Introduction
Essential hypertension (EH) is a disorder
characterized by high blood pressure of unknown cause and is a
major risk factor for cardiovascular and cerebrovascular disease
and a serious public health problem worldwide. The prevalence was
at 26.7% in 2010 in China (1),
this is predicted to increase to 29.2% globally by 2025 (2). EH may be closely associated with
dysregulation of the renin-angiotensin system (RAS). However, the
underlying molecular mechanisms that lead to the dysregulation
remain to be elucidated; however, genetic alterations,
environmental factors, gene-gene, and gene-environment interactions
may be considered key factors (3,4).
The RAS is a master regulator of blood pressure.
Angiotensin II is an important vasoconstrictor in this system,
whereas angiotensin converting enzyme 2 (ACE2), the
discovery of which was considered to be a breakthrough in 2000
(5,6), promotes vasodilation by degrading
angiotensin II, and generating the vasodilators Ang 1–7 (7). Accordingly, increasing the expression
of ACE2, which is located on chromosome Xp22, protects
against increased blood pressure, whereas inhibition or deletion
promotes EH (8). Previous genetic
studies have identified polymorphisms in ACE2 as risk
factors for EH in multiple populations, such as the Han-Chinese and
Caucasian population (9,10).
DNA methylation, a common mechanism of reversible
epigenetic regulation, usually occurs at cytosine residues in
cytosine-phosphate-guanine (CpG) dinucleotides in mammalian cells
(11). Environmental factors can
affect DNA methylation levels in the genome and thus alter gene
expression. Promoter hypermethylation silences genes, whereas
hypomethylation promotes active transcription (12). Therefore, controlling methylation
of relevant genes may provide novel opportunities to treat or
prevent EH. Previous studies have determined that aberrant
methylation of components of the RAS, including angiotensinogen,
ACE, and angiotensin II receptor type 1 (AGTR1) was
associated with the onset and development of EH (13–16).
However, the association between EH and methylation of the
ACE2 promoter remains to be elucidated. Therefore, the
present study aimed to investigate whether aberrant methylation of
the ACE2 promoter contributed to EH and the association with
age, sex and other clinical indicators, as has been determined for
other genes, including adducing 1 (17) and glucokinase (18).
Materials and methods
Sample collection
A total of 192 individuals, 96 patients with EH and
96 healthy controls, were recruited at The Seventh Hospital of
Ningbo (Ningbo, China). Participants were from Han Chinese families
who had been residing in Ningbo for a minimum of three generations
and had no history of diabetes mellitus, secondary hypertension,
myocardial infarction, stroke, renal failure, drug abuse or other
serious diseases. Patients were categorized as hypertensive
according to the ‘diagnostic gold standard’ (19) and had at least three consecutive
measurements of systolic blood pressure (SBP) >140 mm Hg and/or
diastolic blood pressure (DBP) >90 mm Hg (19). In addition, the hypertensive
patients were newly diagnosed patients and had not received therapy
for hypertension. Healthy controls had SBP and DBP <120 mm Hg
and <80 mm Hg respectively, had no family history of
hypertension in first degree relatives and had not received therapy
for hypertension. A calibrated mercury sphygmomanometer with an
adult-sized cuff was used to quantify the blood pressure according
to standard protocols of the American Heart Association (20). Blood pressure was measured in the
supine position twice ≥10 min apart by different trained
technicians. Following a 12 h overnight fast, 5 ml blood samples
were collected from the antecubital vein using vacutainer tubes
containing EDTA and stored at −80°C for DNA extraction. The
protocol of the present study was approved by the Ethics Committee
of Ningbo Seventh Hospital of Ningbo (Ningbo, China) and written
informed consent was obtained from all patients.
Biochemical analyses
Plasma levels of total cholesterol, triglyceride,
alanine transaminase (ALT), uric acid, high-density lipoprotein
(HDL), low-density lipoprotein (LDL), homocysteine (Hcy), and
glucose were quantified enzymatically using an AU2700 automatic
analyzer (Olympus Corporation, Tokyo, Japan). A Lab-Aid 820 nucleic
acid extraction analyzer (Zeesan Biotech Co. Ltd., Xiamen, China)
was used to extract genomic DNA from peripheral blood samples. DNA
concentration was quantified using a NanoDrop 2000 ultramicro
nucleic acid ultraviolet tester (Thermo Fisher Scientific, Inc.,
Waltham, MA, USA).
Pyrosequencing, a sequencing-by-synthesis technique,
was used to determine the methylation levels. The target sequences
were first treated with sodium bisulfite using an EpiTech Bisulfite
kit (Qiagen GmbH, Hilden, Germany) to preferentially convert
unmethylated cytosine residues to thymine and then amplified by
polymerase chain reaction, finally ‘sequenced by synthesis’ using
Pyromark Gold Q96 (Qiagen GmbH) as previously described (17,18,21).
In addition, CpG island (CGI) was identified using MethPrimer
(www.urogene.org/methprimer/)
(22). CpG sites of interest and
PCR primers were selected according to the general rules and advice
of primer design as previously described (23) and the scores were automatically
calculated by the PyroMark Assay Design, version 2.0.1.15 (Qiagen
GmbH). Targets were amplified using a Mastercycler Nexus Gradient
(Eppendorf, Hamburg, Germany) in reactions containing 8 µl
DNase/RNase-free water, 12 µl ZymoTaq Premix (Zymo Research
Corporation, Irvine, CA, USA), 2 µl bisulfite-converted DNA, and
1.5 µl each of forward (F) and reverse (R) primer. Reactions were
initially denatured at 95°C for 10 min, amplified over 45 cycles at
95°C for 30 sec, 52.8°C for 40 sec, and 72°C for 50 sec, and
extended at 72°C for 7 min. Targets were amplified with F
5′-GGGTAGATTAAGAGGTTAGAAG-3′ and R 5′-Biotin-ATT CAC CCC ATT CTC
CTA-3′, and sequenced with primer
5′-TTATTAAAAATATAAAAATATTAG-3′.
