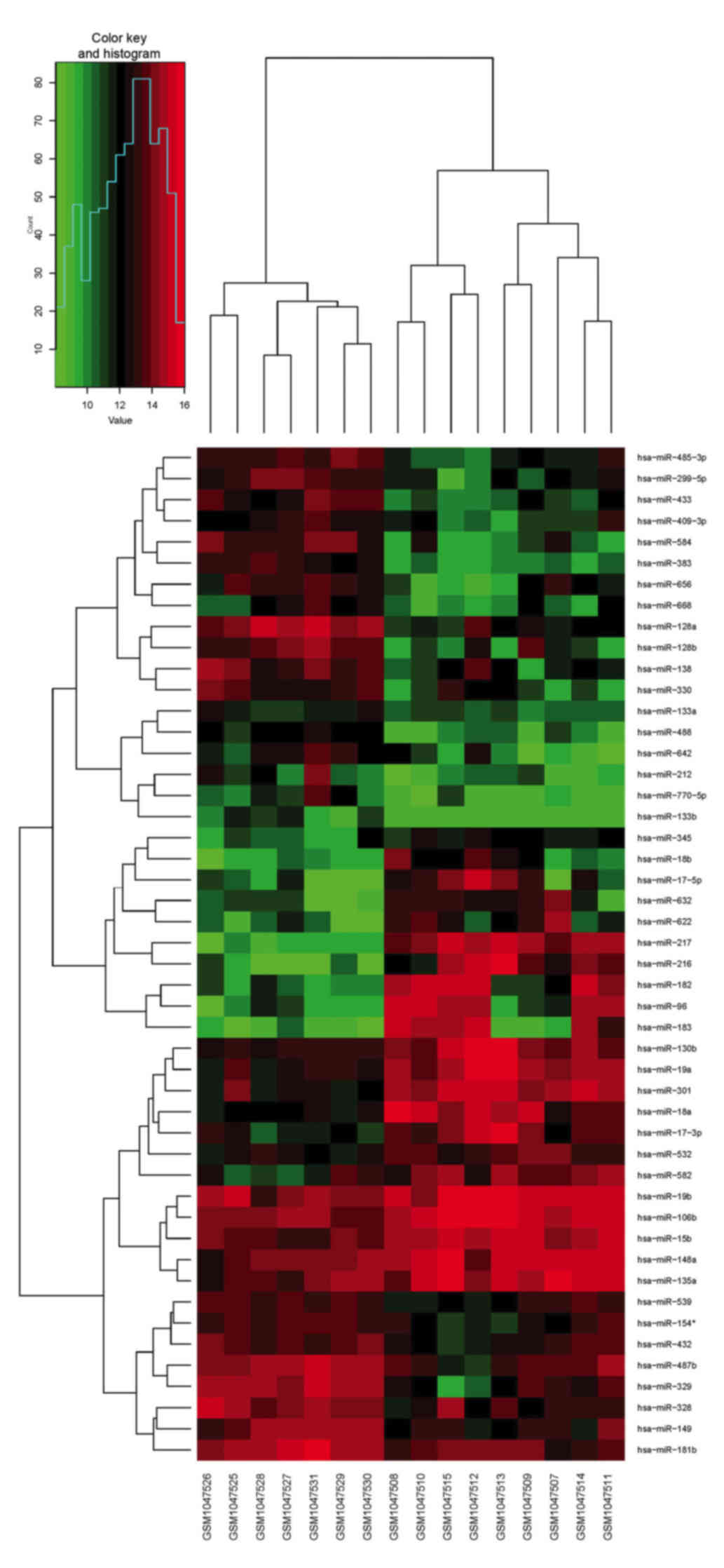
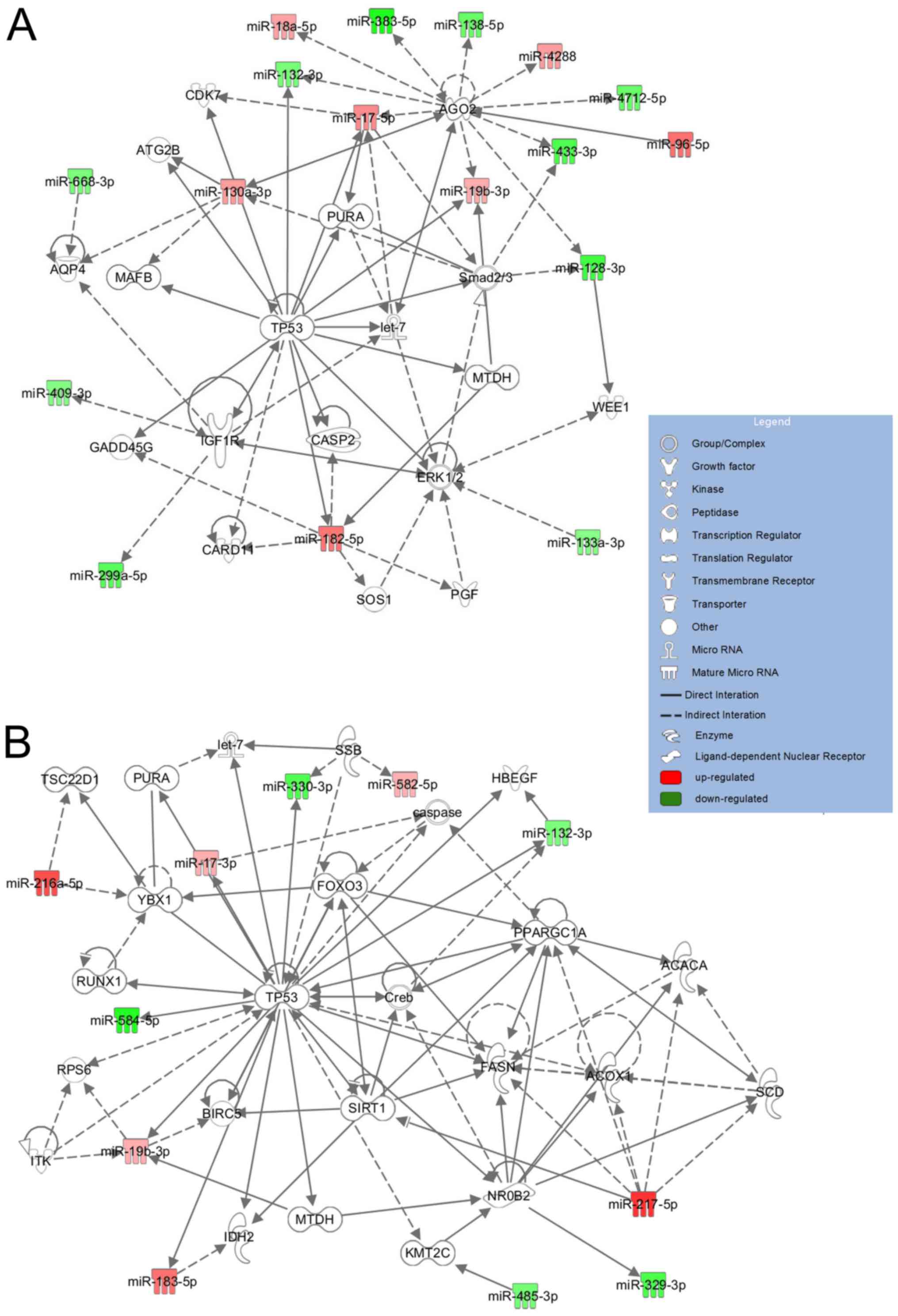
|
1
|
Louis DN, Ohgaki H, Wiestler OD, Cavenee
WK, Burger PC, Jouvet A, Scheithauer BW and Kleihues P: The 2007
WHO classification of tumours of the central nervous system. Acta
Neuropathol. 114:97–109. 2007. View Article : Google Scholar :
|
|
2
|
Roussel MF and Hatten ME: Cerebellum
development and medulloblastoma. Curr Top Dev Biol. 94:235–282.
