Introduction
Hydroxyapatite (HA;
Ca10(PO4)6(OH)2) is a
major component of hard tissues such as tooth and bone (1) and has been used as a biomaterial in
multiple applications, including tissue engineering and bone repair
(2). HA has been used effectively
as a carrier for bioactive molecules, including proteins and DNA,
as its biocompatible and porous properties allow it to deliver
proteins and genes into cells (3).
Previous research has focused on the application of HA as a
treatment for bone-associated diseases (4), but research regarding the use of HA
as a drug delivery system (DDS) in other tissues remains scarce.
One disadvantage associated with the use of HA for DDS is low
dispersal in water, making the formulation of a drug-loaded HA
treatment of particular importance for development of HA as a DDS.
The rise of nanotechnology has provided novel tools and methods for
the study of gene carriers. Among these, calcium phosphate
nanoparticles have emerged as a vector for nonviral gene delivery
(5,6). Hydroxyapatite nanoparticles (HAP)
containing pEGFP-N1 plasmids have been reported to transport DNA
into gastric cancer cells without any considerable cytotoxicity
(7). Tan et al (8) demonstrated that HAP with protamine
enhances the efficiency of gene transfection. Sun et al
(9) used HAP to deliver the
neurotrophin-3 (NT-3) gene into the cochlear neurons of guinea pigs
both in vitro and in vivo, and further demonstrated
that polyethylenimine-induced surface alteration of HAP permitted
specific genetic materials to pass through intact round window
membranes in chinchillas with low toxicity and high transfection
efficiency (10). Yan-Zhong et
al (11) used
arginine-modified nanohydroxyapatite to modify the surface charge
of HAP and demonstrated an enhanced adsorption capacity in human
epithelial cells. CaCl2-modified HAP has also been
previously demonstrated to mediate the transfection of small
interfering (si-)signal transducer and activator of transcription 3
(Stat3) plasmids into murine prostate cancer cells in vivo,
resulting in significant inhibition of cancer growth (12). These previous studies demonstrate
that HAP may be a safe and effective gene vector with potential
clinical applications.
RNA interference (RNAi) is a method of
post-transcriptional gene silencing. Short hairpin RNA (shRNA) has
previously been recognized as a promising novel antitumor strategy
(13), but the efficient delivery
of shRNA remains a challenge for shRNA-based therapies.
Nanomedicine is an emerging field that combines medicine with
nanotechnology, and nanoparticles have been used as diagnostic
probes in targeted therapies. Hydroxyapatite has been used as a
novel biomaterial to treat oral cavities and enhance bone repair,
and as a medicine carrier possesses good tissue compatibility both
in vivo and in vitro (14,15).
Compared with viral vectors, which pose the danger of
immunogenicity and pathogenicity, HAP has high osteoconductivity
and favorable biocompatibility, and/or osteoinductivity without
proinflammatory or immunogenic side effects (16–18).
HAP inhibits tumor cell growth directly (19), and is a useful gene delivery system
due to its efficiency, safety and economy (12).
DNA vector-based Stat3-specific RNAi (sh-Stat3)
silences Stat3 and inhibits prostate tumor development (13). However, this antitumor activity
depends on the efficiency of delivery. The present study examined
the effect of hydroxyapatite-transported sh-Stat3 on the growth of
murine prostate cancer cells. A previous study demonstrated that
sh-Stat3 significantly suppresses Stat3 expression and inhibits
prostate cancer cell growth (12).
In the present study, HAP was used to deliver DNA-vector-based
sh-Stat3 to treat prostate cancer in vitro, and the in
vitro antitumor effects and mechanisms were examined to provide
a theoretical basis for future clinical use.
Materials and methods
Materials
The mouse prostate cancer cell line RM1 was
purchased from the Shanghai Institute of Cellular Research
(Shanghai, China). The cells were cultured in Iscove's modified
Dulbecco's medium (Gibco; Thermo Fisher Scientific, Inc., Waltham,
MA, USA) containing 10% fetal bovine serum (FBS; Gibco; Thermo
Fisher Scientific, Inc.), 100 IU/ml penicillin and 100 µg/ml
streptomycin in a humid atmosphere (37°C, 5% CO2, 95% air).
Plasmids containing sh-Stat3 (sequence GCAGCAGCTGAACAACATG,
corresponding to nucleotides 2,144 to 2,162; Genbank accession no.
