Introduction
In South Korea, lung cancer is expected to
contribute to 17,505 male mortalities in the year 2016 which is the
highest number among all cancers (1). The increasing mortality rates in
patients with lung cancer, has led to an increase in the number of
studies investigating this malignancy. There are two major types of
lung cancer; one of which is small-cell lung cancer, which is
clinically aggressive and often already advanced at diagnosis. The
remaining type is non-small cell lung cancer (NSCLC), which is the
most common type of lung cancer, and is known to exhibit various
pathological features (2,3). The major criteria that distinguishes
these forms are based on their histological characteristics, such
as cell size and the nuclear/cytoplasmic ratio (4). Adenocarcinoma and squamous cell
carcinoma are the dominant phenotypes of NSCLC. Squamous cell
carcinoma is strongly associated with smoking and chronic
inflammation (5).
The epidermal growth factor receptor (EGFR) is
located on the cell surface and functions as a major contributor in
signal transduction pathways that control cell proliferation,
survival and differentation (6).
Mutations in this tyrosine-kinase receptor may induce an
autophosphorylation process, leading to the continuous promotion of
cellular proliferation, decreasing apoptosis and in the end,
malignant transformation occurred (7,8). In
East Asia, the EFGR mutation occurs in approximately 35% of
patients with lung cancer (9). The
mutation frequently occurs in exon 18–21, which is the first exon
that encodes the tyrosine-kinase domain. The two most common
mutations are a deletion in exon 19 and point mutation in exon 21
(10–12). Determining the type of mutation is
important, as it is used to determine patient treatment. A previous
study demonstrated that the presence of an EGFR mutation is
associated with enhanced progression-free survival and a high
response rate to EGFR tyrosine kinase inhibitors (TKIs), when
compared with standard first-line chemotherapy in NSCLC (13). In addition, different mutations may
be associated with different clinical characteristics. As
determined by Fukuoka et al (13), NSCLCs in Chinese patients harboring
mutations in exon 21 of EGFR exhibit a greater number of malignant
features when compared with NSCLCs in patients harboring mutations
in exon 19. Therefore, determining the type of mutation early
during diagnosis is essential.
The detection of EGFR mutations is typically
achieved using invasive cytologic or histologic techniques,
followed by DNA-sequencing (14–16).
However, a less invasive technique, such as the collection of blood
samples, is important for aiding diagnosis, as cytologic or
histologic detection is time-consuming and unsafe (17). Biomarkers, which are measurable
compounds present in biological fluids, may be used for diagnosis,
assessing disease outcome and progression, and for predicting
outcomes of treatment in clinical practice (18–21).
In a previous study, the use of high-resolution
metabolomics (HRM) generated promising results by identifying
prospective biomarkers that effectively discriminated between
healthy subjects and patients with lung cancer (20,22,23).
In the present study, HRM was performed using a recently developed
configuration involving liquid chromatography coupled with mass
spectrometry (LC-MS/MS). Quadrupole time-of-flight (Q-TOF) MS was
employed to investigate and identify significant compounds as
potential biomarkers in human plasma samples from South Korean
patients with lung cancer. The aim of the present study was to
identify biomarkers associated with mutations in exon 19 or 21 of
EGFR, in order to facilitate the early detection and provide a
minimally-invasive diagnosis of NSCLC.
Materials and methods
Sample collection
A total of 15 plasma samples were obtained from
patients with NSCLC lung cancer (age, 55–87; male/female, 7/8)
admitted to Korea University Guro Hospital (Seoul, Republic of
Korea) between January 2014 and September 2014. Of the 15 samples,
5 samples were derived from patients without EGFR mutations (termed
NoEM), 4 samples were derived from patients harboring EGFR
mutations in exon 19 and 6 samples were derived from patients
harboring EGFR mutations in exon 21. Subjects with mutations in
exons 19 or 21 were termed EMLC and were diagnosed based on
PNA-mediated real-time polymerase chain reaction clamping using the
PNAClamp™ EGFR Mutation Detection kit (Panagene, Inc.,
Daejeon, Republic of Korea). Details, such as sex and body mass
index (BMI) are provided in Table
I. The BMI values were analyzed using a Student's t-test, which
demonstrated no significant differences among the NoEM and EMLC
groups. EMLC subjects received EGFR TKI treatments while NoEM
subjects received supportive care. The present study was approved
by the Institutional Review Board of Korea University (approval no.
KUGH14273-002), and written informed consent was obtained from all
patients.
 | Table I.Age, sex, weight and BMI of patients
with lung cancer included in the present study. |
Table I.
Age, sex, weight and BMI of patients
with lung cancer included in the present study.
| Parameters | No EGFR
mutation | EGFR mutation |
|---|
| Number of
subjects | 5 | 10 (4/6) |
| Age (years) | 77.6±8.65 | 65.4±10.85 |
| Sex
(male/female) | 3/2 | 4/6 |
| Weight (kg) | 60.17±10.38 | 52.26±9.40 |
| BMI
(kg/m2) | 25.61±2.36 | 22.32±3.61 |
| Stage of
disease | T2aN0M0; | T2aN2M0; |
|
| T1aN0M1b; | T2aN2M1b; |
|
| T2aN2M1b; | T3N1M0; |
|
| T2N3M1b; | T2aN0M0; |
|
| T2aN2M1a; | T2N3M1b; |
|
|
| T2aN0M0; |
|
|
| T2N0M1b; |
|
|
| T2N0M1b; |
|
|
| T4N2M1a; |
|
|
| No information for
1 subject |
Sample preparation and LC-MS
measurements
Samples (50 µl) were treated with acetonitrile (1:2,
v/v), and centrifuged at 14,000 × g for 5 min at 4°C in order to
separate proteins (24).