2011. View Article : Google Scholar :
|
|
3
|
Gottardo NG and Gajjar A: Current therapy
for medulloblastoma. Curr Treat Options Neurol. 8:319–334. 2006.
View Article : Google Scholar
|
|
4
|
Packer RJ, Rood BR and MacDonald TJ:
Medulloblastoma: Present concepts of stratification into risk
groups. Pediatr Neurosurg. 39:60–67. 2003. View Article : Google Scholar
|
|
5
|
Ferretti E, De Smaele E, Po A, Di
Marcotullio L, Tosi E, Espinola MS, Di Rocco C, Riccardi R,
Giangaspero F, Farcomeni A, et al: MicroRNA profiling in human
medulloblastoma. Int J Cancer. 124:568–577. 2009. View Article : Google Scholar
|
|
6
|
Li KK, Pang JC, Lau KM, Zhou L, Mao Y,
Wang Y, Poon WS and Ng HK: MiR-383 is downregulated in
medulloblastoma and targets peroxiredoxin 3 (PRDX3). Brain pathol.
23:413–425. 2013. View Article : Google Scholar
|
|
7
|
Panwalkar P, Moiyadi A, Goel A, Shetty P,
Goel N, Sridhar E and Shirsat N: MiR-206, a Cerebellum Enriched
miRNA is downregulated in all medulloblastoma subgroups and its
overexpression is necessary for growth inhibition of
medulloblastoma cells. J Mol Neurosci. 56:673–680. 2015. View Article : Google Scholar
|
|
8
|
Weeraratne SD, Amani V, Teider N,
Pierre-Francois J, Winter D, Kye MJ, Sengupta S, Archer T, Remke M,
Bai AH, et al: Pleiotropic effects of miR-183~96~182 converge to
regulate cell survival, proliferation and migration in
medulloblastoma. Acta Neuropathol. 123:539–552. 2012. View Article : Google Scholar
|
|
9
|
Zhang Z, Li S and Cheng SY: The
miR-183-96-182 cluster promotes tumorigenesis in a mouse model of
medulloblastoma. J Biomed Res. 27:486–494. 2013.
|
|
10
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar
|
|
11
|
Esquela-Kerscher A and Slack FJ:
Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer.
6:259–269. 2006. View
Article : Google Scholar
|
|
12
|
Malumbres M: miRNAs versus oncogenes: The
power of social networking. Mol Systems Biol. 8:5692012. View Article : Google Scholar
|
|
13
|
Adams BD, Kasinski AL and Slack FJ:
Aberrant regulation and function of microRNAs in cancer. Curr Biol.
24:R762–R776. 2014. View Article : Google Scholar :
|
|
14
|
Cho YJ, Tsherniak A, Tamayo P, Santagata
S, Ligon A, Greulich H, Berhoukim R, Amani V, Goumnerova L,
Eberhart CG, et al: Integrative genomic analysis of medulloblastoma
identifies a molecular subgroup that drives poor clinical outcome.
J Clin Oncol. 29:1424–1430. 2011. View Article : Google Scholar
|
|
15
|
de Y, ébenes VG, Bartolomé-Izquierdo N,
Nogales-Cadenas R, Pérez-Durán P, Mur SM, Martínez N, Di Lisio L,
Robbiani DF, Pascual-Montano A, Cañamero M, et al: miR-217 is an
oncogene that enhances the germinal center reaction. Blood.
124:229–239. 2014. View Article : Google Scholar
|
|
16
|
Su J, Wang Q, Liu Y and Zhong M: miR-217
inhibits invasion of hepatocellular carcinoma cells through direct
suppression of E2F3. Mol Cell Biochem. 392:289–296. 2014.