NM_003150) were constructed in situ. According to previous
research, the sh-Stat3 most effective at inhibiting cancer growth
is located at the SH2 domain of the mouse and human Stat3 genes
(20). A negative control
scrambled shRNA sequence (sh-scramble; Ambion; Thermo Fisher
Scientific, Inc.) lacking homology to mouse or human gene sequences
was used to determine the frequency of nonspecific effects. HAP
nanoparticles were purchased from Nanjing Emperor Nano Material
Co., Ltd. (Nanjing, China).
Cell culture, transfection and
measurement of cell growth
Cell cultures, HAP and transfection plasmids were
prepared as previously described (12). For transfection, RM1 cells in the
logarithmic growth phase were seeded in 12-well plates (1×105
cells/well). When the cells reached 50% confluence, they were
transfected with sh-Stat3 (HAP/sh-Stat3 group) or sh-scramble
(HAP/sh-scramble group) plasmids, delivered with HAP, for 48–72 h
prior to analysis of mRNA and protein levels, cell apoptosis and
viability. As controls, untransfected cells (mock) and cells
treated with HAP alone (HAP group) were also used.
Cell viability following plasmid treatment was
measured by MTT assay. RM1 cells were seeded in 96-well plates and
incubated at 37°C for 24 and 48 h in the presence of sh-Stat3 or
sh-scramble plasmids. Then, 10 ml of MTT (Sigma-Aldrich; Merck
Millipore, Darmstadt, Germany) at a concentration of 5 mg/ml in
phosphate buffered saline (PBS) was added to each well, and
incubated for 4 h at 37°C. Following removal of the culture medium,
the formazan crystals were solubilized in 100 µl dimethyl sulfoxide
(Sigma-Aldrich; Merck Millipore) and shaken for 15 min. Absorption
was measured at 570 nm on a microplate reader (Bio-Rad
Laboratories, Inc., Hercules, CA, USA). Each assay was performed
nine times.
Apoptosis and cell cycle
assessment
RM1 cells (1×106 cells per well) were treated with
shRNA-Stat3 and scramble shRNA plasmids for up to 48 h, collected,
and resuspended in 100 µl PBS, and incubated at room temperature
for 5–15 min in the dark with 5 µl Annexin V-fluorescein
isothiocyanate (eBioscience; Thermo Fisher Scientific, Inc.), then
with 5 µl propidium iodide (PI; Beckman Coulter, Fullerton, CA,
USA) for 30 min at room temperature in the dark. Flow cytometry was
performed to measure apoptosis rates, using an Epics-XL-MCL flow
cytometer (Beckman Coulter, Inc.). Data was analyzed using FlowJo
v10 software (Tree Star, Inc., Ashland, OR, USA).
Cell cycle distribution was analyzed by measuring
the DNA fragments stained with PI. RM1 cells (1×106)
grown in 6-well plates were harvested and centrifuged 48 h
following transfection. Cells were counted and washed twice with
pre-cooled PBS. Then, cells were fixed and permeabilized overnight
by adding 1 ml of 70% (v/v) pre-cooled ethanol to each tube at 4°C.
Following centrifugation at 3,000 × g for 5 min, the fixatives were
decanted and 1×106 cells were resuspended in 0.5 ml
staining solution, containing 50 µg/ml PI and 100 µg/ml DNase-free
RNase (Sigma-Aldrich; Merck Millipore) and incubated for 30 min at
room temperature in the dark. Finally, cells were analyzed by flow
cytometry using a FACScan™ system (BD Biosciences, San Jose, CA,
USA) and data analysed using CellQuest™ software (version 3.3; BD
Biosciences).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) assay
The mRNA expression level of Stat3 and Stat3
downstream genes was determined using semi quantitative RT-PCR.
Cells treated with sh-Stat3 or sh-scramble were collected following
48 h incubation, and total RNA was extracted using TRIzol reagent
(Invitrogen; Thermo Fisher Scientific, Inc.). Reverse transcription
was performed with 2 µg of total RNA using a commercially available
RT-PCR kit (cat. no. A4051; Promega Corporation, Madison, WI, USA)
according to the manufacturer's instructions, following DNAse I
(cat. no. D5025-15KU; Sigma-Aldrich; Merck Millipore) treatment.