Metabolites were separated using the Agilent 1200 High Performance
Liquid Chromatography (HPLC) System (Agilent Technologies, Inc.,
Santa Clara, CA, USA) with a Higgins Analytical Targa HPLC C18
100×2.1 mm column, 5 µm particle size (Higgins Analytical, Inc.,
Mountain View, CA, USA). Mobile phase A consisted of 0.1% formic
acid in water (HPLC grade, Tedia Company, Inc., Fairfield, Ohio,
USA) and mobile phase B consisted of 0.1% formic acid in
acetonitrile (HPLC grade, Tedia Company, Inc.). The HPLC gradient
was programed as follows: 0–7 min, 5% for B; 7–15 min, gradient
decrease to 2% for B; 15–20 min, hold 40% for B; 20–24 min, 95% for
B; 24–25 min, gradient decrease to 2% for B. The injection volume
was 5 µl, with a flow rate of 0.4 ml/min and a column temperature
of 40°C. Masses of metabolites ranging from 50–1000 m/z were
detected using the Agilent 6530 Accurate Mass Q-TOF-LC/MS (Agilent
Technologies, Inc.) in the positive ionization mode (25). This LC-MS/MS is ideally suited for
metabolic stability and profiling studies, as this system is highly
sensitive for the detection of compounds at low concentrations
(pg/ml), has a resolving power of 40 k, and is able to identify
masses and isotopes for the accurate identification of metabolites.
This system was used to detect the m/z of ions from 50 to 1,000,
with 20,000 resolution (arbitrary units) over 30 min, and LC
operated with data extraction using the apLCMS software version
5.9.6 (http://clinicalmetabolomics.org/welcome/default/software)
(26), which provided a minimum of
3,000 reproducible features; a number of which displayed sufficient
mass accuracy to allow prediction of elemental composition. Ion
intensity, m/z, and retention time was used to define an m/z
feature.
Metabolic profiling with univariate
and multivariate statistical analysis
apLCMS generated the total features from the samples
for subsequent statistical analyses and bioinformatics. Features
from triplicate LC-MS analyses were averaged, log2
transformed and normalized by z-transformation. Univariate analysis
and the false discovery rates (FDR) (27) were calculated to reduce the
incidence of false positives, and Manhattan plots were constructed
using the Limma R package (version, 2.15; https://www.r-project.org/) (28), in order to identify the
significantly different metabolites between NoEM and EMLC groups.
Using the same package, two-way hierarchical cluster analysis (HCA)
was used to separate the metabolic profiles of the two groups based
on their metabolite profiles (29). The Limma package is used for the
analysis of gene expression data generated from microarray or
RNA-Sequencing technologies. It provides the ability to compare a
number of targets simultaneously (21,30).
Receiver operating characteristic (ROC) curves (MedCalc Software
bvba version 16.8, Ostend Belgium) were used to classify the
samples based on significant metabolite levels.
Data annotation and pathway
analysis
Using the list of all m/z features detected
(Lref) and the significant m/z features
(Lsig), the significant metabolites were
annotated using Mummichog software version 1.0.5 (http://clinicalmetabolomics.org/welcome/default/software)
to generate a potential metabolic network model (31). Mummichog software was used to
identify potentially matched metabolites from the m/z features in
Lsig, as well as establish a reference metabolic network for
all modules that may be produced by these metabolites. Random lists
of m/z features were generated from Lref a number of times
to estimate the null distribution of module activities, in order to
compute its statistical significance. A module may be within a
known pathway or in between several pathways. The predicted
