Introduction
Medulloblastoma is the most common pediatric brain
tumor, with an incidence of ~0.5/100,000 children <15 years old
(1). It is classified as grade IV
according to the World Health Organization (WHO) (2). Current treatment consisting of
surgery, radiotherapy and adjuvant chemotherapy has a limited role
in improving the prognosis of patients (3,4).
Novel therapeutic methods based on the specific mechanism of
medulloblastoma carcinogenesis are required to improve treatment
efficiency and avoid the side effects of traditional therapy. An
increasing number of studies have revealed that microRNAs
(miRNAs/miRs) serve a role in tumorigenesis (5–9).
miRNAs are a type of small non-coding RNA (18–24
nucleotides). Gene expression is regulated by thousands of known
miRNAs, which bind to imperfect complementary sites within the 3′
untranslated regions of their target protein-coding mRNAs, and
repressing the expression of these genes at the level of mRNA
stability and translation (10).
Previous research has revealed that miRNAs are involved in the
regulation of the majority of physiological and pathological
process, including development, life span, and metabolism, and
their dysregulation contributes to a number of types of disease,
notably cancer (11). These small
RNAs may function as oncogenes or tumor suppressors by modulating
the levels of critical proteins, and their relevance in human
disease and therapy is currently under investigation (12).
An increasing number of studies have demonstrated
abnormal miRNA expression levels in the central nervous system
(CNS) of cancer patients, suggesting that miRNAs may serve as key
regulatory components in cancer of the CNS (5,10,13).
The present study analyzed the medulloblastoma miRNA expression
profiles for pediatric brain tumors. It was observed that 22 miRNAs
were upregulated and 26 miRNAs were downregulated in the tumor
tissue. Ingenuity Pathway Analysis (IPA) was used to analyze the
regulatory network of the differentially expressed miRNA in
medulloblastoma. It was proposed that the pathways exhibited in the
network may have an important role in the tumorigenesis of
medulloblastoma.
Materials and methods