Statistical analyses
Data were analyzed using PASW Statistics, version
18.0 (IBM SPSS, Armonk, NY, USA). Continuous variables, including
DNA methylation, age, body mass index (BMI), total cholesterol,
triglycerides, glucose, ALT, uric acid, HDL, LDL and Hcy were
compared by Student's t-test or rank-sum test Pearson χ2
or Fisher's exact test were used to analyze the association between
EH and categorical variables such as sex, smoking and alcohol
consumption. Pearson correlation analysis was used to investigate
interactions among the five CpG sites in the ACE2 promoter.
Logistic regression and multiple linear regression were applied to
adjust for confounding factors. Receiver operating characteristic
(ROC) curves were constructed to determine the sensitivity of
ACE2 methylation as a predictor of EH. P<0.05 was
considered to indicate a statistically significant difference.
Generalized multifactor dimensionality reduction
(GMDR) (http://www.ssg.uab.edu/gmdr/) was
used to investigate potential high-order interactions between
ACE2 promoter methylation and risk of EH. In this approach,
high-dimensional data is reduced to a one-dimensional variable with
two levels (high risk or low risk) (24). The method may detect interactions
in small sample sizes, adjust for quantitative and discrete
covariates and may be used dichotomous and continuous phenotypes.
Additionally, this approach does not require a genetic model and is
a non-parametric alternative to linear or logistic regression for
the detection and characterization of interactions between genetic
and environmental attributes (24). In the present study, the data set
