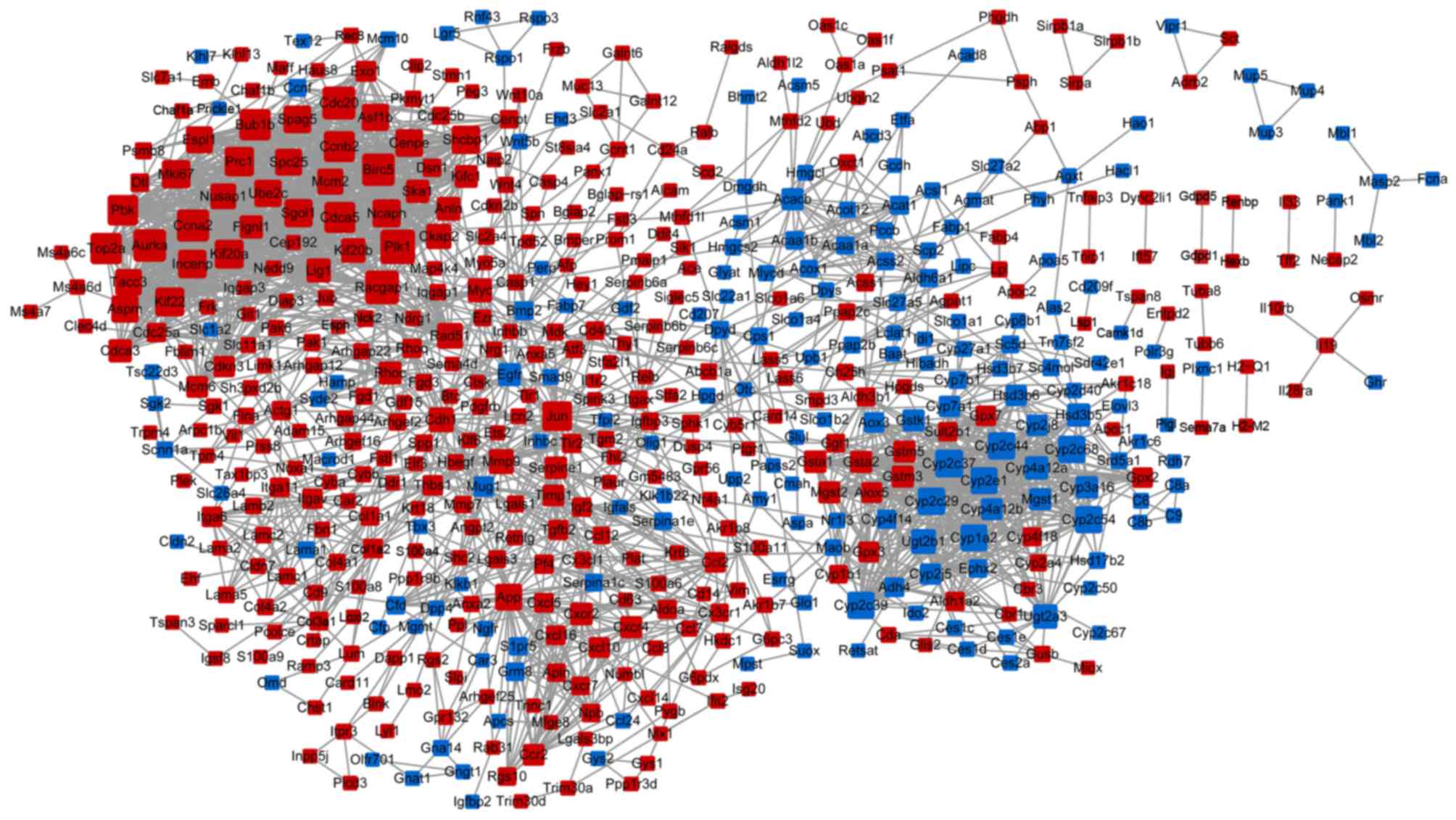
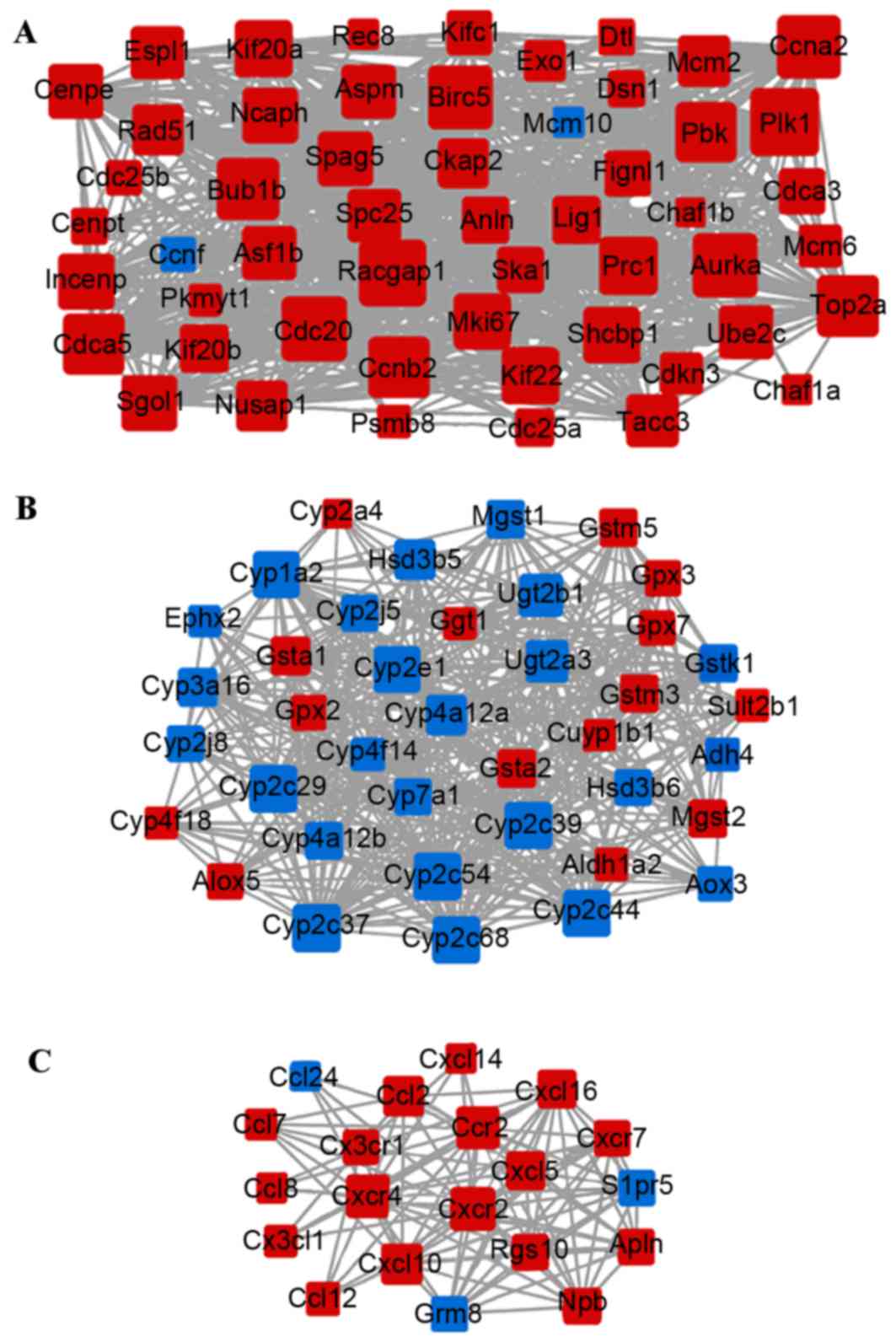
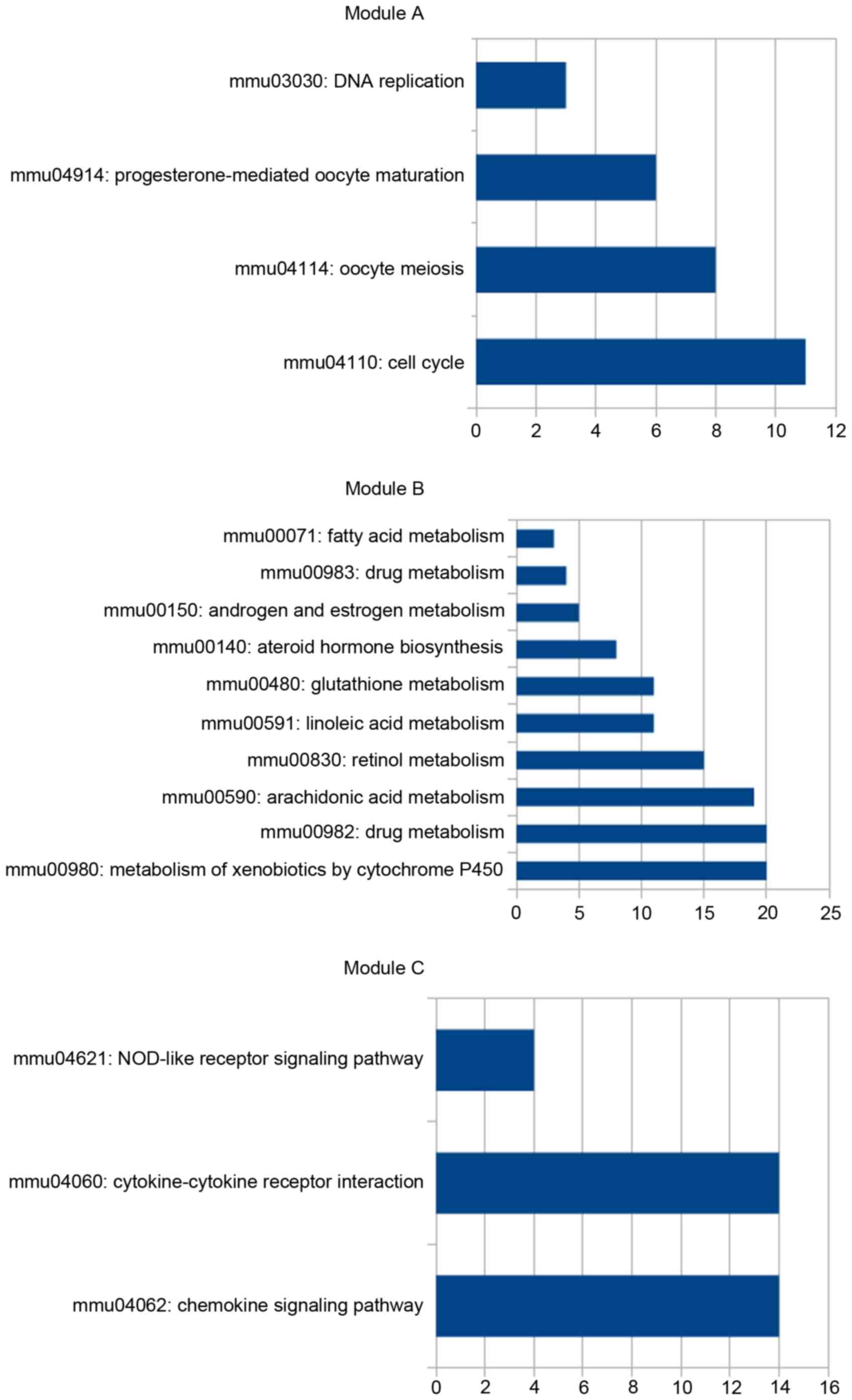
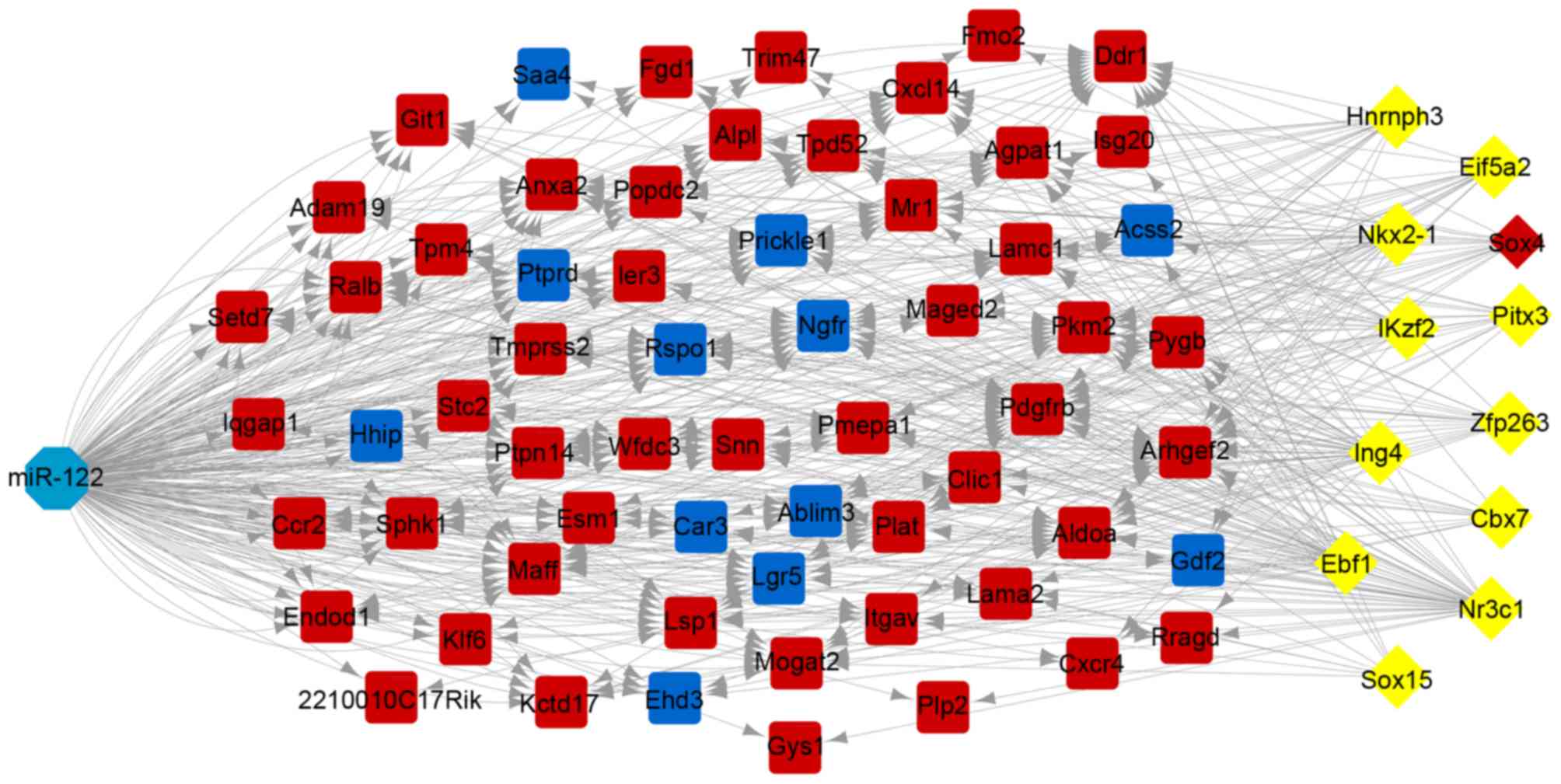
|
1
|
Yang J, Cai X, Lu W, Hu C, Xu X, Yu Q and
Cao P: Evodiamine inhibits STAT3 signaling by inducing phosphatase
shatterproof 1 in hepatocellular carcinoma cells. Cancer Lett.
328:243–251. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Borel F, Konstantinova P and Jansen PL:
Diagnostic and therapeutic potential of miRNA signatures in
patients with hepatocellular carcinoma. J Hepatol. 56:1371–1383.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Al-Mahtab M, Uddin H and Fazle Akbar SM:
Epidemiology and Risk Factors of Hepatocellular Carcinoma in Asia.
J Gastroenterology and Hepatology Res. 3:2014.
|
|
4
|
Paranaguá-Vezozzo DC, Ono SK,
Alvarado-Mora MV, Farias AQ, Cunha-Silva M, França JI, Alves VA,
Sherman M and Carrilho FJ: Epidemiology of HCC in Brazil: Incidence
and risk factors in a ten-year cohort. Ann Hepatol. 13:386–393.