The data were downloaded from the NCBI Gene
Expression Omnibus (GEO) database (GSE42657, www.ncbi.nlm.nih.gov/geo). The series matrix file was
downloaded and a log2 transformation was performed. The data
include 7 control samples (consisting of 3 cerebellum and 4 frontal
lobe controls) and 9 medulloblastoma samples. The data from all
samples was normalized using the limma package in R software
version 3.3.0 (https://www.r-project.org/). The selected samples of
this profile are exhibited in Table
I. The differentially expressed genes (DEGs) were obtained with
the thresholds of |logFC|>1.0 and P<0.01, using linear models
and empirical Bayes methods for assessing differential expression
in microarray experiments in the limma package (Table II). A heat map of the different
groups was produced to visualize the DEGs and cluster the
corresponding groups with the differentially expressed miRNAs from
the tumor tissue, using the R package gplots (Fig. 1).
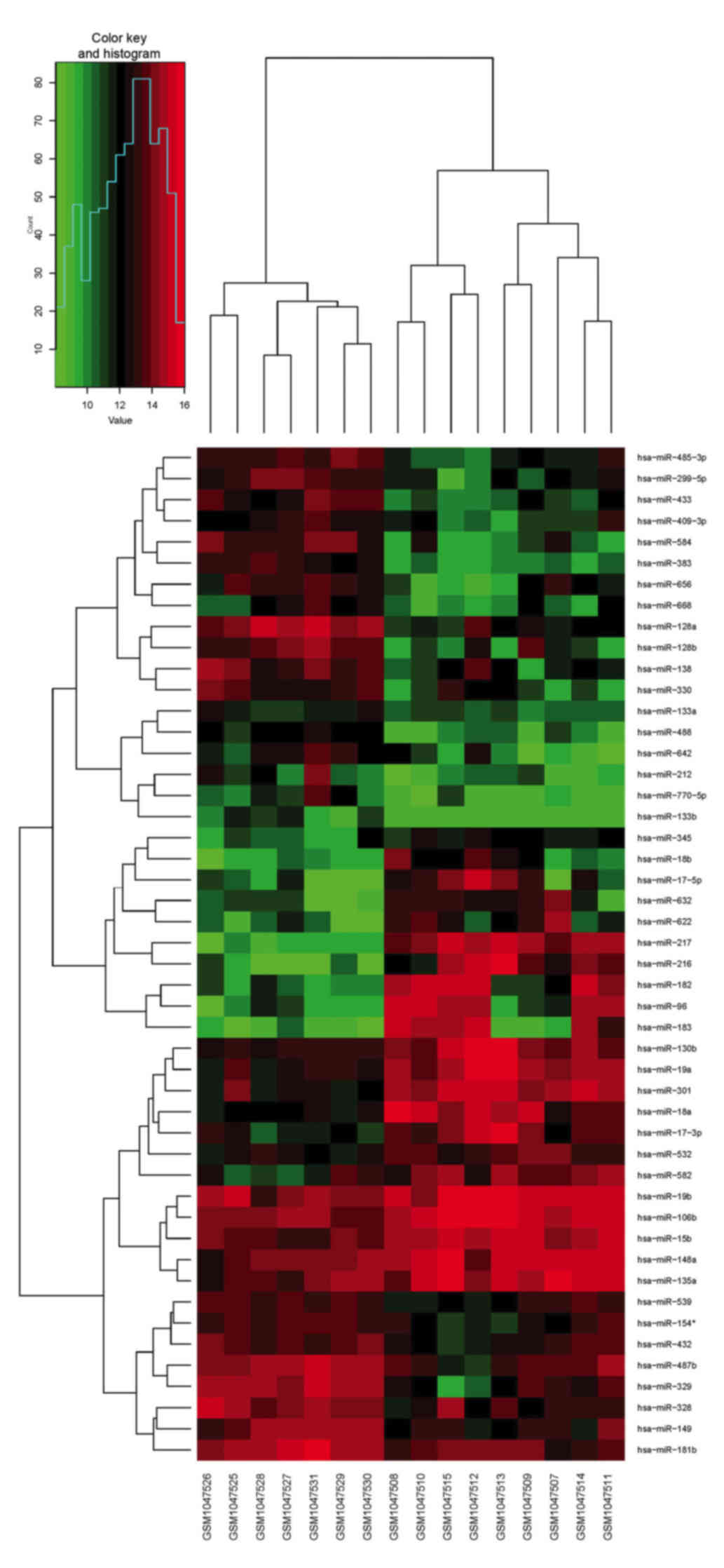 | Figure 1.Differentially expressed miRs in
medulloblastoma tissue compared with normal control tissue,
visualized through clustering of sample data using the gplots
package in R software. Red represents a miR expression level above
the mean; green represents a miR expression level below the mean.
The differentially expressed miRs were validated with the data in
Gene Expression Omnibus database and the PubMed database articles.
miRs miR-17-5p, miR-148a, miR-18a, miR-19a and miR-106a were
co-upregulated in medulloblastoma. miR-383, miR-328, miR-128a/b,
miR-133b, miR-149, miR-181b, miR-138 and miR-154 were
co-downregulated in medulloblastoma. miR, microRNA. |
 | Table I.Sample descriptions. |
Table I.
Sample descriptions.
| Diagnosis | No. of sample | Serial no. of
sample |
|---|
| Normal (frontal
lobe) | 4 | GSM1047526,
GSM1047529-GSM1047531 |
| Normal
(cerebellum) | 3 | GSM1047525,
GSM1047527-GSM1047528 |
| Medulloblastoma | 9 |
GSM1047507-GSM1047515 |
 | Table II.miRs significantly differentially
expressed (P<0.01) in medulloblastoma tissue compared with
normal control samples. |
Table II.
miRs significantly differentially
expressed (P<0.01) in medulloblastoma tissue compared with
normal control samples.