was randomly split into 10 subsets, of which 9 were used for
training and one for testing. N factors were selected from
the training set and combined in n-dimensional space. A number of
parameters were provided to estimate training balanced accuracy,
testing balanced accuracy, sign test P-value, and cross-validation
consistency for each candidate interaction model. From the
candidate models, the one with a sign test P-value of <0.05 and
the highest cross-validation consistency, training, and testing
balanced accuracy was identified to be the most suitable model
(24).
Results
Patient characteristics and analysis
of promoter methylation
A total of 96 patients with EH were recruited, along
with 96 sex- and age-matched (±3 years) healthy controls. The
characteristics of the study population are summarized in Table I.
 | Table I.Characteristics of the study
population (n=192). |
Table I.
Characteristics of the study
population (n=192).
| Characteristic | Healthy (n=96) | EH (n=96) |
t/χ2 | P-value |
|---|
| Age (years) | 56.32±8.23 | 56.72±8.71 | −4.49 |
2.02×10−5b |
| Sex (M/F) | 38/58 | 38/58 | 0 | 1.000 |
| Smoking (Y/N) | 17/79 | 27/69 | 4.05 |
0.041b |
| Drinking (Y/N) | 31/65 | 40/56 | 2.31 | 0.175 |
| BMI
(kg/m2) | 22.20±2.40 | 23.62±3.28 | −3.48 |
0.001b |
| Total cholesterol
(mmol/l) | 5.21±0.88 | 5.38±0.61 | −1.55 | 0.125 |
| Triglycerides
(mmol/l) | 1.21±0.68 | 1.43±0.72 | −2.25 |
0.027b |
| Glucose
(mmol/l) | 4.91±0.79 | 4.90±0.31 | 0.14 | 0.888 |
| ALT (IU/l) | 26.43±16.18 | 28.44±11.95 | −0.96 | 0.340 |
| HDL (mmol/l) | 8.01±6.35 | 2.07±5.61 | 6.53 |
3.32×10−9b |
| LDL (mmol/l) | 3.22±0.86 | 3.31±0.69 | −0.81 | 0.421 |