metabolites listed in modules were colored according to fold
change. This program has been successfully used to facilitate the
identification of metabolite activity networks in immune responses
to viruses (31), and for the
metabolic profiling of fruit flies (32). Based on the results from Manhattan
plot, FDR, HCA, and Mummichog pathway analysis, potential
biomarkers were then selected and a histogram displaying
differences in the relative concentration of different metabolites
in NoEM and EMLC groups were generated using Microsoft Excel
(Microsoft Corporation, Redmond, WA, USA).
Statistical analysis
Differences in the levels of endogenous compounds
between NoEM and EMLC groups were analyzed using the Student's
t-test followed by FDR multiple testing correction. Statistical
analysis was performed using the Limma R package version 2.15
version (https://www.r-project.org/).
P<0.05 was considered to indicate a statistically significant
difference. To reduce the incidence of false positives, P-values
were adjusted by applying a multiple testing correction (the
Benjamini and Hochberg procedure for adjusting the false discovery
rate), thus producing optimum statistical significance.
Results
In the present study, metabolome-wide association
analysis was performed to determine metabolic alterations in NoEM
and EMLC groups of patients with lung cancer. Statistical tests
were performed using the Manhattan plot to identify significant
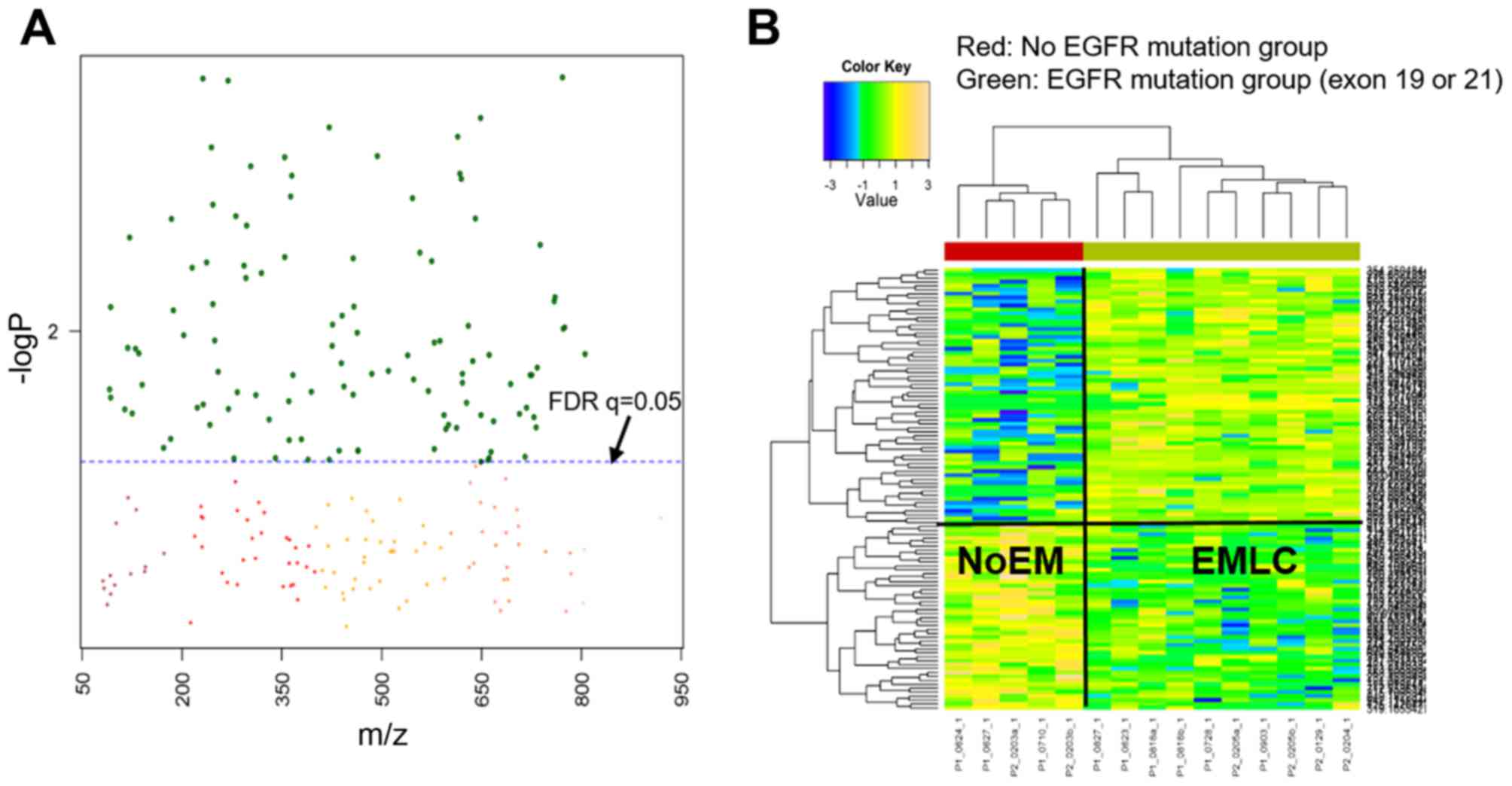
features between the NoEM vs. EMLC groups. As shown in Fig. 1A, the y-axis represents the -log10
of the raw P-values between the NoEM and EMLC groups, while the
x-axis represents the m/z values ranging from 50 to 1,000. The
dashed-line represents the significant FDR threshold (q=0.05),
which distinguishes the significant features from insignificant m/z
values. Therefore, metabolites were considered to be significantly
different in the EMLC and NoEM groups if they were placed above
this threshold.
The number of features that were significantly
different between the EMLC and NoEM groups was 112 out of the 3,939
total detected features (Fig. 1A).
Two-way HCA was performed to identify the correlation between
samples and significant metabolites, with a clear separation of
NoEM and EMLC groups expected. As demonstrated in Fig. 1B, the NoEM group (red panel), is
grouped as one cluster while the EGFR group (green panel) is
grouped as a different cluster. This apparent separation in the
heatmaps suggests that the metabolites are highly differentiated in
each group.
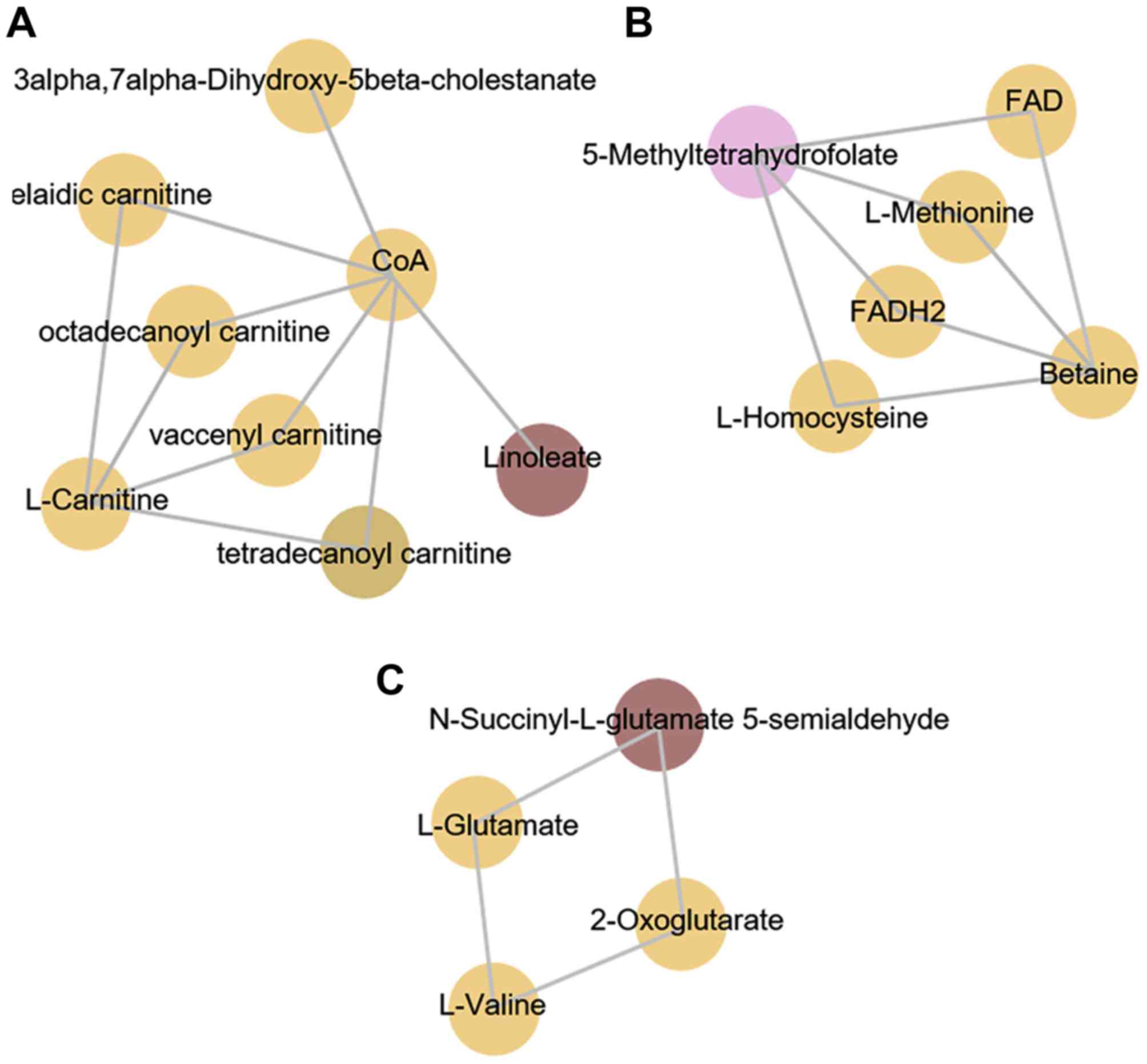
Mummichog analysis
Metabolite enrichment was achieved using the
Mummichog program. This program is used to generate metabolite
networks and associated particular pathways for display as
interactive figures, with color-identified nodules based on fold
changes. Among the metabolites that were significantly different
between the NoEM and EMLC groups, four groups of significant
metabolites that demonstrated a strong correlation to certain
pathways were identified. The first group consisted of metabolites
associated with fatty acid (FA) metabolism and the carnitine
shuttle, which are pathways that are associated with energy
production. As shown in Fig. 2A,
linoleic acid and tetradecanoyl carnitine were observed to be the
significant metabolites, as indicated by the different colored
modules when compared with the other metabolites. The remaining
groups correlated with the following signaling pathways: Urea
cycle/amino group metabolism, amino acids and folate metabolism. As
demonstrated in Fig. 2B and C,
5-methyltetrahydrofolate (5-MTHF) and N-succinyl-L-glutamate-5
semialdehyde (NSGS) were significantly different among the
metabolites.
