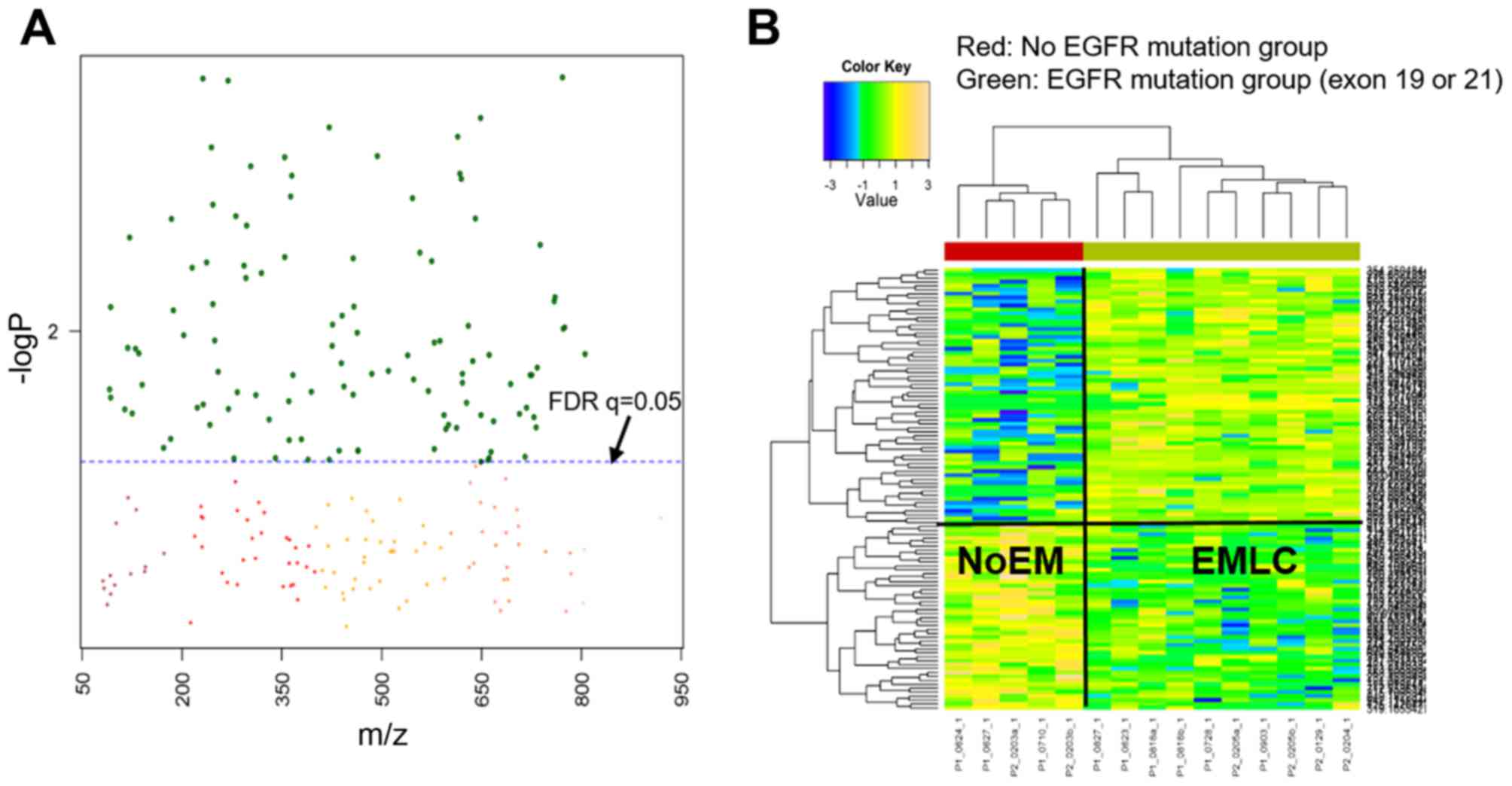
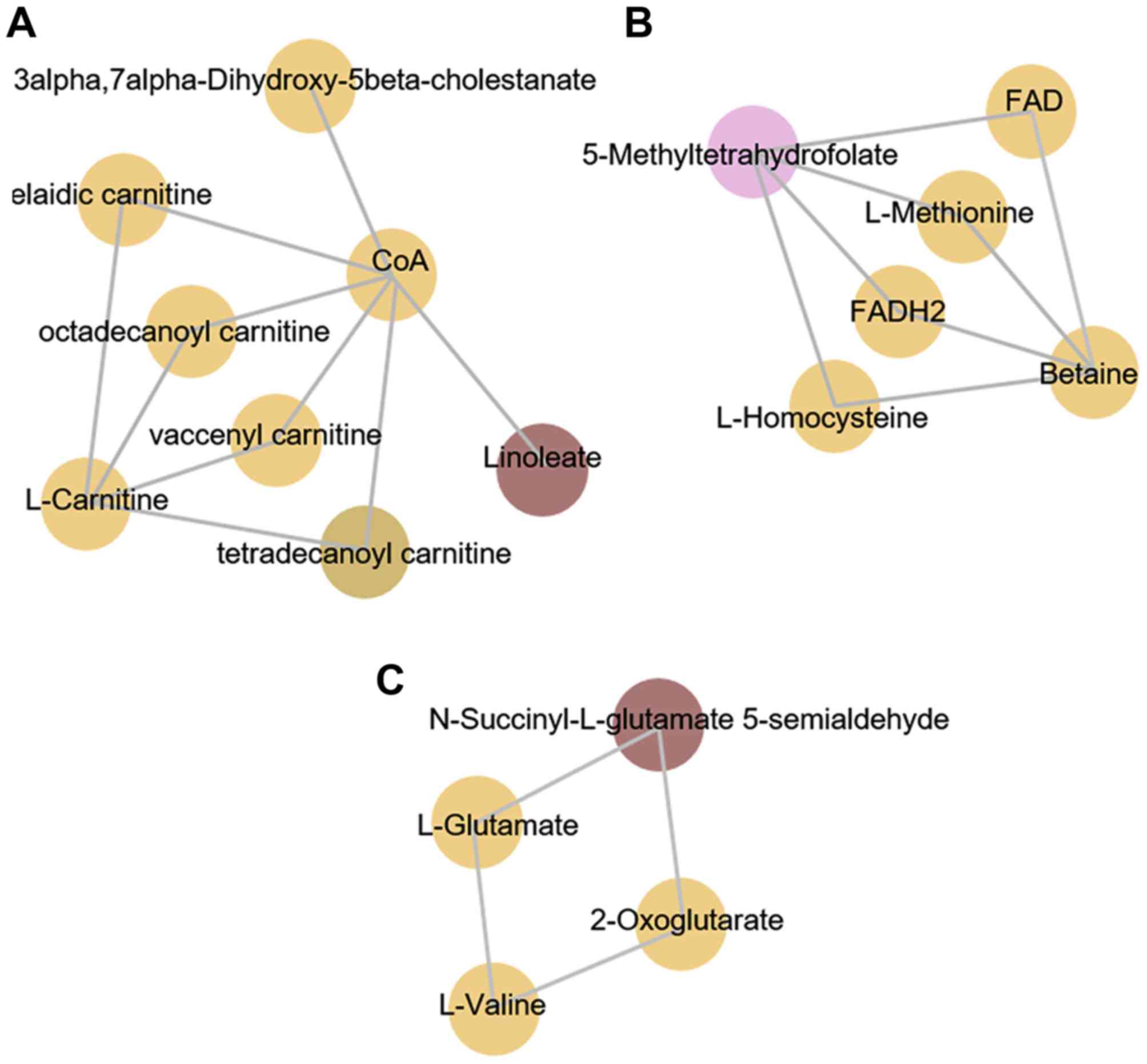