| Uric acid
(mmol/l) | 300.81±73.38 | 325.75±83.07 | −2.68 |
0.009b |
| Hcy (µmol/l) | 9.38±2.04 | 12.33±4.28 | −2.64 |
0.018b |
| CpG1 methylation
(%) | 69.97±2.40 | 69.07±5.09 | 1.42 |
0.147a |
| CpG2 methylation
(%) | 35.20±2.54 | 35.30±1.90 | −0.37 |
0.870a |
| CpG3 methylation
(%) | 23.13±3.75 | 23.18±4.25 | 0.09 |
0.055a |
| CpG4 methylation
(%) | 95.73±9.11 | 97.56±5.65 | −1.61 |
0.020a,b |
| CpG5 methylation
(%) | 11.47±3.67 | 12.75±4.15 | −2.45 |
0.036a,b |
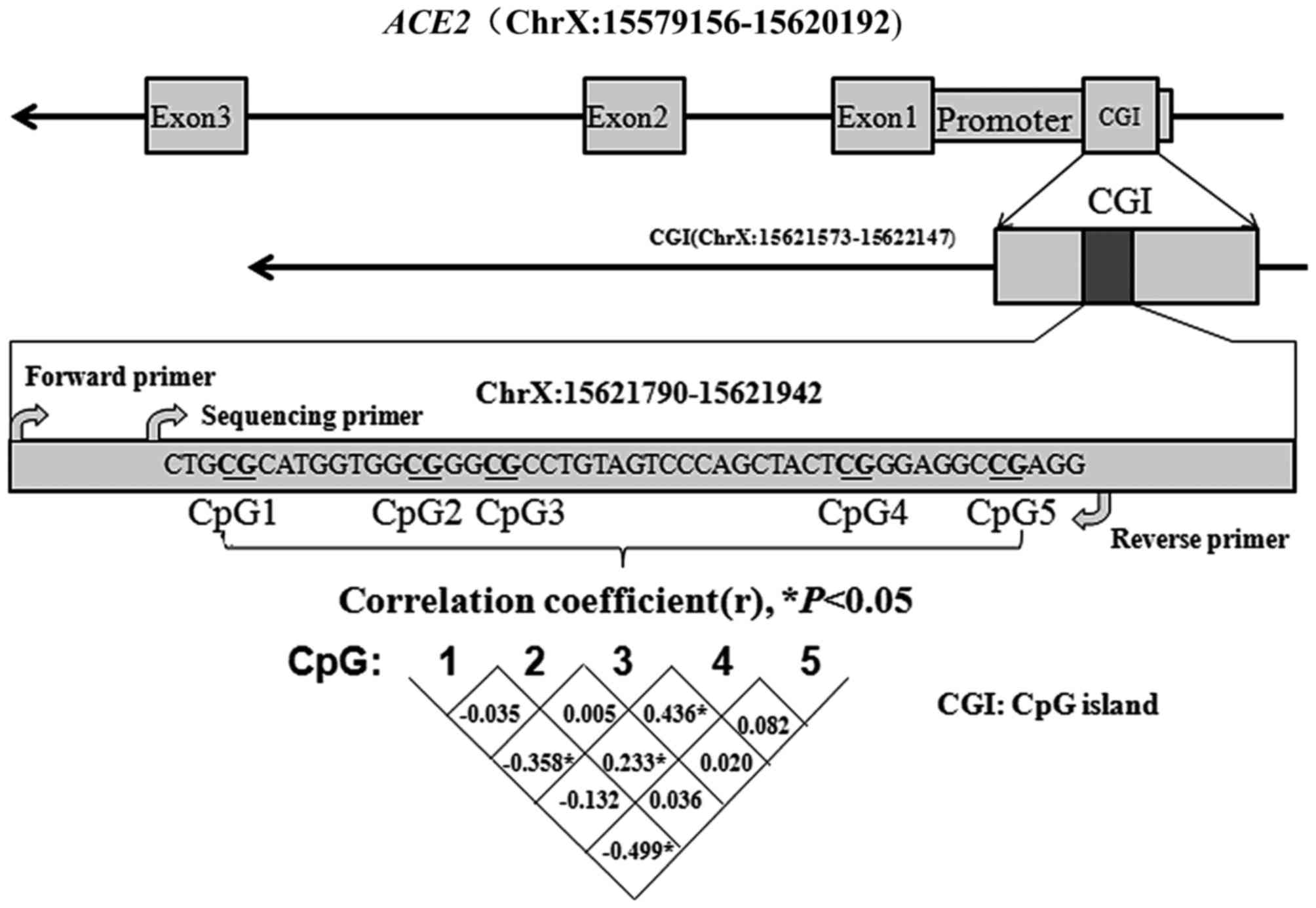
A CpG island (CGI) was identified in the ACE2
promoter using MethPrimer (22).
Subsequently, a fragment containing five CpG dinucleotides in this
island (ChrX:15621573-15622147) was selected (Fig. 1). The correlation among the five
CpG sites is presented Fig. 1
(r<0.5).
Promoter methylation and essential
hypertension
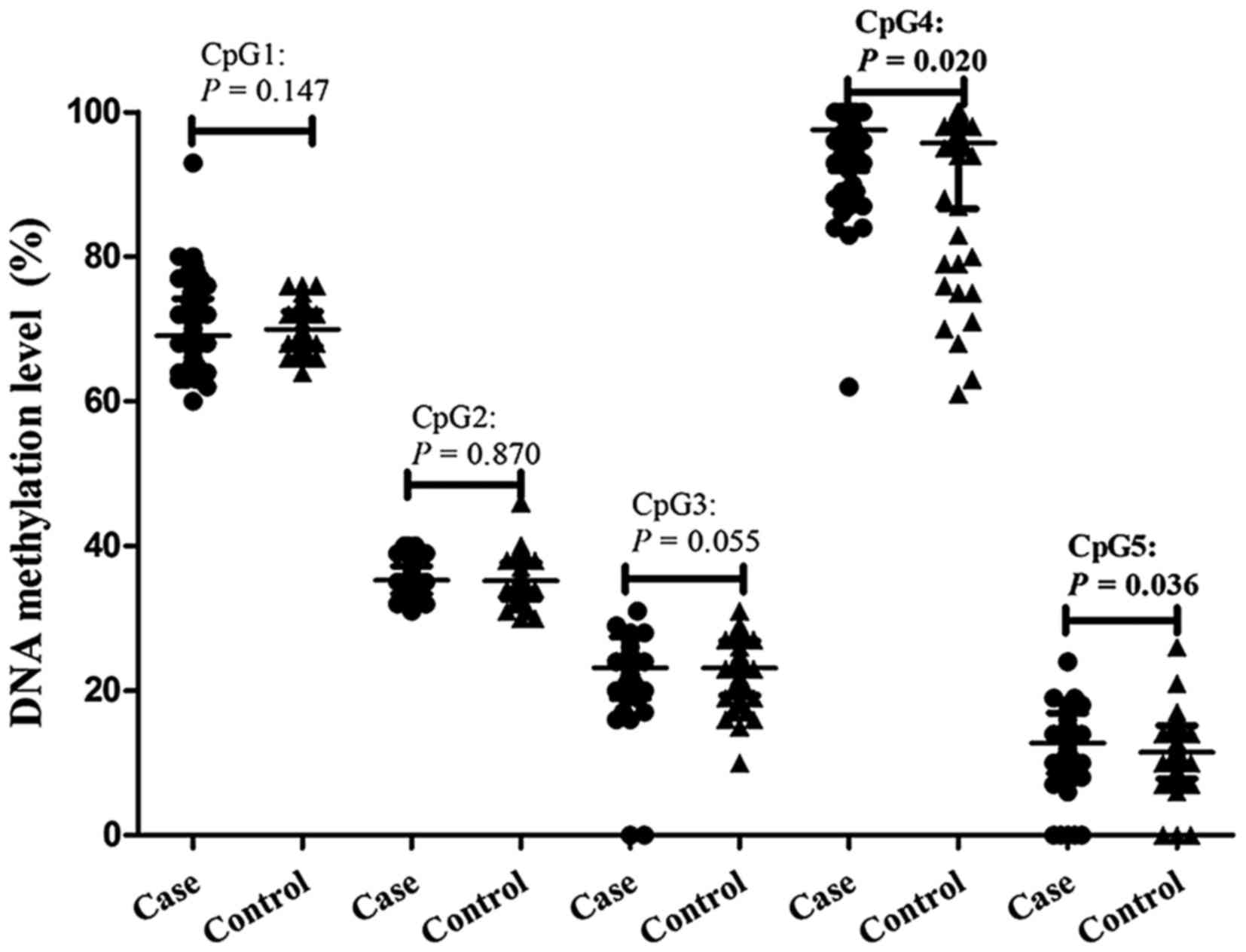
Methylation of ACE2 in CpG4 (adjusted
P=0.020) and CpG5 (adjusted P=0.036) was significantly higher in
cases of EH, with frequency 97.56±5.65% and 12.75±4.15% in patients
with EH and 95.73±9.11% and 11.47±3.67% in healthy controls,
respectively. However, EH was not significantly associated with
methylation of the remaining three CpG sites following adjusting
for age, sex, smoking, alcohol use, BMI, triglycerides, HDL, uric
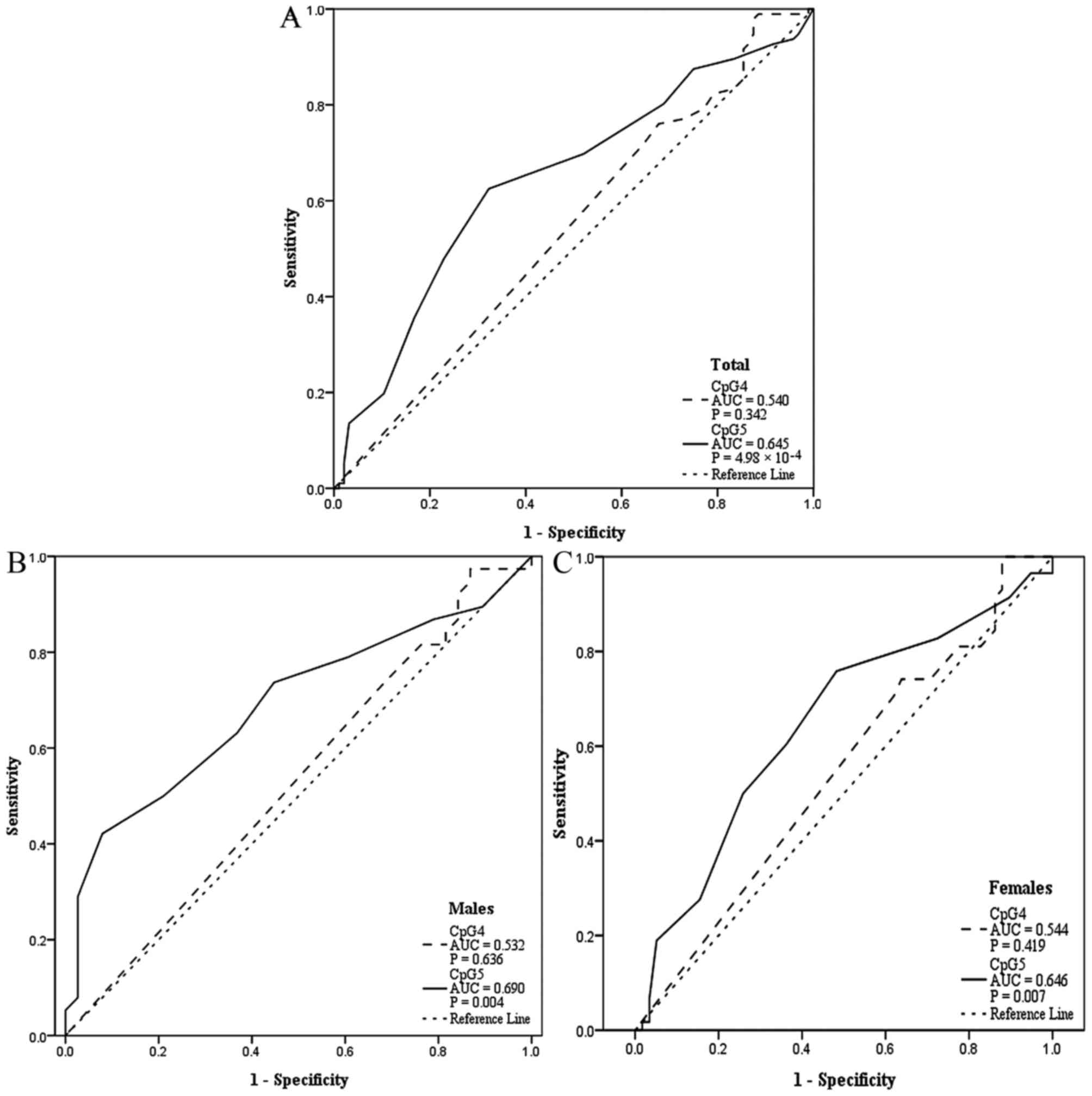
acid, and Hcy (Table I; Fig. 2). In addition, CpG5 methylation was
determined to be a significant predictor of EH based on ROC curves
(Fig. 3), with area under the
curve was 0.645 for all patients (P=4.98×10−4), 0.690
for males (P=0.004), and 0.646 for females (P=0.007).