| Gene names | Log fold change |
|---|
| Upregulated miRs |
|
|
hsa-miR-217 | 5.40 |
|
hsa-miR-301 | 2.54 |
|
hsa-miR-216 | 4.67 |
|
hsa-miR-19a | 2.11 |
|
hsa-miR-15b | 1.16 |
|
hsa-miR-106b | 1.19 |
|
hsa-miR-18a | 2.37 |
|
hsa-miR-130b | 1.56 |
|
hsa-miR-182 | 3.52 |
|
hsa-miR-345 | 1.62 |
|
hsa-miR-17-3p | 2.10 |
|
hsa-miR-96 | 3.71 |
|
hsa-miR-582 | 2.06 |
|
hsa-miR-622 | 2.57 |
|
hsa-miR-532 | 1.12 |
|
hsa-miR-19b | 1.02 |
|
hsa-miR-18b | 2.31 |
|
hsa-miR-632 | 2.52 |
|
hsa-miR-148a | 1.13 |
|
hsa-miR-17-5p | 2.99 |
|
hsa-miR-183 | 3.73 |
|
hsa-miR-135a | 1.19 |
| Downregulated
miRs |
|
|
hsa-miR-383 | −3.05 |
|
hsa-miR-128a | −2.53 |
|
hsa-miR-433 | −2.38 |
|
hsa-miR-488 | −2.22 |
|
hsa-miR-584 | −2.97 |
|
hsa-miR-128b | −2.92 |
|
hsa-miR-485-3p | −1.89 |
|
hsa-miR-329 | −2.43 |
|
hsa-miR-299-5p | −2.22 |
|
hsa-miR-133b | −1.60 |
|
hsa-miR-330 | −2.37 |
|
hsa-miR-181b | −1.17 |
|
hsa-miR-138 | −2.03 |
|
hsa-miR-770-5p | −2.03 |
|
hsa-miR-149 | −1.57 |
|
hsa-miR-133a | −1.00 |
|
hsa-miR-328 | −1.44 |
|
hsa-miR-409-3p | −1.63 |
|
hsa-miR-656 | −2.17 |
|
hsa-miR-642 | −2.37 |
|
hsa-miR-432 | −1.15 |
|
hsa-miR-539 | −1.03 |
|
hsa-miR-668 | −1.79 |
|
hsa-miR-487b | −1.48 |
|
hsa-miR-154* | −1.15 |
|
hsa-miR-212 | −1.81 |
The dataset (48 differentially expressed miRNAs) was
uploaded to the Ingenuity Pathway Analysis software version 2016
(Qiagen, Inc., Valencia, CA, USA), using the microRNA target filter
to find the targeting information. A total of 40 microRNAs
targeting 11,757 mRNAs were identified by the filter. The core
analysis function (rapid assessment of the signaling and metabolic
pathways, molecular networks, and biological processes that are
most markedly perturbed in the dataset of interest) to analyze the
associated network functions of the miRNAs.
Results
Using miRNA microarray data from the GEO database,
48 miRNAs which are differentially expressed in medulloblastoma
were identified, compared with the control group (Table II). The core analysis demonstrated
the molecular network interactions and signaling pathways
associated with 28 differentially expressed miRNAs of
medulloblastoma, and their predicted molecular targets were rebuilt
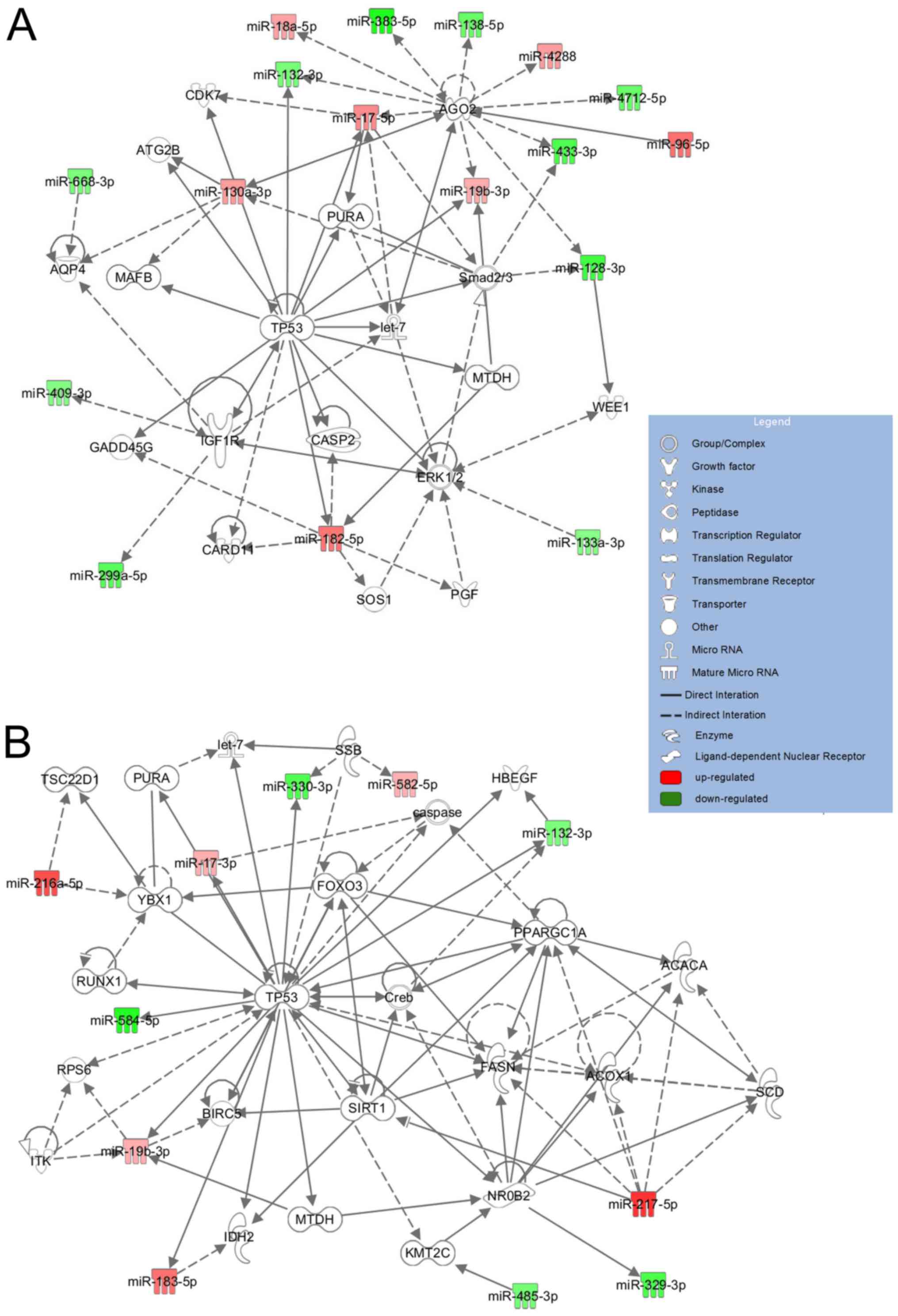
using IPA. The network ‘Organismal Injury and Abnormalities,
Reproductive System Disease, Cancer’ had an IPA score of 41,
focusing on 17 miRNA molecules, and the network ‘Cancer, Organismal
Injury and Abnormalities, Cell Death and Survival’ had an IPA score