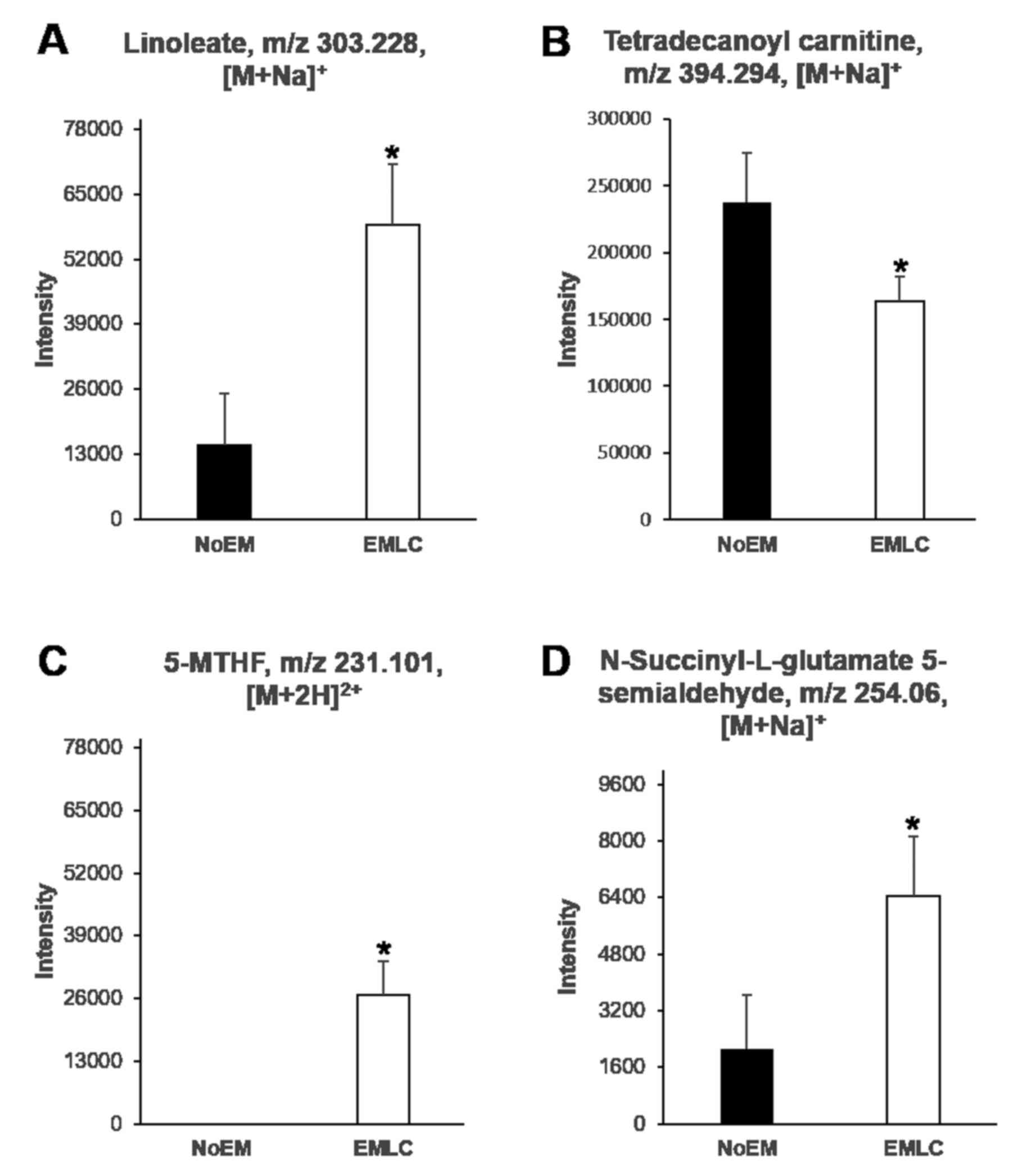
Identification and verification of
potential biomarkers
Differences in the levels of endogenous compounds
were identified between NoEM and EMLC groups. Linoleic acid [303.23
m/z, (M+Na)+], 5-methyl tetrahydrofolate [231.10 m/z,
(M+2H)+] and N-succinyl-L-glutamate-5 semialdehyde
[254.06 m/z, (M+Na)+] were observed to be significantly
elevated in patients harboring EGFR mutations, whereas
tetradecanoyl carnitine [394.29 m/z, (M+Na)+] was
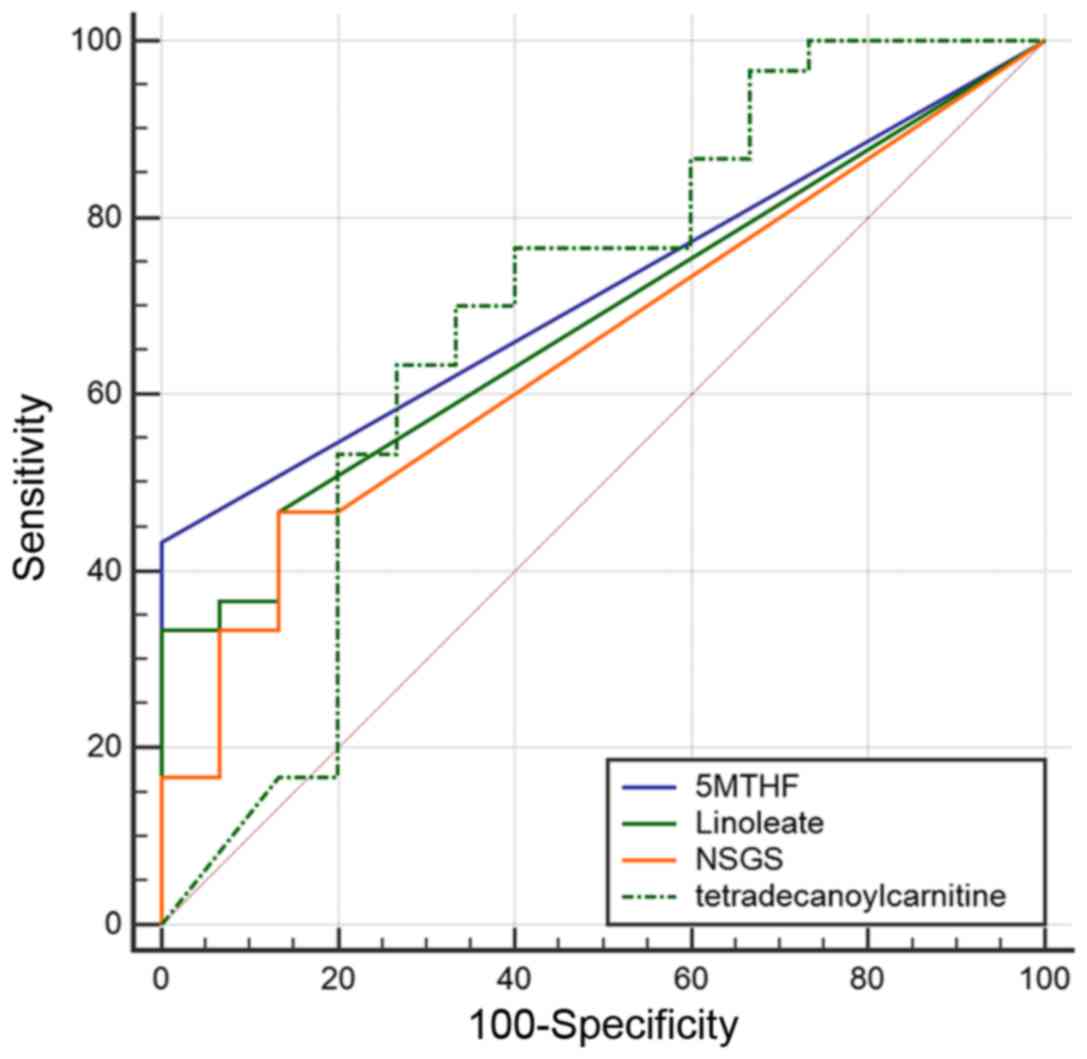
observed to be significantly reduced (Fig. 3). ROC analysis was then performed
to assess the classification of samples based on the detected
levels of significant compounds (Fig.
4). The area under the curve (AUC) for linoleate, 5-MTHF, NSGS
and tetradecanoyl carnitine, was 0.68, 0.72, 0.65, and 0.69,
respectively (Table II). The AUC
values of two compounds with the highest values were verified by
detecting the transition of the specific precursor ion to the
product ion: 394.29–377.26 m/z for tetradecanoyl carnitine and
231.1–170.7 m/z for 5-MTHF (data not shown).
 | Table II.ROC curves for the significant
metabolites identified. |
Table II.
ROC curves for the significant
metabolites identified.
| Metabolite | AUC | Sensitivity
(%) | Specificity
(%) | Positive predictive
value (%) | Negative predictive
value (%) |
|---|
| Linoleate | 0.68 | 46.67 |
86.67 |
87.5 | 44.8 |
| 5-MTHF | 0.72 | 43.33 | 100.00 | 100.0 | 46.9 |
| NSGS | 0.65 | 46.67 |
86.67 |
87.5 | 44.8 |
| Tetradecanoyl
carnitine | 0.69 | 63.33 |
73.33 |
82.6 | 50.0 |
Discussion
The main objective of the present study was to
identify low molecular-weight metabolites that may be used as
biomarkers for lung cancer mutation diagnostic tools. By comparing
the metabolite profiles of plasma samples from patients with no
EGFR mutations with patients harboring EGFR mutations, four