2014.PubMed/NCBI
|
|
5
|
Weinmann A, Koch S, Niederle IM,
Schulze-Bergkamen H, König J, Hoppe-Lotichius M, Hansen T, Pitton
MB, Düber C, Otto G, et al: Trends in epidemiology, treatment, and
survival of hepatocellular carcinoma patients between 1998 and
2009: An analysis of 1066 cases of a German HCC registry. J Clin
Gastroenterol. 48:279–289. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Soresi M, Magliarisi C, Campagna P, Leto
G, Bonfissuto G, Riili A, Carroccio A, Sesti R, Tripi S and
Montalto G: Usefulness of alpha-fetoprotein in the diagnosis of
hepatocellular carcinoma. Anticancer Res. 23:1747–1753.
2003.PubMed/NCBI
|
|
7
|
Capurro M, Wanless IR, Sherman M, Deboer
G, Shi W, Miyoshi E and Filmus J: Glypican-3: A novel serum and
histochemical marker for hepatocellular carcinoma.
Gastroenterology. 125:89–97. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Murata M, Matsuzaki K, Yoshida K, Sekimoto
G, Tahashi Y, Mori S, Uemura Y, Sakaida N, Fujisawa J, Seki T, et
al: Hepatitis B virus X protein shifts human hepatic transforming
growth factor (TGF)-beta signaling from tumor suppression to
oncogenesis in early chronic hepatitis B. Hepatology. 49:1203–1217.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Beta M, Venkatesan N, Vasudevan M,
Vetrivel U, Khetan V and Krishnakumar S: Identification and
insilico analysis of retinoblastoma serum microRNA profile and gene
targets towards prediction of novel serum biomarkers. Bioinform
Biol Insights. 7:21–34. 2013.PubMed/NCBI
|
|
10
|
Kosaka N, Iguchi H and Ochiya T:
Circulating microRNA in body fluid: A new potential biomarker for
cancer diagnosis and prognosis. Cancer Sci. 101:2087–2092. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Augello C, Vaira V, Caruso L, Destro A,
Maggioni M, Park YN, Montorsi M, Santambrogio R, Roncalli M and
Bosari S: MicroRNA profiling of hepatocarcinogenesis identifies
C19MC cluster as a novel prognostic biomarker in hepatocellular
carcinoma. Liver Int. 32:772–782. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sarma NJ, Tiriveedhi V, Subramanian V,
Shenoy S, Crippin JS, Chapman WC and Mohanakumar T: Hepatitis C
virus mediated changes in miRNA-449a modulates inflammatory
biomarker YKL40 through components of the NOTCH signaling pathway.
PLoS One. 7:e508262012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang ZZ, Liu X, Wang DQ, Teng MK, Niu LW,
Huang AL and Liang Z: Hepatitis B virus and hepatocellular
carcinoma at the miRNA level. World J Gastroenterol. 17:3353–3358.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hsu SH, Wang B, Kutay H, Bid H, Shreve J,
Zhang X, Costinean S, Bratasz A, Houghton P and Ghoshal K: Hepatic
loss of miR-122 predisposes mice to hepatobiliary cyst and
hepatocellular carcinoma upon diethylnitrosamine exposure. Am J
Pathol. 183:1719–1730. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lin CJ, Gong HY, Tseng HC, Wang WL and Wu
JL: miR-122 targets an anti-apoptotic gene, Bcl-w, in human
hepatocellular carcinoma cell lines. Biochem Biophys Res Commun.