of 23, focusing on 11 miRNA molecules (Fig. 2). The most impacted biological
processes and diseases regulated by the analyzed miRNAs included
organismal injury and abnormalities, reproductive system disease
and cancer. The molecular network maps demonstrated that three
primary components were identified to be at the central hub of the
most significant network with a score of 41; these were tumor
protein p53 (TP53), argonaute 2 (AGO2) and mitogen activated
kinases 3 and 1 (ERK1/2) (Fig.
2A). TP53, sirtuin 1 (SIRT1) and Y box protein 1 (YBX1) were
located in the center of the second network with a core analysis
score of 23 (Fig. 2B). The first
network included miR-17-5p, miR-130a-3p, miR-182-5p, miR-18a-5p,
miR-19b-3p, miR-4288, miR-96-5p, which were upregulated in the
medulloblastoma samples, and miR-128-3p, miR-132-3p, miR-133a-3p,
miR-299a-5p, miR-409, miR-668-3p, which were downregulated. The
second notable network (Fig. 2B)
involved upregulated miR-217-5p and miR-216a-5p, and downregulated
miR-329, miR-330 and miR-584, which are associated with the
regulation of key regulatory genes in medulloblastoma development,
including TP53, SIRT1 and YBX1.
However, when the cerebellum samples were used as
the control, it was observed that 4 miRNAs were overexpressed and 5
miRNAs were underexpressed in the medulloblastoma (Table III). The differentially expressed
miRNAs were uploaded to the IPA and core analysis was performed.
The network ‘Cell death and Survival, Gastrointestinal Disease,
Hepatic System Disease’ had an IPA score of 9, focusing on 4 miRNA
molecules (Fig. 3).
 | Table III.miRs significantly differentially
expressed (P<0.01) in medulloblastoma tissue compared with
normal cerebellum samples. |
Table III.
miRs significantly differentially
expressed (P<0.01) in medulloblastoma tissue compared with
normal cerebellum samples.
| Gene names | Log fold
change |
|---|
| Upregulated
miRs |
|
|
hsa-miR-217 |
5.22 |
|
hsa-miR-216 |
5.17 |
|
hsa-miR-582 |
3.31 |
|
hsa-miR-15b |
1.28 |
| Downregulated
miRs |
|
|
hsa-miR-383 | −3.39 |
|
hsa-miR-206 | −3.40 |
|
hsa-miR-133b | −2.47 |
|
hsa-miR-128a | −2.61 |
|
hsa-miR-654 | −1.89 |
Discussion
miRNAs function as master regulators and signal
modulators by fine-tuning gene expression in multiple complex
pathways. Disruption of miRNAs may result in a permissive
tumorigenic state (13). A number
of the miRNAs which exhibit dysregulated expression in
medulloepithelioma have been reported to serve various roles in
carcinogenesis (6,7,9).