compounds, including linoleic acid, tetradecanoyl carnitine,
5-MTHF, and NSGS, were identified as potential biomarkers.
Activating mutations in EGFR is a major factor that
contributes to abnormal cell proliferation and malignant
transformation (33). Gene
mutations in cancer cells may lead to uncontrolled proliferation
and disruption of signaling pathways, which activates the uptake
and metabolism of nutrients. Therefore, mutations promote cell
survival and increase cell growth (34,35).
Human cells generate energy by utilizing glucose and
lipid metabolic pathways, which are affected by the irregularities
in cancer cells. First described by Warburg, glucose metabolism
alterations in cancer cells have been investigated in a number of
studies (36–38). In addition, alterations in FA
metabolism in cancer cells have recently garnered increasing
attention (39–41). FAs, such as linoleic acid, are
metabolized via mitochondrial FA oxidation. FA is first activated
via coenzyme A (CoA) esterification, and is subsequently
transported to the mitochondria by the carnitine shuttle (42). This shuttle transfers long-chain
acyl-CoAs as their corresponding carnitine ester, which are then
converted to the FA-acylCoA form prior to β-oxidation for energy
production. This transfiguration process demonstrates the
importance of the carnitine shuttle in FA-dependent energy
formation.
Previous studies have suggested that peroxisome
proliferator-activated receptor (PPAR) serves a vital role as a
regulator of FA oxidation and carnitine metabolism (43–45).
The PPARγ subtype contributes to the suppression of cell
proliferation (45,46). According to Hou et al
(46) dysregulated EGFR expression
promotes cell proliferation by inhibiting PPARγ function. Hence,
mutations in EGFR potentially disrupt the function of PPAR in
regulating FA metabolism.