GMDR was then used to investigate high-order
interactions among the five CpG sites. The best models at various
orders are summarized in Table
II. The five-factor model had the best training balanced
accuracy (0.72), testing balanced accuracy (0.65), and
cross-validation consistency (10/10). The adjusted P-value was 0.01
following the sign test and the training odds ratio (OR) was 7.33
with 95% confidence interval (2.03, 26.49).
 | Table II.GMDR models of high-order interaction
among the five CpG sites in angiotensin I converting enzyme 2
promoter on essential hypertension risk. |
Table II.
GMDR models of high-order interaction
among the five CpG sites in angiotensin I converting enzyme 2
promoter on essential hypertension risk.
| Model | Training balanced
accuracy | Testing balanced
accuracy | Sign test
(P-value) | Cross-validation
consistency |
|---|
| CpG5 | 0.62 | 0.62 | 9
(P=0.011a) | 10/10 |
| CpG3, CpG5 | 0.63 | 0.57 | 7 (P=0.172) |
6/10 |
| CpG2, CpG3,
CpG5 | 0.67 | 0.58 | 7 (P=0.172) |
6/10 |
| CpG1, CpG3, CpG4,
CpG5 | 0.69 | 0.60 | 8 (P=0.055) |
7/10 |
| CpG1, CpG2, CpG3,
CpG4, CpG5 | 0.72 | 0.65 | 9
(P=0.011a) | 10/10 |
Association of clinical variables with
promoter methylation
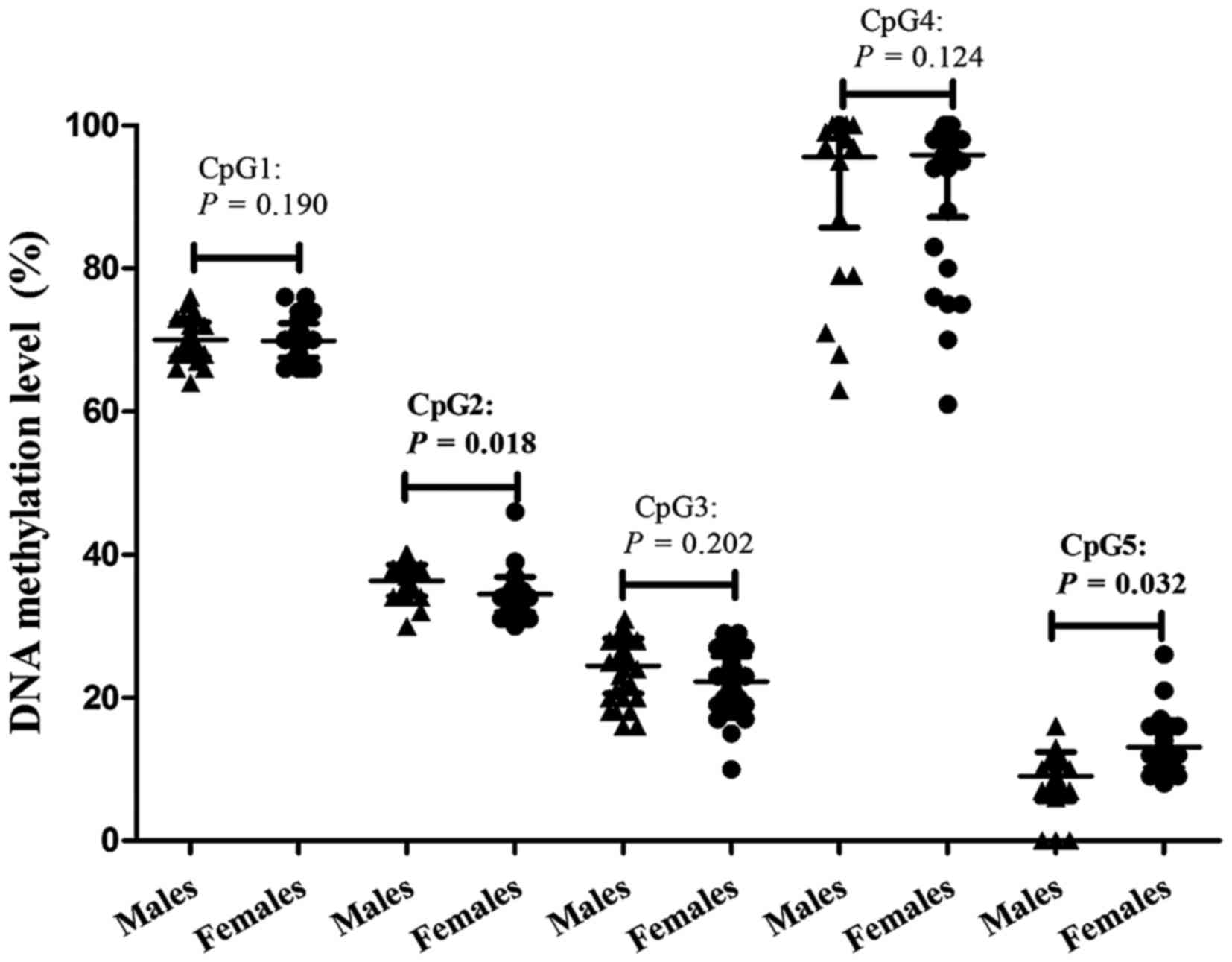
Methylation of CpG2 was significantly higher
(adjusted P=0.018) in healthy males compared with healthy females,
with frequency 36.21±2.21% and 34.71±1.40%, respectively. In
contrast, CpG5 methylation was significantly lower (adjusted
P=0.032) in males (10.97±4.28%) compared with females (13.91±3.66%)
following adjusting for confounding factors (Table III; Fig. 4). As presented in Table I, significant differences between
hypertensive and healthy subjects were also detected in age
(P=2.02×10−5), smoking (P=0.041), BMI (P=0.001),
triglyceride (P=0.027), HDL (P=3.32×10−9), uric acid
(P=0.009) and Hcy (P=0.018). Therefore, a multiple linear
regression was used to test whether these clinical variables were
associated with ACE2 methylation in healthy controls.