Through analysis of a GEO miRNA expression profiling dataset,
miRNAs which are over- and underexpressed in medulloblastoma were
identified, including miR-217, miR-216, miR-183, miR-182 and
miR-96, which are upregulated in tumor tissue. Previous research
demonstrates that miR-183-96-182 regulates cell survival,
proliferation and migration in medulloblastoma, and increased
miR-183-96-182 expression is associated with aggressive clinical
course, high rates of metastasis and poor overall survival
(8,9,14).
miR-217 and miR-216 were overexpressed in
medulloblastoma. However, the role of miR-217 was controversial.
miR-217 is an oncogene that is overexpressed in aggressive human B
cell lymphomas (15). However, it
may be a tumor suppressor in hepatocellular carcinoma (16). In pancreatic intraepithelial
neoplasm, pancreatic ductal adenocarcinomas and clear cell renal
carcinomas, miR-217 was observed to be downregulated (17,18).
Further studies are required to determine whether increased
expression of miR-217 in medulloblastoma represents an oncogenic
effect, or if the dysregulation functions as a potential tumor
suppressor. While miR-216 was underexpressed in various types of
cancer, including breast cancer, pancreatic cancer and
nasopharyngeal carcinoma (19–21),
it may serve as a tumor suppressor to inhibit cell proliferation,
invasion and tumor growth. Further studies are required to
determine whether increased expression of miR-216 in
medulloblastoma represents an oncogenic effect or is a potential
tumor suppressor.
In the present study, marked downregulation of
miR-383, miR-206, miR-138, miR-128a/b and miR-133b was identified.
A previous study suggested that miR-383 is downregulated in
medulloblastoma. miR-383, repressing peroxiredoxin 3 at
transcriptional and translational levels, suppressed the cell
growth of medulloblastoma (6).
Additionally, miR-206 was downregulated in all the four molecular
subgroups of medulloblastoma as well as in cell lines.
Orthodenticle homeobox 2 (OTX2), a target gene of miR-206 which is
overexpressed in all non-sonic-hedgehog-driven medulloblastomas, is
an oncogene in medulloblastoma. Overexpressed OTX2 supported the
growth and proliferation of medulloblastomas. Therefore,
underexpression of miR-206 contributed to the upregulation of OTX2
expression and enhanced growth of medulloblastoma cell lines
(7). Studies have demonstrated a
reduction in the miRNAs miR-383, miR-138, miR-128a/b, miR-133,
miR-124 and let-7g in medulloblastoma. Nevertheless, miR-328,
miR-133b, miR-149, miR-181b and miR-154 were downregulated in the
study of Ferretti et al (5)
and the present study. In addition, the present study revealed that
miR-433, miR-488, miR-584, miR-329, miR-299-5p, miR-330,
miR-770-5p, miR-656 and miR-642 were underexpressed in
medulloblastoma, and further research is required to identify
whether these miRNAs have a specific role in the tumorigenesis of
medulloblastoma.
Based on microarray analysis, miRNAs that
distinguish normal brain from medulloblastoma tissue were
identified. As presented in the heat map, the dysregulated miRNAs
are more closely associated with the medulloblastoma samples
compared with the normal control samples. Additionally, by
uploading the miRNAs from the first network to the IPA, it was
identified that the dysregulated miRNAs were associated with key
regulatory genes in tumorigenesis, including TP53, insulin-like
growth factor 1 receptor, AGO2 and ERK1/2. The overexpressed miRNAs
miR-17-5p and miR-182 (log fold change >2) have interactions
with TP53; studies have demonstrated an association between these
miRNAs and TP53 in tumorigenesis (22,23).
AGOs are associated with miRNAs, due to their function in the RNA
induced silencing complex (RISC), wherein they use small RNA guides
to recognize targets. AGO2 had an interaction with miRNAs that are
dysregulated in medulloblastoma, including miR-17-5p, miR-128,
miR-96, miR-433, miR-383 and miR-138.
The second network, including miR-217 and miR-216,
which are upregulated in medulloblastoma, exhibits associations
with key regulatory genes in medulloblastoma development, including
TP53, SIRT1 and YBX1. SIRT1 is a well understood member of the
sirtuin protein class, preventing cell aging and apoptosis of
normal cells (24,25). The effects of SIRT1 are
controversial; the protein was considered to be a tumor promoter in
neuroblastoma, prostate and skin cancer (26–28),
whereas it was considered to be a tumor suppressor in breast and
colon cancer (29,30). Therefore, SIRT1 may serve a role in
medulloblastoma and correlate with the formation and prognosis of
human medulloblastomas (31).
Recently, Dey et al (32)
proved that YBX1 (YB1) is upregulated across human medulloblastoma
subclasses and is required for medulloblastoma cell proliferation.
Furthermore, the study identified that the SHH:YAP:YB1:IGF2 axis
may be a powerful target for therapeutic intervention in
medulloblastoma.