In the current study, the activity of the carnitine
shuttle was highlighted as a putative deregulated pathway in
patients with EGFR mutations. Based on the results of previous
studies, it is possible that EGFR mutations led to deregulation of
the carnitine shuttle by terminating PPAR function. Four acyl
carnitines were detected, notably tetradecanoyl carnitine, which
was present at lower levels in EMLC patients, while linoleic acid,
a long-chain FA, was observed to be elevated. The levels of
linoleic acid may have been elevated due to disruption of its
transport mechanism to the mitochondria. In addition, regardless of
its role in the cytosol, acyl carnitine is transported across cell
membranes and can therefore be detected in plasma (47).
In the present study, the metabolism of amino acids
and folate was identified as an affected pathway in patients with
lung cancer harboring EGFR mutations. Previous studies have
identified abnormalities in metabolic proteins in lung cancer
patients by determining the difference in amino acid levels in
blood plasma samples (48,49). The level of plasma amino acids may
indicate the sum of various pathologic conditions affecting the
total flux of amino acids in the body (50). In the present study, the detection
of NSGS was significantly higher in the EMLC group of patients when
compared with the NoEM group. This compound is an intermediate in
the glutamate, aspartate and proline metabolic pathways. The
elevated levels of 5-MTHF observed in the EMLC group may have been
associated with overexpression of folate receptor α (FRα) due to
the EGFR mutation (51). FRα is a
glycoprotein located in the cell membrane that binds folic acid
with a high affinity (Kd<1 nM) and mediates its
intracellular transport (52,53).
In addition, FRα binds one-carbon reduced folate derivatives, such
as 5-MTHF, with a lower affinity (Kd, 1–10 nM)
(53,54). Therefore, overexpression of FRα,
and the subsequent binding of folic acid, may have been caused by
the observed increase in extracellular 5-MTHF. Notably, this study
demonstrated extreme differences in the abundance of 5-MTHF between
NoEM and EMLC groups. In addition, the medication history of all
patients was checked to remove bias. Three subjects, one of them
from the NoEM group, received folic acid supplements; however, this
did not influence the results, as zero abundance of 5-MTHF was
detected in the NoEM group. This may be useful for developing
improved treatments for patients with NSCLC, as elevated 5-MTHF
levels may be associated with high levels of FRα expression, which
may improve responses to antifolate chemotherapy (51). In addition, the results of a phase
II clinical trial in China demonstrated that the efficacy of TKIs
(erlotinib) combined with antifolate (capecitabine) treatment were
significantly beneficial in patients with lung cancer with EGFR
mutations who hadn't received cancer treatment prior to the study
(55).
In conclusion, four potential biomarkers were
identified in the plasma samples of patients with lung cancer that
harbor EGFR mutations. Two of these biomarkers were associated with
energy production signaling pathways, specifically FA metabolism.
These results may provide opportunities for the development of
novel diagnostic tools for EGFR mutation detection in lung cancer.
Future studies with a larger population of subjects are required to
validate the results, and provide a rationale for the clinical
practicality of this approach.
Acknowledgements
The present study was supported by the National
Research Foundation of Korea (grant nos. NRF-2014R1A1A2053787 and
NRF-2017R1A2B4003890) and the Korea Health Industry Development
Institute (grant no. HI14C2686). The authors would like to thank Dr
Karan Uppal from the Emory University School of Medicine (Atlanta,
GA, USA) for providing the R-package used to run the
metabolome-wide association study. Mr. Aryo D. Pamungkas gratefully
acknowledges the Indonesia Endowment Fund for Education (LPDP) for
the financial support of his master degree scholarship.
Glossary
Abbreviations
Abbreviations:
|
HRM
|
high-resolution metabolomics
|
|
EGFR
|
epidermal growth factor receptor
|
|
5-MTHF
|
5-methyltetrahydro folate
|
References
|
1
|
Jung KW, Won YJ, Oh CM, Kong HJ, Cho H,
Lee JK, Lee DH and Lee KH: Prediction of cancer incidence and
mortality in Korea, 2016. Cancer Res Treat. 48:451–457. 2016.
View Article : Google Scholar :
|
|
2
|
Collins LG, Haines C, Perkel R and Enck
RE: Lung cancer: Diagnosis and management. Am Fam Physician.
75:56–63. 2007.
|
|
3
|
Chen Z, Fillmore CM, Hammerman PS, Kim CF
and Wong KK: Nonsmallcell lung cancers: A heterogeneous set of
diseases. Nat Rev Cancer. 14:535–546. 2014. View Article : Google Scholar
|
|
4
|
Travis WD: Update on small cell carcinoma
and its differentiation from squamous cell carcinoma and other
non-small cell carcinomas. Modern Pathol. 25:S18–S30. 2012.
View Article : Google Scholar
|
|
5
|
Langer CJ, Besse B, Gualberto A, Brambilla
E and Soria JC: The evolving role of histology in the management of
advanced non-small-cell lung cancer. J Clin Oncol. 28:5311–5320.
2010. View Article : Google Scholar
|
|
6
|
Wieduwilt MJ and Moasser MM: The epidermal
growth factor receptor family: Biology driving targeted
therapeutics. Cell Mol Life Sci. 65:1566–1584. 2008. View Article : Google Scholar :
|
|
7
|
Scagliotti GV, Selvaggi G, Novello S and
Hirsch FR: The biology of epidermal growth factor receptor in lung
cancer. Clin Cancer Res. 10:4227s–4232s. 2004. View Article : Google Scholar
|
|
8
|
da Cunha Santos G, Shepherd FA and Tsao
MS: EGFR mutations and lung cancer. Annu Rev Pathol. 6:49–69. 2011.
View Article : Google Scholar
|
|
9
|
Paez JG, Janne PA, Lee JC, Tracy S,
Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, et
al: EGFR mutations in lung cancer: Correlation with clinical
response to gefitinib therapy. Science. 304:1497–1500. 2004.
View Article : Google Scholar
|
|
10
|
Yamamoto H, Toyooka S and Mitsudomi T:
Impact of EGFR mutation analysis in non-small cell lung cancer.
Lung Cancer. 63:315–321. 2009. View Article : Google Scholar
|
|
11
|
Shigematsu H and Gazdar AF: Somatic
mutations of epidermal growth factor receptor signaling pathway in
lung cancers. Int J Cancer. 118:257–262. 2006. View Article : Google Scholar
|
|
12
|
Siegelin MD and Borczuk AC: Epidermal
growth factor receptor mutations in lung adenocarcinoma. Lab
Invest. 94:129–137. 2014. View Article : Google Scholar
|
|
13
|
Fukuoka M, Wu YL, Thongprasert S,
Sunpaweravong P, Leong SS, Sriuranpong V, Chao TY, Nakagawa K, Chu
DT, Saijo N, et al: Biomarker analyses and final overall survival
results from a phase III, randomized, open-label, first-line study
of gefitinib versus carboplatin/paclitaxel in clinically selected
patients with advanced non-small-cell lung cancer in Asia (IPASS).