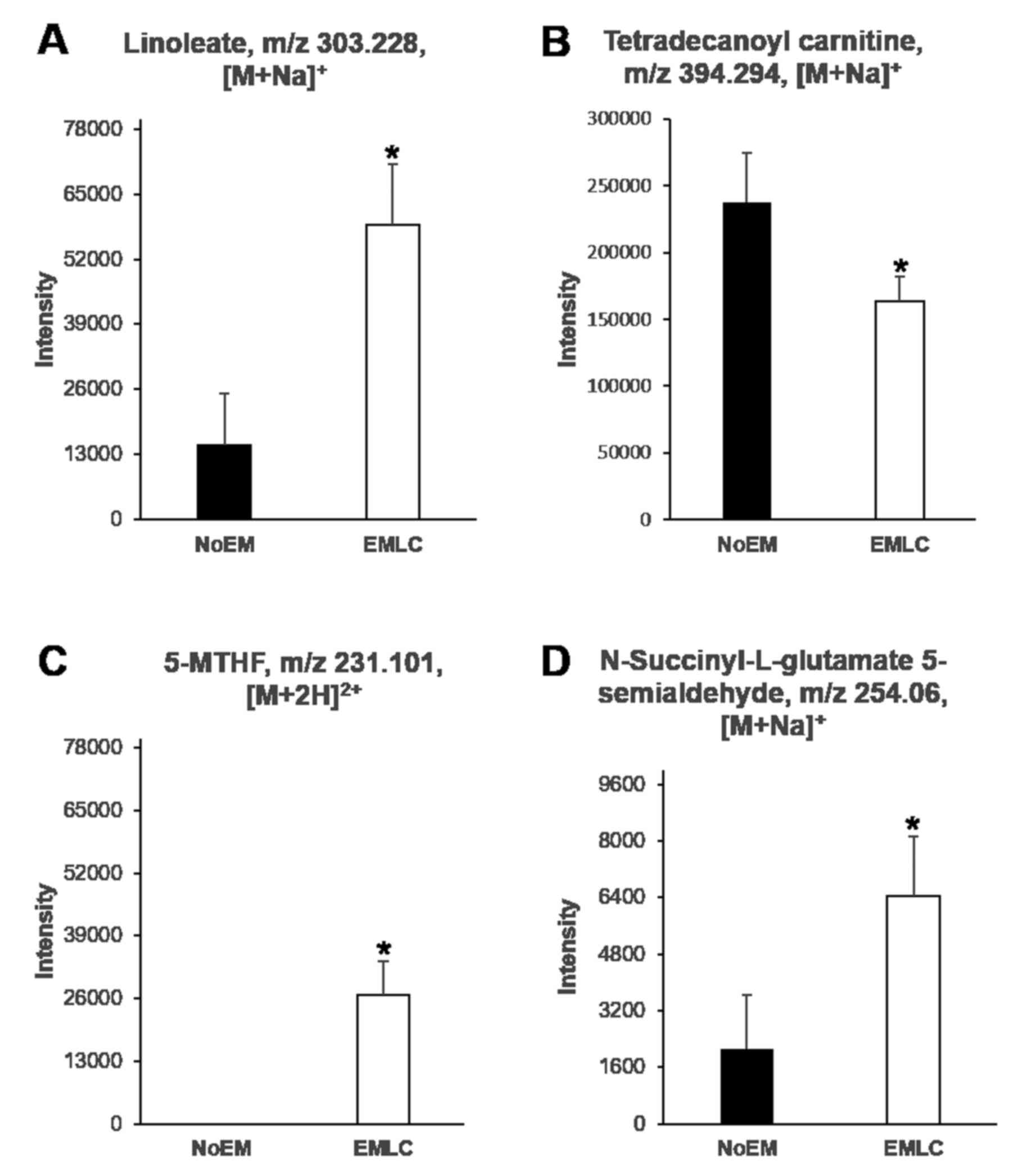
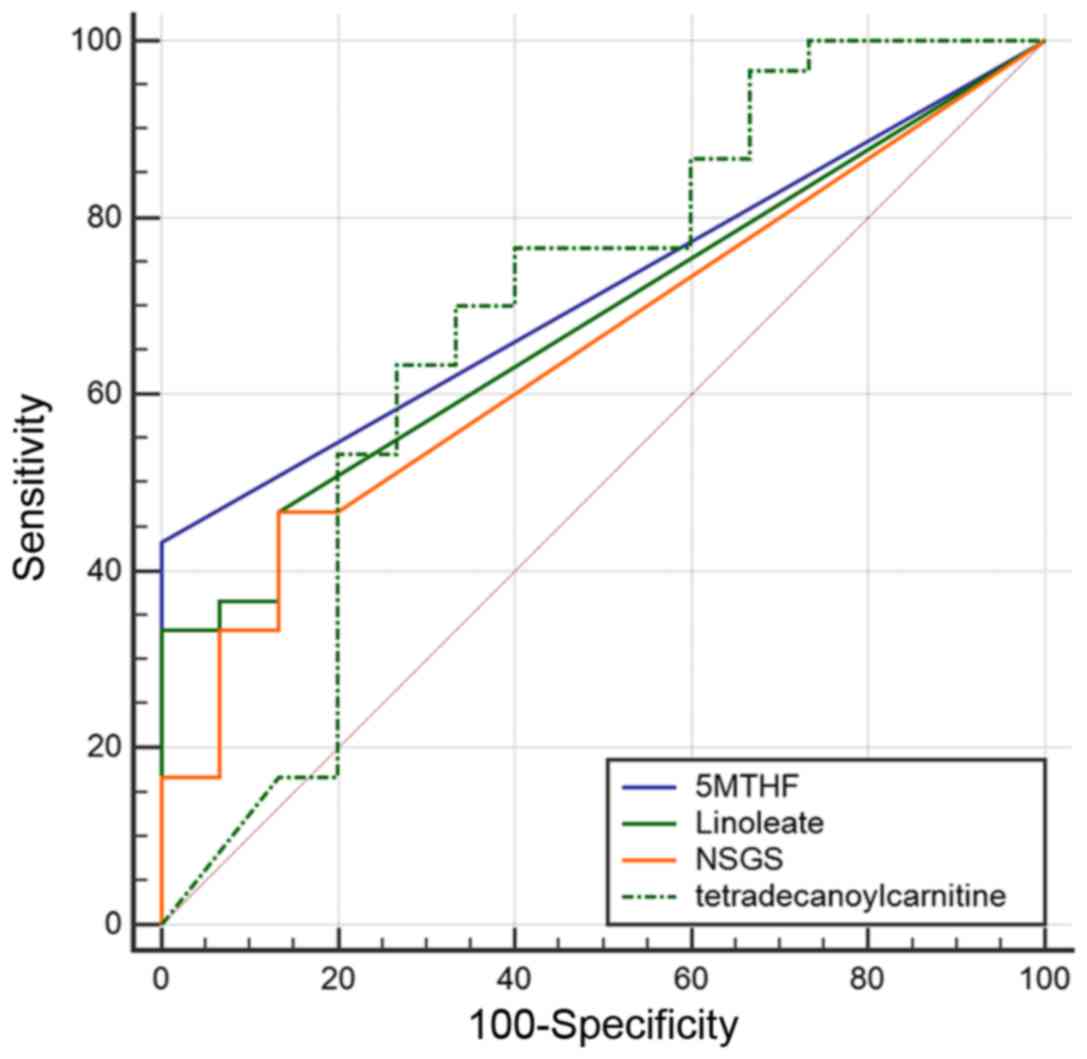
|
1
|
Jung KW, Won YJ, Oh CM, Kong HJ, Cho H,
Lee JK, Lee DH and Lee KH: Prediction of cancer incidence and
mortality in Korea, 2016. Cancer Res Treat. 48:451–457. 2016.