When the medulloblastoma samples were compared with
the normal cerebellum, 9 dysregulated miRNAs were identified. The 9
miRNAs were uploaded to the IPA in order to perform the core
analysis. The network involved 4 downregulated miRNAs and was
associated with important regulators of tumorigenesis, including
MET proto-oncogene (MET), AKT serine-threonine kinase 1 and sterol
regulatory element-binding factor 1 (SREBF1). Previous evidence has
demonstrated the association between aberrant MET signaling and
medulloblastoma development (33,34).
Previous studies have revealed that overexpressed miR-206 may
inhibit MET in lung cancer (35,36),
however the function of downregulated miR-206 in medulloblastoma is
unknown. SREBF1 is a transcription factor, regulating lipogenesis,
which is also involved in tumorigenesis (37). These network components alone are
not sufficient to initiate medulloblastoma formation; the
interactions between these dysregulated factors and additional
genetic insults promote tumorigenesis.
In conclusion, based on the miRNA array data in the
GEO database, miRNAs which are differentially expressed in
medulloblastoma samples were identified. Certain dysregulated
miRNAs have been confirmed; however, further research is required
to verify the miRNAs which have not been previously identified.
Additionally, the IPA core analysis, presenting the interaction of
miRNAs and counterpart targets, provides a method to understand the
tumorigenesis of medulloblastoma. The results of the present study
may provide a potent target for therapeutic intervention or
diagnosis in medulloblastomas.
Acknowledgements
The authors of the preset study would like to
acknowledge the submitters of all these GEO data arrays and all the
participants of this study. The present study was supported by the
Lanzhou Science and Technology Bureau Project (grant nos. 2013-3-27
and 2015-3-86), Gansu Province Health Industry Research Project
(grant nos. GSWSKY-2015-58 and GSWSKY-2015-58) and the doctoral
research fund of Lanzhou University Second Hospital (grant no.
ynbskyjj2015-1-02).
References
|
1
|
Louis DN, Ohgaki H, Wiestler OD, Cavenee
WK, Burger PC, Jouvet A, Scheithauer BW and Kleihues P: The 2007
WHO classification of tumours of the central nervous system. Acta
Neuropathol. 114:97–109. 2007. View Article : Google Scholar :
|
|
2
|
Roussel MF and Hatten ME: Cerebellum
development and medulloblastoma. Curr Top Dev Biol. 94:235–282.
2011. View Article : Google Scholar :
|
|
3
|
Gottardo NG and Gajjar A: Current therapy
for medulloblastoma. Curr Treat Options Neurol. 8:319–334. 2006.
View Article : Google Scholar
|
|
4
|
Packer RJ, Rood BR and MacDonald TJ:
Medulloblastoma: Present concepts of stratification into risk
groups. Pediatr Neurosurg. 39:60–67. 2003. View Article : Google Scholar
|
|
5
|
Ferretti E, De Smaele E, Po A, Di
Marcotullio L, Tosi E, Espinola MS, Di Rocco C, Riccardi R,
Giangaspero F, Farcomeni A, et al: MicroRNA profiling in human
medulloblastoma. Int J Cancer. 124:568–577. 2009. View Article : Google Scholar
|
|
6
|
Li KK, Pang JC, Lau KM, Zhou L, Mao Y,
Wang Y, Poon WS and Ng HK: MiR-383 is downregulated in
medulloblastoma and targets peroxiredoxin 3 (PRDX3). Brain pathol.
23:413–425. 2013. View Article : Google Scholar
|
|
7
|
Panwalkar P, Moiyadi A, Goel A, Shetty P,
Goel N, Sridhar E and Shirsat N: MiR-206, a Cerebellum Enriched
miRNA is downregulated in all medulloblastoma subgroups and its
overexpression is necessary for growth inhibition of
medulloblastoma cells. J Mol Neurosci. 56:673–680. 2015. View Article : Google Scholar
|
|
8
|
Weeraratne SD, Amani V, Teider N,
Pierre-Francois J, Winter D, Kye MJ, Sengupta S, Archer T, Remke M,
Bai AH, et al: Pleiotropic effects of miR-183~96~182 converge to
regulate cell survival, proliferation and migration in
medulloblastoma. Acta Neuropathol. 123:539–552. 2012. View Article : Google Scholar
|
|
9
|
Zhang Z, Li S and Cheng SY: The
miR-183-96-182 cluster promotes tumorigenesis in a mouse model of
medulloblastoma. J Biomed Res. 27:486–494. 2013.
|
|
10
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar
|
|
11
|
Esquela-Kerscher A and Slack FJ:
Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer.