View Article : Google Scholar
|
|
17
|
Li H, Zhao J, Zhang JW, Huang QY, Huang
JZ, Chi LS, Tang HJ, Liu GQ, Zhu DJ and Ma WM: MicroRNA-217,
down-regulated in clear cell renal cell carcinoma and associated
with lower survival, suppresses cell proliferation and migration.
Neoplasma. 60:511–515. 2013. View Article : Google Scholar
|
|
18
|
Xue Y, Tayoun AN Abou, Abo KM, Pipas JM,
Gordon SR, Gardner TB, RJ Jr, Suriawinata AA Barth and Tsongalis
GJ: MicroRNAs as diagnostic markers for pancreatic ductal
adenocarcinoma and its precursor, pancreatic intraepithelial
neoplasm. Cancer Genet. 206:217–221. 2013. View Article : Google Scholar
|
|
19
|
Deng M, Tang H, Zhou Y, Zhou M, Xiong W,
Zheng Y, Ye Q, Zeng X, Liao Q, Guo X, et al: miR-216b suppresses
tumor growth and invasion by targeting KRAS in nasopharyngeal
carcinoma. J Cell Sci. 124:2997–3005. 2011. View Article : Google Scholar
|
|
20
|
Rachagani S, Macha MA, Menning MS, Dey P,
Pai P, Smith LM, Mo YY and Batra SK: Changes in microRNA (miRNA)
expression during pancreatic cancer development and progression in
a genetically engineered KrasG12D; Pdx1-Cre mouse (KC) model.
Oncotarget. 6:40295–40309. 2015.
|
|
21
|
Faraji F, Hu Y, Goldberger N, Wu G..Buetow
KH, Zhang J and Hunter KW: Abstract A6: microRNA sequencing of AKXD
recombinant inbred panel identifies miR-216b as a candidate
metastasis suppressor in a murine model of breast cancer. Cancer
Res. 72:A62012. View Article : Google Scholar
|
|
22
|
Kanaan Z, Rai SN, Eichenberger MR, Barnes
C, Dworkin AM, Weller C, Cohen E, Roberts H, Keskey B, Petras RE,
et al: Differential microRNA expression tracks neoplastic
progression in inflammatory bowel disease-associated colorectal
cancer. Hum Mutat. 33:551–560. 2012. View Article : Google Scholar :
|
|
23
|
Lin LT, Chiou SH and Lee YJ: Abstract
3548: Study of the tumor suppressive machinery of arsenic
trioxide-induced glioblastoma multiforme inhibition via
microRNA-182-Sestrin2 regulatory circuit. Cancer Res. 74:35482014.
View Article : Google Scholar
|
|
24
|
Kim D, Nguyen MD, Dobbin MM, Fischer A,
Sananbenesi F, Rodgers JT, Delalle I, Baur JA, Sui G, Armour SM, et
al: SIRT1 deacetylase protects against neurodegeneration in models
for Alzheimer's disease and amyotrophic lateral sclerosis. EMBO.
26:3169–3179. 2007. View Article : Google Scholar
|
|
25
|
Law IK, Liu L, Xu A, Lam KS, Vanhoutte PM,
Che CM, Leung PT and Wang Y: Identification and characterization of
proteins interacting with SIRT1 and SIRT3: Implications in the
anti-aging and metabolic effects of sirtuins. Proteomics.
9:2444–2456. 2009. View Article : Google Scholar
|
|
26
|
Chu F, Chou PM, Zheng X, Mirkin BL and
Rebbaa A: Control of multidrug resistance gene mdr1 and cancer
resistance to chemotherapy by the longevity gene sirt1. Cancer Res.