View Article : Google Scholar :
|
|
2
|
Collins LG, Haines C, Perkel R and Enck
RE: Lung cancer: Diagnosis and management. Am Fam Physician.
75:56–63. 2007.
|
|
3
|
Chen Z, Fillmore CM, Hammerman PS, Kim CF
and Wong KK: Nonsmallcell lung cancers: A heterogeneous set of
diseases. Nat Rev Cancer. 14:535–546. 2014. View Article : Google Scholar
|
|
4
|
Travis WD: Update on small cell carcinoma
and its differentiation from squamous cell carcinoma and other
non-small cell carcinomas. Modern Pathol. 25:S18–S30. 2012.
View Article : Google Scholar
|
|
5
|
Langer CJ, Besse B, Gualberto A, Brambilla
E and Soria JC: The evolving role of histology in the management of
advanced non-small-cell lung cancer. J Clin Oncol. 28:5311–5320.
2010. View Article : Google Scholar
|
|
6
|
Wieduwilt MJ and Moasser MM: The epidermal
growth factor receptor family: Biology driving targeted
therapeutics. Cell Mol Life Sci. 65:1566–1584. 2008. View Article : Google Scholar :
|
|
7
|
Scagliotti GV, Selvaggi G, Novello S and
Hirsch FR: The biology of epidermal growth factor receptor in lung
cancer. Clin Cancer Res. 10:4227s–4232s. 2004. View Article : Google Scholar
|
|
8
|
da Cunha Santos G, Shepherd FA and Tsao
MS: EGFR mutations and lung cancer. Annu Rev Pathol. 6:49–69. 2011.
View Article : Google Scholar
|
|
9
|
Paez JG, Janne PA, Lee JC, Tracy S,
Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, et
al: EGFR mutations in lung cancer: Correlation with clinical
response to gefitinib therapy. Science. 304:1497–1500. 2004.
View Article : Google Scholar
|
|
10
|
Yamamoto H, Toyooka S and Mitsudomi T:
Impact of EGFR mutation analysis in non-small cell lung cancer.
Lung Cancer. 63:315–321. 2009. View Article : Google Scholar
|
|
11
|
Shigematsu H and Gazdar AF: Somatic
mutations of epidermal growth factor receptor signaling pathway in
lung cancers. Int J Cancer. 118:257–262. 2006. View Article : Google Scholar
|
|
12
|
Siegelin MD and Borczuk AC: Epidermal
growth factor receptor mutations in lung adenocarcinoma. Lab
Invest. 94:129–137. 2014. View Article : Google Scholar
|
|
13
|
Fukuoka M, Wu YL, Thongprasert S,
Sunpaweravong P, Leong SS, Sriuranpong V, Chao TY, Nakagawa K, Chu
DT, Saijo N, et al: Biomarker analyses and final overall survival
results from a phase III, randomized, open-label, first-line study
of gefitinib versus carboplatin/paclitaxel in clinically selected
patients with advanced non-small-cell lung cancer in Asia (IPASS).
J Clin Oncol. 29:2866–2874. 2011. View Article : Google Scholar
|
|
14
|
Metzger B, Chambeau L, Begon DY, Faber C,
Kayser J, Berchem G, Pauly M, Boniver J, Delvenne P, Dicato M and
Wenner T: The human epidermal growth factor receptor (EGFR) gene in
European patients with advanced colorectal cancer harbors
infrequent mutations in its tyrosine kinase domain. BMC Med Genet.
12:1442011. View Article : Google Scholar :
|
|
15
|
Khoo C, Rogers TM, Fellowes A, Bell A and
Fox S: Molecular methods for somatic mutation testing in lung
adenocarcinoma: EGFR and beyond. Transl Lung Cancer Res. 4:126–141.