6:259–269. 2006. View
Article : Google Scholar
|
|
12
|
Malumbres M: miRNAs versus oncogenes: The
power of social networking. Mol Systems Biol. 8:5692012. View Article : Google Scholar
|
|
13
|
Adams BD, Kasinski AL and Slack FJ:
Aberrant regulation and function of microRNAs in cancer. Curr Biol.
24:R762–R776. 2014. View Article : Google Scholar :
|
|
14
|
Cho YJ, Tsherniak A, Tamayo P, Santagata
S, Ligon A, Greulich H, Berhoukim R, Amani V, Goumnerova L,
Eberhart CG, et al: Integrative genomic analysis of medulloblastoma
identifies a molecular subgroup that drives poor clinical outcome.
J Clin Oncol. 29:1424–1430. 2011. View Article : Google Scholar
|
|
15
|
de Y, ébenes VG, Bartolomé-Izquierdo N,
Nogales-Cadenas R, Pérez-Durán P, Mur SM, Martínez N, Di Lisio L,
Robbiani DF, Pascual-Montano A, Cañamero M, et al: miR-217 is an
oncogene that enhances the germinal center reaction. Blood.
124:229–239. 2014. View Article : Google Scholar
|
|
16
|
Su J, Wang Q, Liu Y and Zhong M: miR-217
inhibits invasion of hepatocellular carcinoma cells through direct
suppression of E2F3. Mol Cell Biochem. 392:289–296. 2014.
View Article : Google Scholar
|
|
17
|
Li H, Zhao J, Zhang JW, Huang QY, Huang
JZ, Chi LS, Tang HJ, Liu GQ, Zhu DJ and Ma WM: MicroRNA-217,
down-regulated in clear cell renal cell carcinoma and associated
with lower survival, suppresses cell proliferation and migration.
Neoplasma. 60:511–515. 2013. View Article : Google Scholar
|
|
18
|
Xue Y, Tayoun AN Abou, Abo KM, Pipas JM,
Gordon SR, Gardner TB, RJ Jr, Suriawinata AA Barth and Tsongalis
GJ: MicroRNAs as diagnostic markers for pancreatic ductal
adenocarcinoma and its precursor, pancreatic intraepithelial
neoplasm. Cancer Genet. 206:217–221. 2013. View Article : Google Scholar
|
|
19
|
Deng M, Tang H, Zhou Y, Zhou M, Xiong W,
Zheng Y, Ye Q, Zeng X, Liao Q, Guo X, et al: miR-216b suppresses
tumor growth and invasion by targeting KRAS in nasopharyngeal
carcinoma. J Cell Sci. 124:2997–3005. 2011. View Article : Google Scholar
|
|
20
|
Rachagani S, Macha MA, Menning MS, Dey P,
Pai P, Smith LM, Mo YY and Batra SK: Changes in microRNA (miRNA)
expression during pancreatic cancer development and progression in
a genetically engineered KrasG12D; Pdx1-Cre mouse (KC) model.
Oncotarget. 6:40295–40309. 2015.
|
|
21
|
Faraji F, Hu Y, Goldberger N, Wu G..Buetow
KH, Zhang J and Hunter KW: Abstract A6: microRNA sequencing of AKXD
recombinant inbred panel identifies miR-216b as a candidate
metastasis suppressor in a murine model of breast cancer. Cancer
Res. 72:A62012. View Article : Google Scholar
|
|
22
|
Kanaan Z, Rai SN, Eichenberger MR, Barnes
C, Dworkin AM, Weller C, Cohen E, Roberts H, Keskey B, Petras RE,
et al: Differential microRNA expression tracks neoplastic
progression in inflammatory bowel disease-associated colorectal
cancer. Hum Mutat. 33:551–560. 2012. View Article : Google Scholar :
|
|
23
|
Lin LT, Chiou SH and Lee YJ: Abstract
3548: Study of the tumor suppressive machinery of arsenic
trioxide-induced glioblastoma multiforme inhibition via
microRNA-182-Sestrin2 regulatory circuit. Cancer Res. 74:35482014.
View Article : Google Scholar
|
|
24
|
Kim D, Nguyen MD, Dobbin MM, Fischer A,
Sananbenesi F, Rodgers JT, Delalle I, Baur JA, Sui G, Armour SM, et
al: SIRT1 deacetylase protects against neurodegeneration in models
for Alzheimer's disease and amyotrophic lateral sclerosis. EMBO.
26:3169–3179. 2007. View Article : Google Scholar
|
|
25
|
Law IK, Liu L, Xu A, Lam KS, Vanhoutte PM,
Che CM, Leung PT and Wang Y: Identification and characterization of
proteins interacting with SIRT1 and SIRT3: Implications in the
anti-aging and metabolic effects of sirtuins. Proteomics.