375:315–320. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xu J, Zhu X, Wu L, Yang R, Yang Z, Wang Q
and Wu F: MicroRNA-122 suppresses cell proliferation and induces
cell apoptosis in hepatocellular carcinoma by directly targeting
Wnt/β-catenin pathway. Liver Int. 32:752–760. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hsu SH, Wang B, Kota J, Yu J, Costinean S,
Kutay H, Yu L, Bai S, La Perle K, Chivukula RR, et al: Essential
metabolic, anti-inflammatory, and anti-tumorigenic functions of
miR-122 in liver. J Clin Invest. 122:2871–2883. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Smyth GK: Limma: Linear models for
microarray dataBioinformatics and computational biology solutions
using R and Bioconductor. Springer; pp. 397–420. 2005, View Article : Google Scholar
|
|
19
|
Haynes W: Benjamini-Hochberg
MethodEncyclopedia of Systems Biology. Springer; pp. 78. 2013,
View Article : Google Scholar
|
|
20
|
Dennis G Jr, Sherman BT, Hosack DA, Yang
J, Gao W, Lane HC and Lempicki RA: DAVID: Database for annotation,
visualization and integrated discovery. Genome Biol. 4:P32003.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
da W Huang, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009.PubMed/NCBI
|
|
22
|
da W Huang, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gene Ontology Consortium, ; Blake JA,
Dolan M, Drabkin H, Hill DP, Li N, Sitnikov D, Bridges S, Burgess
S, Buza T, McCarthy F, et al: Gene Ontology annotations and
resources. Nucleic Acids Res. 41:D530–D535. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kanehisa M, Goto S, Sato Y, Furumichi M
and Tanabe M: KEGG for integration and interpretation of
large-scale molecular data sets. Nucleic Acids Res. 40:D109–D114.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43:D447–D452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Smoot ME, Ono K, Ruscheinski J, Wang PL
and Ideker T: Cytoscape 2.8: New features for data integration and
network visualization. Bioinformatics. 27:431–432. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wan FC, Cui YP, Wu JT, Wang JM-Z, Liu Q
and Gao ZL: The PPI network and cluster ONE analysis to explain the
mechanism of bladder cancer. Eur Rev Med Pharmacol Sci. 17:618–623.
2013.PubMed/NCBI
|
|
28
|
Xiao F, Zuo Z, Cai G, Kang S, Gao X and Li
T: miRecords: An integrated resource for microRNA-target
interactions. Nucleic Acids Res. 37:D105–D110. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lewis BP, Shih IH, Jones-Rhoades MW,
Bartel DP and Burge CB: Prediction of mammalian microRNA targets.
Cell. 115:787–798. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Betel D, Wilson M, Gabow A, Marks DS and
Sander C: The microRNA. org resource: Targets and expression.
Nucleic Acids Res. 36:D149–D153. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Janky R, Verfaillie A, Imrichová H, Van de
Sande B, Standaert L, Christiaens V, Hulselmans G, Herten K,
Sanchez M Naval, Potier D, et al: iRegulon: from a gene list to a
gene regulatory network using large motif and track collections.
PLoS Comput Biol. 10:e10037312014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cline MS, Smoot M, Cerami E, Kuchinsky A,
Landys N, Workman C, Christmas R, Avila-Campilo I, Creech M, Gross
B, et al: Integration of biological networks and gene expression
data using Cytoscape. Nat Protoc. 2:2366–2382. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ma L, Liu J, Shen J, Liu L, Wu J, Li W,
Luo J, Chen Q and Qian C: Expression of miR-122 mediated by
adenoviral vector induces apoptosis and cell cycle arrest of cancer
cells. Cancer Biol Ther. 9:554–561. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Raemaekers T, Ribbeck K, Beaudouin J,
Annaert W, Van Camp M, Stockmans I, Smets N, Bouillon R, Ellenberg
J and Carmeliet G: NuSAP, a novel microtubule-associated protein
involved in mitotic spindle organization. J Cell Biol.
162:1017–1029. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Cuzick J, Swanson GP, Fisher G, Brothman
AR, Berney DM, Reid JE, Mesher D, Speights VO, Stankiewicz E,
Foster CS, et al: Prognostic value of an RNA expression signature
derived from cell cycle proliferation genes in patients with
prostate cancer: A retrospective study. Lancet Oncol. 12:245–255.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhou C, Chen H, Han L, Wang A and Chen LA:
Identification of featured biomarkers in different types of lung
cancer with DNA microarray. Mol Biol Rep. 41:6357–6363. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Pan HW, Chou HY, Liu SH, Peng SY, Liu CL
and Hsu HC: Role of L2DTL, cell cycle-regulated nuclear and
centrosome protein, in aggressive hepatocellular carcinoma. Cell
Cycle. 5:2676–2687. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li T, Wan B, Huang J and Zhang X:
Comparison of gene expression in hepatocellular carcinoma, liver
development, and liver regeneration. Mol Genet Genomics.