2015.
|
|
16
|
Jung CY: Biopsy and mutation detection
strategies in non-small cell lung cancer. Tuberc Respir Dis
(Seoul). 75:181–187. 2013. View Article : Google Scholar :
|
|
17
|
Lindeman NI, Cagle PT, Beasley MB, Chitale
DA, Dacic S, Giaccone G, Jenkins RB, Kwiatkowski DJ, Saldivar JS,
Squire J, et al: Molecular testing guideline for selection of lung
cancer patients for EGFR and ALK tyrosine kinase inhibitors:
Guideline from the College of American Pathologists, International
Association for the Study of Lung Cancer, and Association for
Molecular Pathology. J Thorac Oncol. 8:823–859. 2013. View Article : Google Scholar :
|
|
18
|
Hulka BS and Wilcosky T: Biological
markers in epidemiologic research. Arch Environ Health. 43:83–89.
1988. View Article : Google Scholar
|
|
19
|
Mayeux R: Biomarkers: Potential uses and
limitations. NeuroRx. 1:182–188. 2004. View Article : Google Scholar :
|
|
20
|
Miyamoto S, Taylor SL, Barupal DK, Taguchi
A, Wohlgemuth G, Wikoff WR, Yoneda KY, Gandara DR, Hanash SM, Kim K
and Fiehn O: Systemic metabolomic changes in blood samples of lung
cancer patients identified by gas chromatography time-of-flight
mass spectrometry. Metabolites. 5:192–210. 2015. View Article : Google Scholar :
|
|
21
|
Park YH, Shi YP, Liang B, Medriano CA,
Jeon YH, Torres E, Uppal K, Slutsker L and Jones DP:
High-resolution metabolomics to discover potential
parasite-specific biomarkers in a Plasmodium falciparum
erythrocytic stage culture system. Malar J. 14:1222015. View Article : Google Scholar :
|
|
22
|
Peralbo-Molina A, Calderón-Santiago M,
Priego-Capote F, JuradoGámez B and de Castro MD Luque: Metabolomics
analysis of exhaled breath condensate for discrimination between
lung cancer patients and risk factor individuals. J Breath Res.
10:0160112016. View Article : Google Scholar
|
|
23
|
Pamungkas AD, Park C, Lee S, Jee SH and
Park YH: High resolution metabolomics to discriminate compounds in
serum of male lung cancer patients in South Korea. Respir Res.
17:1002016. View Article : Google Scholar :
|
|
24
|
Johnson JM, Yu T, Strobel FH and Jones DP:
A practical approach to detect unique metabolic patterns for
personalized medicine. Analyst. 135:2864–2870. 2010. View Article : Google Scholar :
|
|
25
|
De Sotto R, Medriano C, Cho Y, Seok KS,
Park Y and Kim S: Significance of metabolite extraction method for
evaluating sulfamethazine toxicity in adult zebrafish using
metabolomics. Ecotoxicol Environ Saf. 127:127–134. 2016. View Article : Google Scholar
|
|
26
|
Yu T, Park Y, Johnson JM and Jones DP:
apLCMSadaptive processing of highresolution LC/MS data.
Bioinformatics. 25:193019362009. View Article : Google Scholar
|
|
27
|
Benjamini Y, Drai D, Elmer G, Kafkafi N
and Golani I: Controlling the false discovery rate in behavior
genetics research. Behav Brain Res. 125:2792842001. View Article : Google Scholar
|
|
28
|
Cribbs SK, Park Y, Guidot DM, Martin GS,
Brown LA, Lennox J and Jones DP: Metabolomics of bronchoalveolar
lavage differentiate healthy HIV-1-infected subjects from controls.