9:2444–2456. 2009. View Article : Google Scholar
|
|
26
|
Chu F, Chou PM, Zheng X, Mirkin BL and
Rebbaa A: Control of multidrug resistance gene mdr1 and cancer
resistance to chemotherapy by the longevity gene sirt1. Cancer Res.
65:10183–10187. 2005. View Article : Google Scholar
|
|
27
|
Hida Y, Kubo Y, Murao K and Arase S:
Strong expression of a longevity-related protein, SIRT1, in Bowen's
disease. Arch Dermatol Res. 299:103–106. 2007. View Article : Google Scholar
|
|
28
|
Yu W, Fu YC, Zhou XH, Chen CJ, Wang X, Lin
RB and Wang W: Effects of resveratrol on H(2)O(2)-induced apoptosis
and expression of SIRTs in H9c2 cells. J Cell Biochem. 107:741–747.
2009. View Article : Google Scholar
|
|
29
|
Firestein R, Blander G, Michan S,
Oberdoerffer P, Ogino S, Campbell J, Bhimavarapu A, Luikenhuis S,
de Cabo R, Fuchs C, et al: The SIRT1 deacetylase suppresses
intestinal tumorigenesis and colon cancer growth. PLoS One.
3:e20202008. View Article : Google Scholar :
|
|
30
|
Wang RH, Zheng Y, Kim HS, Xu X, Cao L,
Luhasen T, Lee MH, Xiao C, Vassilopoulos A, Chen W, et al:
Interplay among BRCA1, SIRT1, and Survivin during BRCA1-associated
tumorigenesis. Mol Cell. 32:11–20. 2008. View Article : Google Scholar :
|
|
31
|
Ma JX, Li H, Chen XM, Yang XH, Wang Q, Wu
ML, Kong QY, Li ZX and Liu J: Expression patterns and potential
roles of SIRT1 in human medulloblastoma cells in vivo and in vitro.
Neuropathology. 33:7–16. 2013. View Article : Google Scholar
|
|
32
|
Dey A, Robitaille M, Remke M, Maier C,
Malhotra A, Gregorieff A, Wrana JL, Taylor MD, Angers S and Kenney
AM: YB-1 is elevated in medulloblastoma and drives proliferation in
Sonic hedgehog-dependent cerebellar granule neuron progenitor cells
and medulloblastoma cells. Oncogene. 35:4256–4268. 2016. View Article : Google Scholar :
|
|
33
|
Guessous F, Yang YZ, Johnson E,
Marcinkiewicz L, Smith M, Zhang Y, Kofman A, Schiff D, Christensen
J and Abounader R: Cooperation between c-met and focal adhesion
kinase family members in medulloblastoma and implications for
therapy. Mol Cancer Ther. 11:288–297. 2012. View Article : Google Scholar
|
|
34
|
Li Y, Lal B, Kwon S, Fan X, Saldanha U,
Reznik TE, Kuchner EB, Eberhart C, Laterra J and Abounader R: The
scatter factor/hepatocyte growth factor: c-met pathway in human
embryonal central nervous system tumor malignancy. Cancer Res.
65:9355–9362. 2005. View Article : Google Scholar
|
|
35
|
Chen QY, Jiao DM, Wu YQ, Chen J, Wang J,
Tang XL, Mou H, Hu HZ, Song J, Yan J, et al: MiR-206 inhibits
HGF-induced epithelial-mesenchymal transition and angiogenesis in
non-small cell lung cancer via c-Met/PI3k/Akt/mTOR pathway.
Oncotarget. 7:18247–18261. 2016.
|
|
36
|
Chen QY, Jiao DM, Yan L, Wu YQ, Hu HZ,
Song J, Yan J, Wu LJ, Xu LQ and Shi JG: Comprehensive gene and
microRNA expression profiling reveals miR-206 inhibits MET in lung
cancer metastasis. Mol Biosyst. 11:2290–2302. 2015. View Article : Google Scholar
|
|
37
|
Sun Y, He WW, Luo M, Zhou Y, Chang G, Ren
W, Wu K, Li X, Shen J, Zhao X and Hu Y: SREBP1 regulates
tumorigenesis and prognosis of pancreatic cancer through targeting
lipid metabolism. Tumor Biol. 36:4133–4141. 2015. View Article : Google Scholar
|
|
38
|
Jinek M and Doudna JA: A three-dimensional
view of the molecular machinery of RNA interference. Nature.
457:405–412. 2009. View Article : Google Scholar
|