However, no significant difference was identified (data not
shown).
 | Table III.Angiotensin I converting enzyme 2 CpG
methylation in healthy males (n=38) and females (n=58). |
Table III.
Angiotensin I converting enzyme 2 CpG
methylation in healthy males (n=38) and females (n=58).
| Characteristic | Males | Females |
t/χ2 | P-value |
|---|
| Age (years) | 55.55±10.79 | 56.83±6.06 | −0.66 | 0.51 |
| Smoking (Y/N) | 17/21 | 0/58 | 31.53 |
1.96×10−8b |
| Drinking (Y/N) | 21/17 | 10/48 | 15.18 |
9.77×10−5b |
| BMI
(kg/m2) | 23.11±2.35 | 21.54±2.05 | 3.46 |
0.001b |
| Total cholesterol
(mmol/l) | 5.07±1.07 | 5.27±0.76 | −1.06 |
0.293 |
| Triglycerides
(mmol/l) | 1.44±0.87 | 1.06±0.47 | 2.46 |
0.017b |
| Glucose
(mmol/l) | 4.86±1.20 | 4.94±0.33 | 0.43 |
0.669 |
| ALT (IU/l) | 27.05±14.29 | 25.98±17.29 | 0.32 |
0.752 |
| HDL (mmol/l) | 5.16±6.58 | 9.85±5.44 | −3.79 |
2.62×10−4b |
| LDL (mmol/l) | 3.20±1.05 | 3.21±0.73 | −0.07 |
0.942 |
| Uric acid
(mmol/l) | 352.36±2.18 | 266.23±50.48 | 6.88 |
6.52×10−10b |
| Hcy (µmol/l) | 12.07±8.18 | 9.21±1.17 | 2.14 |
0.039b |
| CpG1 methylation
(%) | 70.92±5.51 | 67.86±4.44 | 2.87 |
0.190a |
| CpG2 methylation
(%) | 36.21±2.21 | 34.71±1.40 | 3.73 |
0.018a,b |
| CpG3 methylation
(%) | 23.42±3.06 | 23.02±4.89 | 0.45 |
0.202a |
| CpG4 methylation
(%) | 97.39±7.09 | 97.67±4.52 | −0.24 |
0.124a |
| CpG5 methylation
(%) | 10.97±4.28 | 13.91±3.66 | −3.60 |
0.032a,b |
Discussion
Previous studies have demonstrated that ACE2
polymorphisms are associated with risk of EH (9,10).
Therefore, it is possible that aberrant methylation of the
ACE2 promoter may also contribute to this risk. The results
of the present study indicated that CpG4 and CpG5 in the
ACE2 promoter were hypermethylated in patients with EH and a
significant interaction among the five CpG sites was observed.
Furthermore, the present study determined that methylation of CpG2
and CpG5 was significantly different between males and females. The
observations of the present study elucidated the underlying
mechanism of the pathogenesis of EH.
ACE2 counterbalances the effect of RAS by degrading
the vasoconstrictor angiotensin II, and generating the vasodilators
Ang 1–7 (7). Since its discovery
in 2000 (5,6), ACE2 has been identified as a
candidate gene that may be responsible for the development of EH
and to the best of our knowledge, the present study was the first
to examine the association between EH and the methylation status of
the ACE2 promoter. Promoter hypermethylation inactivates
transcription, whereas hypomethylation promotes active
transcription (12). A previous
study determined that promoter hypomethylation upregulated
AGTR1 expression, a key gene in RAS that was closely
associated with EH (25).
Therefore, hypermethylation of CpG4 and CpG5 in the ACE2
promoter may reduce expression, promoting EH pathogenesis. However,
as no expression analysis was performed in the current study, the
observations are only correlative and not causal. Ongoing
expression analysis is required to confirm the present
findings.
As EH is a multifactorial disease, gene-gene and
gene-environment interactions contribute to its onset and
progression. However, due to the ‘curse of dimensionality,’
traditional statistical methods are unsuitable to detect these
potential interactions. Non-parametric methods that do not require
genetic models have been previously used to identify high-order
interactions efficiently. One such method is GMDR, which
accommodates qualitative and quantitative phenotypes, adjusts for
discrete and continuous covariates and enhances prediction accuracy
(24). Using this method, the
present study detected a significant five-order interaction among
the five CpG sites in the ACE2 promoter, an interaction that
may contribute to the risk of EH. It is of note that there may be a
7.33-fold increased risk of developing EH in individuals with
hypermethylation of all five CpG sites (OR=7.33). Nevertheless,
this interaction is purely theoretical at present, based on
statistical analyses, and it is only descriptive of variations in
the population (24). The
physiological relevance of such an interaction, if any, remains to
be elucidated and should be investigated in future experiments.
It is of note, that as the ACE2 gene is
located on the X chromosome and the prevalence and progression of
EH, and the methylation of hypertension-associated genes have been
determined to display sex differences (17,26).
In order to maintain equal gene expression between males and
females, one female X chromosome is randomly inactivated, a process
termed X-inactivation (27). The
inactive female X chromosome has higher methylation levels compared
with the active female X chromosome in promoter CpG islands
(28). However, the ACE2
gene location on Xp22 encompasses an area where genes are reported
to escape from X-inactivation (29), which may lead to the methylation
differences of ACE2 CpG2 and CpG5 between the two sexes
observed in the current study. In addition, sex-specific hormones
that modify DNA methylation (30)
and sex differences in non-heritable risk factors for EH, including
alcohol consumption, smoking, physical activity and a high-sodium
diet, may also alter ACE2 methylation levels (31–34).
Additionally, it is possible that site-specific differences, as
observed between males and females in CpG2 and CpG5 methylation,
may be due to heterogeneity in methylation of different CpG sites
in the same promoter (35–38). This heterogeneity is biologically
relevant; however, the mechanisms that drive site-specific
methylation remain to be elucidated. It is of note that no
association between ACE2 methylation and other clinical
variables such as age and BMI was observed, therefore further
investigation is required to confirm this result.