Aids Res Hum Retroviruses. 30:579–585. 2014. View Article : Google Scholar :
|
|
29
|
Uppal K, Soltow QA, Strobel FH, Pittard
WS, Gernert KM, Yu T and Jones DP: xMSanalyzer: Automated pipeline
for improved feature detection and downstream analysis of
large-scale, non-targeted metabolomics data. BMC Bioinformatics.
14:152013. View Article : Google Scholar :
|
|
30
|
Neujahr DC, Uppal K, Force SD, Fernandez
F, Lawrence C, Pickens A, Bag R, Lockard C, Kirk AD, Tran V, et al:
Bile acid aspiration associated with lung chemical profile linked
to other biomarkers of injury after lung transplantation. Am J
Transplant. 14:841–848. 2014. View Article : Google Scholar
|
|
31
|
Li S, Park Y, Duraisingham S, Strobel FH,
Khan N, Soltow QA, Jones DP and Pulendran B: Predicting network
activity from high throughput metabolomics. PLoS Comput Biol.
9:e10031232013. View Article : Google Scholar :
|
|
32
|
Hoffman JM, Soltow QA, Li S, Sidik A,
Jones DP and Promislow DE: Effects of age, sex, and genotype on
highsensitivity metabolomic profiles in the fruit fly, Drosophila
melanogaster. Aging Cell. 13:5966042014. View Article : Google Scholar
|
|
33
|
Kim YT, Kim TY, Lee DS, Park SJ, Park JY,
Seo SJ, Choi HS, Kang HJ, Hahn S, Kang CH, et al: Molecular changes
of epidermal growth factor receptor (EGFR) and KRAS and their
impact on the clinical outcomes in surgically resected
adenocarcinoma of the lung. Lung Cancer. 59:111–118. 2008.
View Article : Google Scholar
|
|
34
|
Hsu PP and Sabatini DM: Cancer cell
metabolism: Warburg and beyond. Cell. 134:7037072008. View Article : Google Scholar
|
|
35
|
DeBerardinis RJ, Lum JJ, Hatzivassiliou G
and Thompson CB: The biology of cancer: Metabolic reprogramming
fuels cell growth and proliferation. Cell Metab. 7:11202008.
View Article : Google Scholar
|
|
36
|
Warburg O: On the origin of cancer cells.
Science. 123:3093141956. View Article : Google Scholar
|
|
37
|
Samudio I, Harmancey R, Fiegl M,
Kantarjian H, Konopleva M, Korchin B, Kaluarachchi K, Bornmann W,
Duvvuri S, Taegtmeyer H, et al: Pharmacologic inhibition of fatty
acid oxidation sensitizes human leukemia cells to apoptosis
induction. J Clin Invest. 120:142–156. 2010. View Article : Google Scholar
|
|
38
|
Heiden MG Vander, Cantley LC and Thompson
CB: Understanding the Warburg effect: The metabolic requirements of
cell proliferation. Science. 324:102910332009.
|
|
39
|
Santos CR and Schulze A: Lipid metabolism
in cancer. FEBS J. 279:261026232012. View Article : Google Scholar
|
|
40
|
Currie E, Schulze A, Zechner R, Walther TC
and Farese RV Jr: Cellular fatty acid metabolism and cancer. Cell
Metab. 18:1531612013. View Article : Google Scholar
|
|
41
|
Nieman KM, Kenny HA, Penicka CV, Ladanyi
A, Buell-Gutbrod R, Zillhardt MR, Romero IL, Carey MS, Mills GB,
Hotamisligil GS, et al: Adipocytes promote ovarian cancer
metastasis and provide energy for rapid tumor growth. Nat Med.
17:1498–1503. 2011. View
Article : Google Scholar :
|
|
42
|
Ramsay RR, Gandour RD and van der Leij FR:
Molecular enzymology of carnitine transfer and transport. Biochim
Biophys Acta. 1546:21432001.
|
|
43
|
van Vlies N, Ferdinandusse S, Turkenburg
M, Wanders RJ and Vaz FM: PPAR alphaactivation results in enhanced
carnitine biosynthesis and OCTN2mediated hepatic carnitine
accumulation. Biochim Biophys Acta. 1767:113411422007.
|
|
44
|
Brandt JM, Djouadi F and Kelly DP: Fatty
acids activate transcription of the muscle carnitine
palmitoyltransferase I gene in cardiac myocytes via the peroxisome
proliferatoractivated receptor alpha. J Biol Chem.
