantonaki N, Biesterfeld S,
Wehler T, et al: Dissemination of hepatocellular carcinoma is
mediated via chemokine receptor CXCR4. Br J Cancer. 95:210–217.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Chen Y, Ramjiawan RR, Reiberger T, Ng MR,
Hato T, Huang Y, Ochiai H, Kitahara S, Unan EC, Reddy TP, et al:
CXCR4 inhibition in tumor microenvironment facilitates
anti-programmed death receptor-1 immunotherapy in sorafenib-treated
hepatocellular carcinoma in mice. Hepatology. 61:1591–1602. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Chew V, Tow C, Teo M, Wong HL, Chan J,
Gehring A, Loh M, Bolze A, Quek R, Lee VK, et al: Inflammatory
tumour microenvironment is associated with superior survival in
hepatocellular carcinoma patients. J Hepatol. 52:370–379. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Bandiera S, Pfeffer S, Baumert TF and
Zeisel MB: MiR-122-a key factor and therapeutic target in liver
disease. J Hepatol. 62:448–457. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Navab R, Strumpf D, Bandarchi B, Zhu CQ,
Pintilie M, Ramnarine VR, Ibrahimov E, Radulovich N, Leung L,
Barczyk M, et al: Prognostic gene-expression signature of
carcinoma-associated fibroblasts in non-small cell lung cancer.
Proc Natl Acad Sci USA. 108:pp. 7160–7165. 2011; View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Lee HJ, Jang M, Kim H, Kwak W, Park W,
Hwang JY, Lee CK, Jang GW, Park MN, Kim HC, et al: Comparative
transcriptome analysis of adipose tissues reveals that ECM-receptor
interaction is involved in the depot-specific adipogenesis in
cattle. PLoS One. 8:e662672013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Fukui T, Shaykhiev R, Agosto-Perez F,
Mezey JG, Downey RJ, Travis WD and Crystal RG: Lung adenocarcinoma
subtypes based on expression of human airway basal cell genes. Eur
Respir J. 42:1332–1344. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Hou Y: Molecular basis of the effect of
midkine on tumor growth in human gastric cancer cell BGC823.
Biomedicine and pharmacotherapy. 62:4222008. View Article : Google Scholar
|
|
49
|
Xia L, Huang W, Tian D, Zhang L, Qi X,
Chen Z, Shang X, Nie Y and Wu K: Forkhead box Q1 promotes
hepatocellular carcinoma metastasis by transactivating ZEB2 and
VersicanV1 expression. Hepatology. 59:958–973. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Hur W, Rhim H, Jung CK, Kim JD, Bae SH,
Jang JW, Yang JM, Oh ST, Kim DG, Wang HJ, et al: SOX4
overexpression regulates the p53-mediated apoptosis in
hepatocellular carcinoma: Clinical implication and functional
analysis in vitro. Carcinogenesis. 31:1298–1307. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Liao YL, Sun YM, Chau GY, Chau YP, Lai TC,
Wang JL, Horng JT, Hsiao M and Tsou AP: Identification of SOX4
target genes using phylogenetic footprinting-based prediction from
expression microarrays suggests that overexpression of SOX4
potentiates metastasis in hepatocellular carcinoma. Oncogene.
27:5578–5589. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Chang Q, Chen J, Beezhold KJ, Castranova
V, Shi X and Chen F: JNK1 activation predicts the prognostic
outcome of the human hepatocellular carcinoma. Mol Cancer. 8:1–14.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Elyakim E, Sitbon E, Faerman A, Tabak S,
Montia E, Belanis L, Dov A, Marcusson EG, Bennett CF, Chajut A, et
al: hsa-miR-191 is a candidate oncogene target for hepatocellular
carcinoma therapy. Cancer Res. 70:8077–8087. 2010. View Article : Google Scholar : PubMed/NCBI
|