The present study had numerous strengths, and was
able to draw conclusions by adjusting for confounding factors
through the use of logistic and multiple linear regression and by
overcoming the ‘curse of dimensionality’ through GMDR models.
However, the following limitations have been identified: i)
Cause-effect association between methylation of the ACE2
promoter and EH remains to be determined, as the survey was a
case-control study; ii) only a fragment of the CpG island in the
ACE2 promoter was analyzed; iii) the statistical analysis
controlled for certain confounding factors, however, it is possible
that other confounding factors that influence ACE2
methylation may have not been accounted for; iv) peripheral blood
is a surrogate tissue for epigenetic studies, although previous
studies have indicated that CpG methylation patterns are similar
between peripheral blood and other tissues (39,40),
as DNA methylation, may vary across tissues, similar analysis of
ACE2 methylation in other tissues may be required; and v) no
expression analysis was performed in the present study. Therefore,
the observations of the current study can only be regarded as
correlative. Ongoing expression analysis is required to confirm the
results of the present study.
In conclusion, the observations of the present study
provided evidence of the association between EH and
hypermethylation of CpG4 and CpG5 in the ACE2 promoter and
the interactions among CpG1-CpG5. It is of note, that methylation
of ACE2 CpG5 may have predictive potential as a tool to
estimate risk of EH in patients. Additionally, sex may affect
ACE2 methylation. These observations further understanding
of the pathogenesis of EH and may aid in the improvement of the
diagnosis and treatment of patients with EH.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant no. 81373094), the K. C.
Wong Magna Fund in Ningbo University, Ningbo Scientific Innovation
Team for Environmental Hazardous Factor Control and Prevention
(grant no. 2016C51001), Zhejiang Province Social development
Research Project (grant no. 2016C33178), Ningbo Social Development
Research Project (grant no. 2014C50051), Outstanding (Postgraduate)
Dissertation Growth Foundation of Ningbo University (grant no.
py2014015), and the Scientific Research Innovation Foundation of
Ningbo University (grant no. G15070).
References
|
1
|
Li D, Lv J, Liu F, Liu P, Yang X, Feng Y,
Chen G and Hao M: Hypertension burden and control in mainland
China: Analysis of nationwide data 2003–2012. Int J Cardiol.
184:637–644. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kearney PM, Whelton M, Reynolds K, Muntner
P, Whelton PK and He J: Global burden of hypertension: Analysis of
worldwide data. Lancet. 365:217–223. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pausova Z, Tremblay J and Hamet P:
Gene-environment interactions in hypertension. Curr Hypertens Rep.
1:42–50. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhu X, Chang YP, Yan D, Weder A, Cooper R,
Luke A, Kan D and Chakravarti A: Associations between hypertension
and genes in the renin-angiotensin system. Hypertension.
41:1027–1034. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Donoghue M, Hsieh F, Baronas E, Godbout K,
Gosselin M, Stagliano N, Donovan M, Woolf B, Robison K, Jeyaseelan
R, et al: A novel angiotensin-converting enzyme-related
carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1–9.
Circ Res. 87:E1–E9. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tipnis SR, Hooper NM, Hyde R, Karran E,
Christie G and Turner AJ: A human homolog of angiotensin-converting
enzyme. Cloning and functional expression as a
captopril-insensitive carboxypeptidase. J Biol Chem.
275:33238–33243. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tallant EA and Clark MA: Molecular
mechanisms of inhibition of vascular growth by angiotensin-(1–7).
Hypertension. 42:574–579. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yagil Y and Yagil C: Hypothesis: ACE2
modulates blood pressure in the mammalian organism. Hypertension.
41:871–873. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lu N, Yang Y, Wang Y, Liu Y, Fu G, Chen D,
Dai H, Fan X, Hui R and Zheng Y: ACE2 gene polymorphism and
essential hypertension: An updated meta-analysis involving 11,051
subjects. Mol Biol Rep. 39:6581–6589. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Patel SK, Wai B, Ord M, MacIsaac RJ, Grant
S, Velkoska E, Panagiotopoulos S, Jerums G, Srivastava PM and
Burrell LM: Association of ACE2 genetic variants with blood
pressure, left ventricular mass, and cardiac function in Caucasians
with type 2 diabetes. Am J Hypertens. 25:216–222. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Razin A, Webb C, Szyf M, Yisraeli J,
Rosenthal A, Naveh-Many T, Sciaky-Gallili N and Cedar H: Variations
in DNA methylation during mouse cell differentiation in vivo and in
vitro. Proc Natl Acad Sci USA. 81:pp. 2275–2279. 1984; View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Deaton AM and Bird A: CpG islands and the
regulation of transcription. Genes Dev. 25:1010–1022. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bogdarina I, Welham S, King PJ, Burns SP
and Clark AJ: Epigenetic modification of the renin-angiotensin
system in the fetal programming of hypertension. Circ Res.
100:520–526. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Rangel M, dos Santos JC, Ortiz PH, Hirata
M, Jasiulionis MG, Araujo RC, Ierardi DF and Mdo C Franco:
Modification of epigenetic patterns in low birth weight children:
Importance of hypomethylation of the ACE gene promoter. PLoS One.
9:e1061382014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang F, Demura M, Cheng Y, Zhu A,
Karashima S, Yoneda T, Demura Y, Maeda Y, Namiki M, Ono K, et al:
Dynamic CCAAT/enhancer binding protein-associated changes of DNA
methylation in the angiotensinogen gene. Hypertension. 63:281–288.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fan R, Mao S, Zhong F, Gong M, Yin F, Hao
L and Zhang L: Association of AGTR1 promoter methylation levels
with essential hypertension risk: A matched case-control study.