283:485–492. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lin ZY, Chuang WL and Chuang YH:
Amphotericin B up-regulates angiogenic genes in hepatocellular
carcinoma cell lines. Eur J Clin Invest. 39:239–245. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Huang F and Geng XP: Chemokines and
hepatocellular carcinoma. World J Gastroenterol. 16:1832–1836.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Schimanski CC, Bahre R, Gockel I, Müller
A, Frerichs K, Hörner V, Teufel A, Simiantonaki N, Biesterfeld S,
Wehler T, et al: Dissemination of hepatocellular carcinoma is
mediated via chemokine receptor CXCR4. Br J Cancer. 95:210–217.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Chen Y, Ramjiawan RR, Reiberger T, Ng MR,
Hato T, Huang Y, Ochiai H, Kitahara S, Unan EC, Reddy TP, et al:
CXCR4 inhibition in tumor microenvironment facilitates
anti-programmed death receptor-1 immunotherapy in sorafenib-treated
hepatocellular carcinoma in mice. Hepatology. 61:1591–1602. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Chew V, Tow C, Teo M, Wong HL, Chan J,
Gehring A, Loh M, Bolze A, Quek R, Lee VK, et al: Inflammatory
tumour microenvironment is associated with superior survival in
hepatocellular carcinoma patients. J Hepatol. 52:370–379. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Bandiera S, Pfeffer S, Baumert TF and
Zeisel MB: MiR-122-a key factor and therapeutic target in liver
disease. J Hepatol. 62:448–457. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Navab R, Strumpf D, Bandarchi B, Zhu CQ,
Pintilie M, Ramnarine VR, Ibrahimov E, Radulovich N, Leung L,
Barczyk M, et al: Prognostic gene-expression signature of
carcinoma-associated fibroblasts in non-small cell lung cancer.
Proc Natl Acad Sci USA. 108:pp. 7160–7165. 2011; View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Lee HJ, Jang M, Kim H, Kwak W, Park W,
Hwang JY, Lee CK, Jang GW, Park MN, Kim HC, et al: Comparative
transcriptome analysis of adipose tissues reveals that ECM-receptor
interaction is involved in the depot-specific adipogenesis in
cattle. PLoS One. 8:e662672013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Fukui T, Shaykhiev R, Agosto-Perez F,
Mezey JG, Downey RJ, Travis WD and Crystal RG: Lung adenocarcinoma
subtypes based on expression of human airway basal cell genes. Eur
Respir J. 42:1332–1344. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Hou Y: Molecular basis of the effect of
midkine on tumor growth in human gastric cancer cell BGC823.
Biomedicine and pharmacotherapy. 62:4222008. View Article : Google Scholar
|
|
49
|
Xia L, Huang W, Tian D, Zhang L, Qi X,
Chen Z, Shang X, Nie Y and Wu K: Forkhead box Q1 promotes
hepatocellular carcinoma metastasis by transactivating ZEB2 and
VersicanV1 expression. Hepatology. 59:958–973. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Hur W, Rhim H, Jung CK, Kim JD, Bae SH,
Jang JW, Yang JM, Oh ST, Kim DG, Wang HJ, et al: SOX4
overexpression regulates the p53-mediated apoptosis in
hepatocellular carcinoma: Clinical implication and functional
analysis in vitro. Carcinogenesis. 31:1298–1307. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Liao YL, Sun YM, Chau GY, Chau YP, Lai TC,
Wang JL, Horng JT, Hsiao M and Tsou AP: Identification of SOX4
target genes using phylogenetic footprinting-based prediction from
expression microarrays suggests that overexpression of SOX4
potentiates metastasis in hepatocellular carcinoma. Oncogene.
27:5578–5589. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Chang Q, Chen J, Beezhold KJ, Castranova
V, Shi X and Chen F: JNK1 activation predicts the prognostic
outcome of the human hepatocellular carcinoma. Mol Cancer. 8:1–14.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Elyakim E, Sitbon E, Faerman A, Tabak S,
Montia E, Belanis L, Dov A, Marcusson EG, Bennett CF, Chajut A, et
al: hsa-miR-191 is a candidate oncogene target for hepatocellular
carcinoma therapy. Cancer Res. 70:8077–8087. 2010. View Article : Google Scholar : PubMed/NCBI
|