65:10183–10187. 2005. View Article : Google Scholar
|
|
27
|
Hida Y, Kubo Y, Murao K and Arase S:
Strong expression of a longevity-related protein, SIRT1, in Bowen's
disease. Arch Dermatol Res. 299:103–106. 2007. View Article : Google Scholar
|
|
28
|
Yu W, Fu YC, Zhou XH, Chen CJ, Wang X, Lin
RB and Wang W: Effects of resveratrol on H(2)O(2)-induced apoptosis
and expression of SIRTs in H9c2 cells. J Cell Biochem. 107:741–747.
2009. View Article : Google Scholar
|
|
29
|
Firestein R, Blander G, Michan S,
Oberdoerffer P, Ogino S, Campbell J, Bhimavarapu A, Luikenhuis S,
de Cabo R, Fuchs C, et al: The SIRT1 deacetylase suppresses
intestinal tumorigenesis and colon cancer growth. PLoS One.
3:e20202008. View Article : Google Scholar :
|
|
30
|
Wang RH, Zheng Y, Kim HS, Xu X, Cao L,
Luhasen T, Lee MH, Xiao C, Vassilopoulos A, Chen W, et al:
Interplay among BRCA1, SIRT1, and Survivin during BRCA1-associated
tumorigenesis. Mol Cell. 32:11–20. 2008. View Article : Google Scholar :
|
|
31
|
Ma JX, Li H, Chen XM, Yang XH, Wang Q, Wu
ML, Kong QY, Li ZX and Liu J: Expression patterns and potential
roles of SIRT1 in human medulloblastoma cells in vivo and in vitro.
Neuropathology. 33:7–16. 2013. View Article : Google Scholar
|
|
32
|
Dey A, Robitaille M, Remke M, Maier C,
Malhotra A, Gregorieff A, Wrana JL, Taylor MD, Angers S and Kenney
AM: YB-1 is elevated in medulloblastoma and drives proliferation in
Sonic hedgehog-dependent cerebellar granule neuron progenitor cells
and medulloblastoma cells. Oncogene. 35:4256–4268. 2016. View Article : Google Scholar :
|
|
33
|
Guessous F, Yang YZ, Johnson E,
Marcinkiewicz L, Smith M, Zhang Y, Kofman A, Schiff D, Christensen
J and Abounader R: Cooperation between c-met and focal adhesion
kinase family members in medulloblastoma and implications for
therapy. Mol Cancer Ther. 11:288–297. 2012. View Article : Google Scholar
|
|
34
|
Li Y, Lal B, Kwon S, Fan X, Saldanha U,
Reznik TE, Kuchner EB, Eberhart C, Laterra J and Abounader R: The
scatter factor/hepatocyte growth factor: c-met pathway in human
embryonal central nervous system tumor malignancy. Cancer Res.
65:9355–9362. 2005. View Article : Google Scholar
|
|
35
|
Chen QY, Jiao DM, Wu YQ, Chen J, Wang J,
Tang XL, Mou H, Hu HZ, Song J, Yan J, et al: MiR-206 inhibits
HGF-induced epithelial-mesenchymal transition and angiogenesis in
non-small cell lung cancer via c-Met/PI3k/Akt/mTOR pathway.
Oncotarget. 7:18247–18261. 2016.
|
|
36
|
Chen QY, Jiao DM, Yan L, Wu YQ, Hu HZ,
Song J, Yan J, Wu LJ, Xu LQ and Shi JG: Comprehensive gene and
microRNA expression profiling reveals miR-206 inhibits MET in lung
cancer metastasis. Mol Biosyst. 11:2290–2302. 2015. View Article : Google Scholar
|
|
37
|
Sun Y, He WW, Luo M, Zhou Y, Chang G, Ren
W, Wu K, Li X, Shen J, Zhao X and Hu Y: SREBP1 regulates
tumorigenesis and prognosis of pancreatic cancer through targeting
lipid metabolism. Tumor Biol. 36:4133–4141. 2015. View Article : Google Scholar
|
|
38
|
Jinek M and Doudna JA: A three-dimensional
view of the molecular machinery of RNA interference. Nature.
457:405–412. 2009. View Article : Google Scholar
|