Cytogenet Genome Res. 147:95–102. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang LN, Liu PP, Wang L, Yuan F, Xu L,
Xin Y, Fei LJ, Zhong QL, Huang Y, Xu L, et al: Lower ADD1 gene
promoter DNA methylation increases the risk of essential
hypertension. PLoS One. 8:e634552013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Fan R, Wang WJ, Zhong QL, Duan SW, Xu XT,
Hao LM, Zhao J and Zhang LN: Aberrant methylation of the GCK gene
body is associated with the risk of essential hypertension. Mol Med
Rep. 12:2390–2394. 2015.PubMed/NCBI
|
|
19
|
European Society of Hypertension-European
Society of Cardiology Guidelines Committee, . 2003 European Society
of Hypertension-European Society of Cardiology guidelines for the
management of arterial hypertension. J Hypertens. 21:1011–1053.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Perloff D, Grim C, Flack J, Frohlich ED,
Hill M, McDonald M and Morgenstern BZ: Human blood pressure
determination by sphygmomanometry. Circulation. 88:2460–2470. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bassil CF, Huang Z and Murphy SK:
Bisulfite pyrosequencing. Methods Mol Biol. 1049:95–107. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li LC and Dahiya R: MethPrimer: Designing
primers for methylation PCRs. Bioinformatics. 18:1427–1431. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Mikeska T, Felsberg J, Hewitt CA and
Dobrovic A: Analysing DNA methylation using bisulphite
pyrosequencing. Methods Mol Biol. 791:33–53. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lou XY, Chen GB, Yan L, Ma JZ, Zhu J,
Elston RC and Li MD: A generalized combinatorial approach for
detecting gene-by-gene and gene-by-environment interactions with
application to nicotine dependence. Am J Hum Genet. 80:1125–1137.
2007. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Pei F, Wang X, Yue R, Chen C, Huang J,
Huang J, Li X and Zeng C: Differential expression and DNA
methylation of angiotensin type 1A receptors in vascular tissues
during genetic hypertension development. Mol Cell Biochem. 402:1–8.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Jiang D, Zheng D, Wang L, Huang Y, Liu H,
Xu L, Liao Q, Liu P, Shi X, Wang Z, et al: Elevated PLA2G7 gene
promoter methylation as a gender-specific marker of aging increases
the risk of coronary heart disease in females. PLoS One.
8:e597522013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Berletch JB, Yang F, Xu J, Carrel L and
Disteche CM: Genes that escape from X inactivation. Hum Genet.
130:237–245. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hellman A and Chess A: Gene body-specific
methylation on the active X chromosome. Science. 315:1141–1143.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Carrel L and Willard HF: X-inactivation
profile reveals extensive variability in X-linked gene expression
in females. Nature. 434:400–404. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sebag IA, Gillis MA, Calderone A, Kasneci
A, Meilleur M, Haddad R, Noiles W, Patel B and Chalifour LE: Sex
hormone control of left ventricular structure/function: Mechanistic
insights using echocardiography, expression, and DNA methylation
analyses in adult mice. Am J Physiol Heart Circ Physiol.
301:H1706–H1715. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Christensen BC, Houseman EA, Marsit CJ,
Zheng S, Wrensch MR, Wiemels JL, Nelson HH, Karagas MR, Padbury JF,
Bueno R, et al: Aging and environmental exposures alter
tissue-specific DNA methylation dependent upon CpG island context.
PLoS Genet. 5:e10006022009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Breitling LP, Yang R, Korn B, Burwinkel B
and Brenner H: Tobacco-smoking-related differential DNA
methylation: 27K discovery and replication. Am J Hum Genet.
88:450–457. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Philibert RA, Plume JM, Gibbons FX, Brody
GH and Beach SR: The impact of recent alcohol use on genome wide
DNA methylation signatures. Front Genet. 3:542012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ronn T, Volkov P, Davegårdh C, Dayeh T,
Hall E, Olsson AH, Nilsson E, Tornberg A, Nitert M Dekker, Eriksson
KF, et al: A six months exercise intervention influences the
genome-wide DNA methylation pattern in human adipose tissue. PLoS
Genet. 9:e10035722013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Alexeeff SE, Baccarelli AA, Halonen J,
Coull BA, Wright RO, Tarantini L, Bollati V, Sparrow D, Vokonas P
and Schwartz J: Association between blood pressure and DNA
methylation of retrotransposons and pro-inflammatory genes. Int J
Epidemiol. 42:270–280. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ishida K, Kobayashi T, Ito S, Komatsu Y,
Yokoyama T, Okada M, Abe A, Murasawa A and Yoshie H: Interleukin-6
gene promoter methylation in rheumatoid arthritis and chronic
periodontitis. J Periodontol. 83:917–925. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Pogribny IP, Pogribna M, Christman JK and
James SJ: Single-site methylation within the p53 promoter region
reduces gene expression in a reporter gene construct: Possible in
vivo relevance during tumorigenesis. Cancer Res. 60:588–594.
2000.PubMed/NCBI
|
|
38
|
Zou B, Chim CS, Zeng H, Leung SY, Yang Y,
Tu SP, Lin MC, Wang J, He H, Jiang SH, et al: Correlation between
the single-site CpG methylation and expression silencing of the
XAF1 gene in human gastric and colon cancers. Gastroenterology.
131:1835–1843. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Fan S and Zhang X: CpG island methylation
pattern in different human tissues and its correlation with gene
expression. Biochem Biophys Res Commun. 383:421–425. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Mirza S, Sharma G, Parshad R, Srivastava
A, Gupta SD and Ralhan R: Clinical significance of promoter
hypermethylation of ERβ and RARβ2 in tumor and serum DNA in Indian
breast cancer patients. Ann Surg Oncol. 19:3107–3115. 2012.
View Article : Google Scholar : PubMed/NCBI
|