The primers used for PCR are displayed in Table I. Each reaction (20 µl samples) was
carried out under the following cycling conditions: Initialization
for 10 min at 95°C and then 40 cycles of amplification, with 15 sec
at 95°C for denaturation and 1 min at 60°C for annealing and
elongation. A standard curve was plotted for each primer probe,
established by using a serial dilution of pooled cDNA from cells.
All standards and samples were conducted in triplicate, normalised
to β-actin, and relative fold changes in mRNA expression were
calculated using the formula 2-ΔΔCq (21).
 | Table I.Primer sets used for semi
quantitative reverse transcription polymerase chain reaction
analysis. |
Table I.
Primer sets used for semi
quantitative reverse transcription polymerase chain reaction
analysis.
| Target gene | Sequence
(5′-3′) | Accession
number |
|---|
| β-actin | Forward:
CTGAGAGGGAAATCGTGCGT |
|
|
| Reverse:
AACCGCTCGTTGCCAATAGT | NM_007393 |
| Stat3 | Forward:
ACCAGCAATATAGCCGATTCC |
|
|
| Reverse:
CCATTGGCTTCTCAAGATACC | NM_011486 |
| Bcl-2 | Forward:
TCGCAGAGATGTCCAGTCA |
|
|
| Reverse:
CACCGAACTCAAAGAAGGC | NM_009741 |
| Bax | Forward:
AGGGTTTCATCCAGGATCGAGC |
|
|
| Reverse:
AGGCGGTGAGGACTCCAGCC | NM_007527 |
| Caspase3 | Forward:
TGGACTGTGGCATTGAGAC |
|
|
| Reverse:
AGGAATAGTAACCAGGTGCTG | NM_009810 |
| VEGF | Forward:
ATCTTCAAGCCGTCCTGTG |
|
|
| Reverse:
TGGTGATGTTGCTCTCTGAC | NM_009505 |
| Cyclin D1 | Forward:
TCATTTCCAACCCACCCT |
|
|
| Reverse:
GGCTTCAATCTGTTCCTGG | NM_007631 |
Western blot analysis
Cell lysis, protein quantification and western blot
assays were performed as previously described (22). Primary antibodies (1:1,000
dilution) targeting Stat3 (cat. no. sc-482), phospho
(p-)Tyr705-Stat3 (cat. no. sc-7993), Cyclin D1 (cat. no. sc-450),
vascular endothelial growth factor (VEGF; cat. no. sc-152) and
β-actin (cat. no. sc-32251) were obtained from Santa Cruz
Biotechnology, Inc. (Dallas, TX, USA). Primary antibodies targeting
B cell lymphoma 2 apoptosis regulator (Bcl-2; cat. no. 2876s),
Bcl-2 associated X apoptosis regulator (Bax; cat. no. 2772s) and
cleaved (c-)caspase3 (cat. no. 9661L) were obtained from Cell
Signalling Technology, Inc. (Danvers, MA, USA). Protein bands were
visualized using SuperSignal West Pico chemiluminescent substrate
(Pierce; Thermo Fisher Scientific, Inc.), and membranes were
subjected to X-ray autoradiography. Band intensities were
determined with the Quantity One software (v4.2.1; Bio-Rad
Laboratories, Inc.). All experiments were performed in
triplicate.
Statistical analysis
Quantitative data were expressed as the mean ±
standard error of the mean. Statistical analysis was performed with
SPSS version 13.0 (SPSS Inc., Chicago, IL, USA), and one-way
analysis of variance followed by Tukey's test was used to compare
the differences between groups. P<0.05 was considered to
indicate a statistically significant difference.
Results
Stat3 expression
To calculate the ability of HAP-delivered plasmids
to silence Stat3 expression, RT-qPCR and western blot assays were
used to analyze Stat3 mRNA and protein expression levels,
respectively, in RM1 cells. The results demonstrated that Stat3
mRNA expression was significantly decreased in
HAP/sh-Stat3-transfected RM1 cells compared with cells transfected
with HAP/sh-scramble (P<0.01; Table II). Stat3 and p-Stat3 protein
levels were also significantly decreased in
HAP/sh-Stat3-transfected RM1 cells compared with RM1 cells
transfected with HAP/sh-scramble (P<0.01; Fig. 1).
 | Table II.Reverse transcription-quantitative
polymerase chain reaction analysis of Stat3, Bcl-2, Bax, Caspase3,
VEGF and Cyclin D1 mRNA expression levels, relative to the Mock
group. |
Table II.