273:23786237921998. View Article : Google Scholar
|
|
45
|
Sharma S, Sun X, Rafikov R, Kumar S, Hou
Y, Oishi PE, Datar SA, Raff G, Fineman JR and Black SM: PPAR-γ
regulates carnitine homeostasis and mitochondrial function in a
lamb model of increased pulmonary blood flow. PLoS One.
7:e415552012. View Article : Google Scholar :
|
|
46
|
Hou Y, Gao J, Xu H, Xu Y, Zhang Z, Xu Q
and Zhang C: PPARγ E3 ubiquitin ligase regulates MUC1-C oncoprotein
stability. Oncogene. 33:5619–5625. 2014. View Article : Google Scholar
|
|
47
|
Schooneman MG, Vaz FM, Houten SM and
Soeters MR: Acylcarnitines: Reflecting or inflicting insulin
resistance? Diabetes. 62:182013. View Article : Google Scholar
|
|
48
|
Zhao QH, Cao Y, Wang Y, Hu C, Hu A, Ruan
L, Bo Q, Liu Q, Chen W, Tao F, et al: Plasma and tissue free amino
acid profiles and their concentration correlation in patients with
lung cancer. Asia Pac J Clin Nutr. 23:429–436. 2014.
|
|
49
|
Shingyoji M, Iizasa T, Higashiyama M,
Imamura F, Saruki N, Imaizumi A, Yamamoto H, Daimon T, Tochikubo O,
Mitsushima T, et al: The significance and robustness of a plasma
free amino acid (PFAA) profile-based multiplex function for
detecting lung cancer. BMC Cancer. 13:772013. View Article : Google Scholar :
|
|
50
|
Abumrad NN and Miller B: The physiologic
and nutritional significance of plasmafree amino acid levels. JPEN
J Parenter Enteral Nutr. 7:1631701983. View Article : Google Scholar
|
|
51
|
Nunez MI, Behrens C, Woods DM, Lin H,
Suraokar M, Kadara H, Hofstetter W, Kalhor N, Lee JJ, Franklin W,
et al: High expression of folate receptor alpha in lung cancer
correlates with adenocarcinoma histology and EGFR [corrected]
mutation. J Thorac Oncol. 7:833–840. 2012. View Article : Google Scholar :
|
|
52
|
Weitman SD, Weinberg AG, Coney LR,
Zurawski VR, Jennings DS and Kamen BA: Cellular localization of the
folate receptor: Potential role in drug toxicity and folate
homeostasis. Cancer Res. 52:670867111992.
|
|
53
|
Rijnboutt S, Jansen G, Posthuma G, Hynes
JB, Schornagel JH and Strous GJ: Endocytosis of GPIlinked membrane
folate receptoralpha. J Cell Biol. 132:35471996. View Article : Google Scholar
|
|
54
|
Kamen BA and Smith AK: A review of folate
receptor alpha cycling and 5methyltetrahydrofolate accumulation
with an emphasis on cell models in vitro. Adv Drug Deliver Rev.
56:108510972004. View Article : Google Scholar
|
|
55
|
Zhao HY, Chen GY, Huang Y, Li XL, Feng JF,
Shi MQ, Cheng Y, Ma LX, Zhang YP, Gu CP, et al: Erlotinib plus
capecitabine as first-line treatment for older Chinese patients
with advanced adenocarcinoma of the lung (C-TONG0807): An
open-label, single arm, multicenter phase II study. Medicine
(Baltimore). 94:e2492015. View Article : Google Scholar :
|