J Clin Oncol. 29:2866–2874. 2011. View Article : Google Scholar
|
|
14
|
Metzger B, Chambeau L, Begon DY, Faber C,
Kayser J, Berchem G, Pauly M, Boniver J, Delvenne P, Dicato M and
Wenner T: The human epidermal growth factor receptor (EGFR) gene in
European patients with advanced colorectal cancer harbors
infrequent mutations in its tyrosine kinase domain. BMC Med Genet.
12:1442011. View Article : Google Scholar :
|
|
15
|
Khoo C, Rogers TM, Fellowes A, Bell A and
Fox S: Molecular methods for somatic mutation testing in lung
adenocarcinoma: EGFR and beyond. Transl Lung Cancer Res. 4:126–141.
2015.
|
|
16
|
Jung CY: Biopsy and mutation detection
strategies in non-small cell lung cancer. Tuberc Respir Dis
(Seoul). 75:181–187. 2013. View Article : Google Scholar :
|
|
17
|
Lindeman NI, Cagle PT, Beasley MB, Chitale
DA, Dacic S, Giaccone G, Jenkins RB, Kwiatkowski DJ, Saldivar JS,
Squire J, et al: Molecular testing guideline for selection of lung
cancer patients for EGFR and ALK tyrosine kinase inhibitors:
Guideline from the College of American Pathologists, International
Association for the Study of Lung Cancer, and Association for
Molecular Pathology. J Thorac Oncol. 8:823–859. 2013. View Article : Google Scholar :
|
|
18
|
Hulka BS and Wilcosky T: Biological
markers in epidemiologic research. Arch Environ Health. 43:83–89.
1988. View Article : Google Scholar
|
|
19
|
Mayeux R: Biomarkers: Potential uses and
limitations. NeuroRx. 1:182–188. 2004. View Article : Google Scholar :
|
|
20
|
Miyamoto S, Taylor SL, Barupal DK, Taguchi
A, Wohlgemuth G, Wikoff WR, Yoneda KY, Gandara DR, Hanash SM, Kim K
and Fiehn O: Systemic metabolomic changes in blood samples of lung
cancer patients identified by gas chromatography time-of-flight
mass spectrometry. Metabolites. 5:192–210. 2015. View Article : Google Scholar :
|
|
21
|
Park YH, Shi YP, Liang B, Medriano CA,
Jeon YH, Torres E, Uppal K, Slutsker L and Jones DP:
High-resolution metabolomics to discover potential
parasite-specific biomarkers in a Plasmodium falciparum
erythrocytic stage culture system. Malar J. 14:1222015. View Article : Google Scholar :
|
|
22
|
Peralbo-Molina A, Calderón-Santiago M,
Priego-Capote F, JuradoGámez B and de Castro MD Luque: Metabolomics
analysis of exhaled breath condensate for discrimination between
lung cancer patients and risk factor individuals. J Breath Res.
10:0160112016. View Article : Google Scholar
|
|
23
|
Pamungkas AD, Park C, Lee S, Jee SH and
Park YH: High resolution metabolomics to discriminate compounds in
serum of male lung cancer patients in South Korea. Respir Res.
17:1002016. View Article : Google Scholar :
|
|
24
|
Johnson JM, Yu T, Strobel FH and Jones DP:
A practical approach to detect unique metabolic patterns for
personalized medicine. Analyst. 135:2864–2870. 2010. View Article : Google Scholar :
|
|
25
|
De Sotto R, Medriano C, Cho Y, Seok KS,
Park Y and Kim S: Significance of metabolite extraction method for
evaluating sulfamethazine toxicity in adult zebrafish using
metabolomics. Ecotoxicol Environ Saf. 127:127–134. 2016. View Article : Google Scholar
|
|
26
|
Yu T, Park Y, Johnson JM and Jones DP:
apLCMSadaptive processing of highresolution LC/MS data.
Bioinformatics. 25:193019362009. View Article : Google Scholar
|
|
27
|
Benjamini Y, Drai D, Elmer G, Kafkafi N
and Golani I: Controlling the false discovery rate in behavior
genetics research. Behav Brain Res. 125:2792842001. View Article : Google Scholar
|
|
28
|
Cribbs SK, Park Y, Guidot DM, Martin GS,
Brown LA, Lennox J and Jones DP: Metabolomics of bronchoalveolar
lavage differentiate healthy HIV-1-infected subjects from controls.
Aids Res Hum Retroviruses. 30:579–585. 2014. View Article : Google Scholar :
|
|
29
|
Uppal K, Soltow QA, Strobel FH, Pittard
WS, Gernert KM, Yu T and Jones DP: xMSanalyzer: Automated pipeline
for improved feature detection and downstream analysis of
large-scale, non-targeted metabolomics data. BMC Bioinformatics.
14:152013. View Article : Google Scholar :
|
|
30
|
Neujahr DC, Uppal K, Force SD, Fernandez
F, Lawrence C, Pickens A, Bag R, Lockard C, Kirk AD, Tran V, et al:
Bile acid aspiration associated with lung chemical profile linked
to other biomarkers of injury after lung transplantation. Am J
Transplant. 14:841–848. 2014. View Article : Google Scholar
|
|
31
|
Li S, Park Y, Duraisingham S, Strobel FH,
Khan N, Soltow QA, Jones DP and Pulendran B: Predicting network
activity from high throughput metabolomics. PLoS Comput Biol.
9:e10031232013. View Article : Google Scholar :
|
|
32
|
Hoffman JM, Soltow QA, Li S, Sidik A,
Jones DP and Promislow DE: Effects of age, sex, and genotype on
highsensitivity metabolomic profiles in the fruit fly, Drosophila
melanogaster. Aging Cell. 13:5966042014. View Article : Google Scholar
|
|
33
|
Kim YT, Kim TY, Lee DS, Park SJ, Park JY,
Seo SJ, Choi HS, Kang HJ, Hahn S, Kang CH, et al: Molecular changes
of epidermal growth factor receptor (EGFR) and KRAS and their
impact on the clinical outcomes in surgically resected
adenocarcinoma of the lung. Lung Cancer. 59:111–118. 2008.