Reverse transcription-quantitative
polymerase chain reaction analysis of Stat3, Bcl-2, Bax, Caspase3,
VEGF and Cyclin D1 mRNA expression levels, relative to the Mock
group.
|
| Transcripts |
|---|
|
|
|
|---|
| Group (n=3) | Stat3 | Bcl-2 | Bax | Caspase3 | VEGF | Cyclin D1 |
|---|
| Mock | 1 | 1 | 1 | 1 | 1 | 1 |
| HAP | 0.84±0.07 | 0.77±0.06 | 1.03±0.19 | 1.49±0.17 | 0.96±0.01 |
0.41±0.01a |
|
HAP/sh-scramble | 0.58±0.07 |
0.14±0.02a | 1.70±0.24 |
3.16±0.23a | 0.81±0.02 |
0.34±0.01a |
| HAP/sh-Stat3 |
0.25±0.02b |
0.14±0.01 |
4.2±0.38b |
9.05±0.74b |
0.11±0.009b |
0.06±0.005b |
HAP-delivered sh-Stat3 treatment
decreases cell viability in RM1 cells
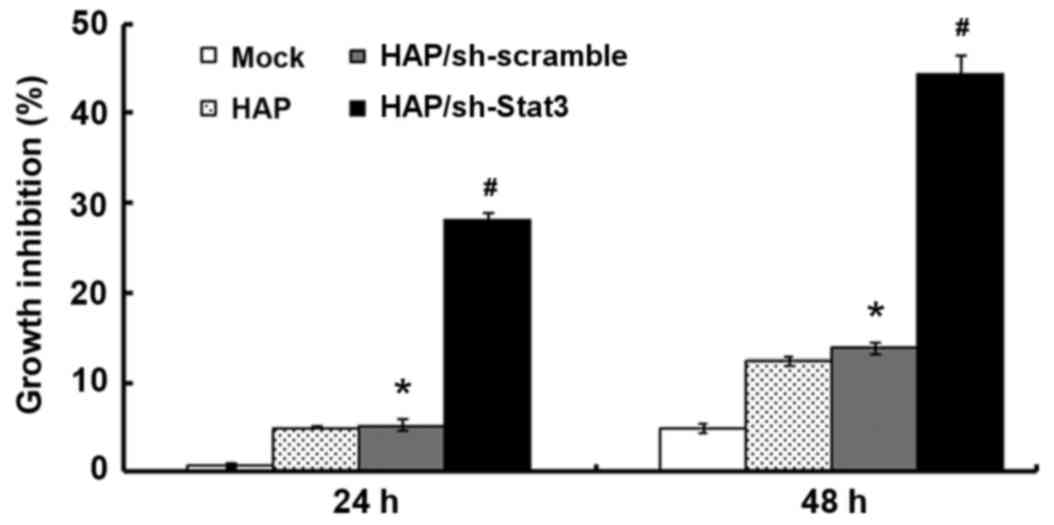
Cell viability in the HAP/sh-Stat3 group was
significantly inhibited compared with the HAP/sh-scramble and Mock
groups at 24 h (P<0.01 and P<0.01, respectively; Fig. 2) and at 48 h (P<0.01 and
P<0.01, respectively; Fig. 2).
However, HAP, HAP/sh-scramble and HAP/sh-Stat3 transfection all
reduced cell viability at 48 h compared with the Mock group; with
growth inhibition rates of 12.41±0.74, 13.98±3.12 and 44.40±1.13%,
respectively (Fig. 2).