View Article : Google Scholar
|
|
34
|
Hsu PP and Sabatini DM: Cancer cell
metabolism: Warburg and beyond. Cell. 134:7037072008. View Article : Google Scholar
|
|
35
|
DeBerardinis RJ, Lum JJ, Hatzivassiliou G
and Thompson CB: The biology of cancer: Metabolic reprogramming
fuels cell growth and proliferation. Cell Metab. 7:11202008.
View Article : Google Scholar
|
|
36
|
Warburg O: On the origin of cancer cells.
Science. 123:3093141956. View Article : Google Scholar
|
|
37
|
Samudio I, Harmancey R, Fiegl M,
Kantarjian H, Konopleva M, Korchin B, Kaluarachchi K, Bornmann W,
Duvvuri S, Taegtmeyer H, et al: Pharmacologic inhibition of fatty
acid oxidation sensitizes human leukemia cells to apoptosis
induction. J Clin Invest. 120:142–156. 2010. View Article : Google Scholar
|
|
38
|
Heiden MG Vander, Cantley LC and Thompson
CB: Understanding the Warburg effect: The metabolic requirements of
cell proliferation. Science. 324:102910332009.
|
|
39
|
Santos CR and Schulze A: Lipid metabolism
in cancer. FEBS J. 279:261026232012. View Article : Google Scholar
|
|
40
|
Currie E, Schulze A, Zechner R, Walther TC
and Farese RV Jr: Cellular fatty acid metabolism and cancer. Cell
Metab. 18:1531612013. View Article : Google Scholar
|
|
41
|
Nieman KM, Kenny HA, Penicka CV, Ladanyi
A, Buell-Gutbrod R, Zillhardt MR, Romero IL, Carey MS, Mills GB,
Hotamisligil GS, et al: Adipocytes promote ovarian cancer
metastasis and provide energy for rapid tumor growth. Nat Med.
17:1498–1503. 2011. View
Article : Google Scholar :
|
|
42
|
Ramsay RR, Gandour RD and van der Leij FR:
Molecular enzymology of carnitine transfer and transport. Biochim
Biophys Acta. 1546:21432001.
|
|
43
|
van Vlies N, Ferdinandusse S, Turkenburg
M, Wanders RJ and Vaz FM: PPAR alphaactivation results in enhanced
carnitine biosynthesis and OCTN2mediated hepatic carnitine
accumulation. Biochim Biophys Acta. 1767:113411422007.
|
|
44
|
Brandt JM, Djouadi F and Kelly DP: Fatty
acids activate transcription of the muscle carnitine
palmitoyltransferase I gene in cardiac myocytes via the peroxisome
proliferatoractivated receptor alpha. J Biol Chem.
273:23786237921998. View Article : Google Scholar
|
|
45
|
Sharma S, Sun X, Rafikov R, Kumar S, Hou
Y, Oishi PE, Datar SA, Raff G, Fineman JR and Black SM: PPAR-γ
regulates carnitine homeostasis and mitochondrial function in a
lamb model of increased pulmonary blood flow. PLoS One.
7:e415552012. View Article : Google Scholar :
|
|
46
|
Hou Y, Gao J, Xu H, Xu Y, Zhang Z, Xu Q
and Zhang C: PPARγ E3 ubiquitin ligase regulates MUC1-C oncoprotein
stability. Oncogene. 33:5619–5625. 2014. View Article : Google Scholar
|
|
47
|
Schooneman MG, Vaz FM, Houten SM and
Soeters MR: Acylcarnitines: Reflecting or inflicting insulin
resistance? Diabetes. 62:182013. View Article : Google Scholar
|
|
48
|
Zhao QH, Cao Y, Wang Y, Hu C, Hu A, Ruan
L, Bo Q, Liu Q, Chen W, Tao F, et al: Plasma and tissue free amino
acid profiles and their concentration correlation in patients with
lung cancer. Asia Pac J Clin Nutr. 23:429–436. 2014.
|
|
49
|
Shingyoji M, Iizasa T, Higashiyama M,
Imamura F, Saruki N, Imaizumi A, Yamamoto H, Daimon T, Tochikubo O,
Mitsushima T, et al: The significance and robustness of a plasma
free amino acid (PFAA) profile-based multiplex function for
detecting lung cancer. BMC Cancer. 13:772013. View Article : Google Scholar :
|
|
50
|
Abumrad NN and Miller B: The physiologic
and nutritional significance of plasmafree amino acid levels. JPEN
J Parenter Enteral Nutr. 7:1631701983. View Article : Google Scholar
|
|
51
|
Nunez MI, Behrens C, Woods DM, Lin H,
Suraokar M, Kadara H, Hofstetter W, Kalhor N, Lee JJ, Franklin W,
et al: High expression of folate receptor alpha in lung cancer
correlates with adenocarcinoma histology and EGFR [corrected]
mutation. J Thorac Oncol. 7:833–840. 2012. View Article : Google Scholar :
|
|
52
|
Weitman SD, Weinberg AG, Coney LR,
Zurawski VR, Jennings DS and Kamen BA: Cellular localization of the
folate receptor: Potential role in drug toxicity and folate
homeostasis. Cancer Res. 52:670867111992.
|
|
53
|
Rijnboutt S, Jansen G, Posthuma G, Hynes
JB, Schornagel JH and Strous GJ: Endocytosis of GPIlinked membrane
folate receptoralpha. J Cell Biol. 132:35471996. View Article : Google Scholar
|
|
54
|
Kamen BA and Smith AK: A review of folate
receptor alpha cycling and 5methyltetrahydrofolate accumulation
with an emphasis on cell models in vitro. Adv Drug Deliver Rev.
56:108510972004. View Article : Google Scholar
|
|
55
|
Zhao HY, Chen GY, Huang Y, Li XL, Feng JF,
Shi MQ, Cheng Y, Ma LX, Zhang YP, Gu CP, et al: Erlotinib plus
capecitabine as first-line treatment for older Chinese patients
with advanced adenocarcinoma of the lung (C-TONG0807): An
open-label, single arm, multicenter phase II study. Medicine
(Baltimore). 94:e2492015. View Article : Google Scholar :
|