HAP-delivered sh-Stat3 treatment
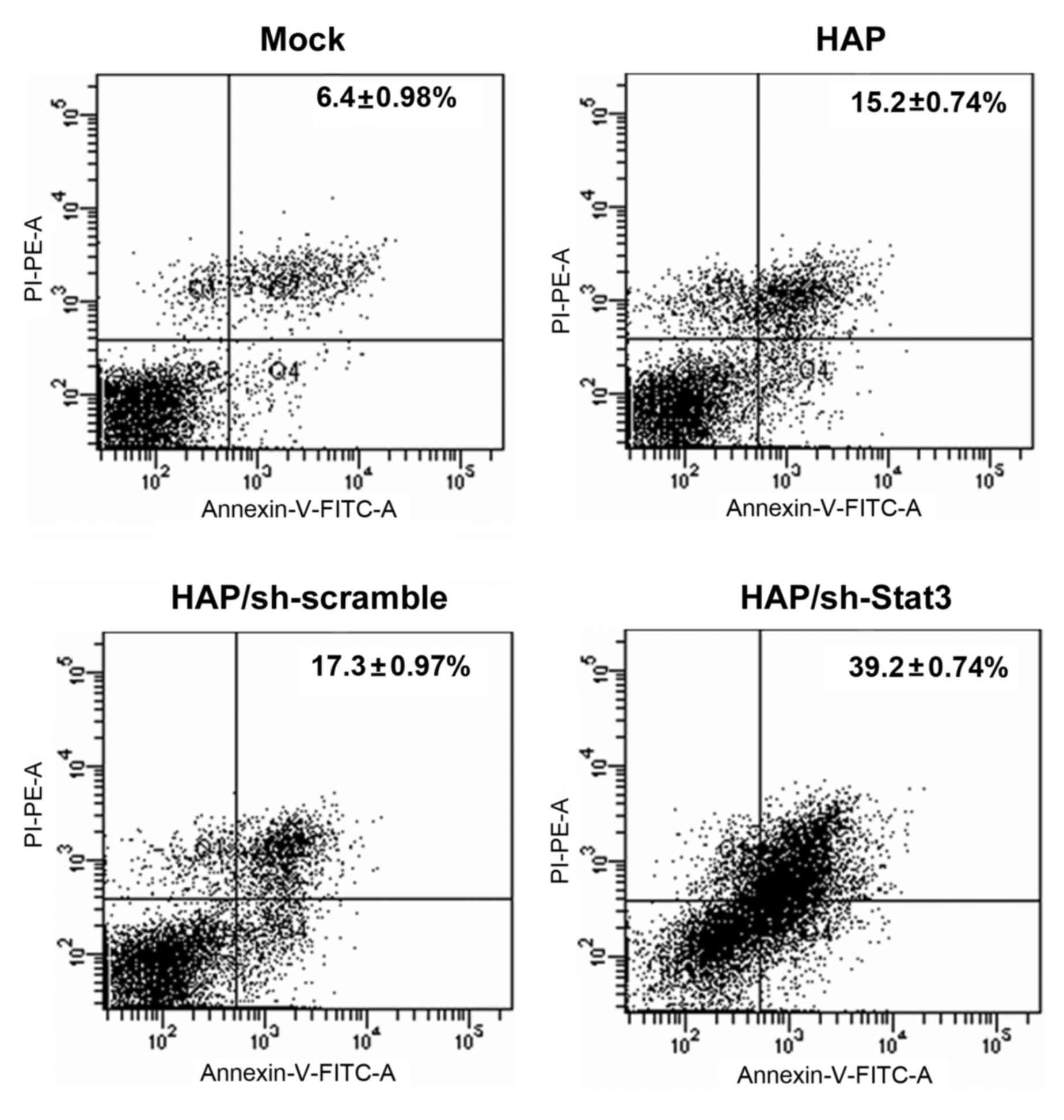
results in G1 arrest and increased apoptosis in RM1 cells
Treatment with HAP-delivered sh-Stat3 induced
significantly increased levels of apoptosis in RM1 cells compared
with HAP-sh-scramble cells, HAP-only cells and Mock control cells
(P<0.01, P<0.01 and P<0.01, respectively; Fig. 3 and Table III). To determine whether this
was associated with specific modifications in the cell cycle, cell
cycle analysis was performed in RM1 cells transfected with sh-Stat3
or sh-scramble. HAP/sh-Stat3-treated cells were significantly
accumulated in the G1 phase compared with Mock treated cells
(P<0.01; Table III),
indicating that sh-Stat3 treatment promotes G1 arrest. Overall,
these findings demonstrate that Stat3 treatment decreases the
viability and survival rate of RM1 prostate cancer cells.
 | Table III.Effect of HAP-delivered sh-Stat3
plasmids on apoptosis and the cell cycle in RM1 cells. |
Table III.
Effect of HAP-delivered sh-Stat3
plasmids on apoptosis and the cell cycle in RM1 cells.
| Group (n=3) | Apoptotic cells
(sum of Q2+Q4 quadrants in Fig. 3)
% | G0-G1, % | S, % |
|---|
| Mock | 6.4±0.98 | 44.5±3.14 | 55.5±3.14 |
| HAP |
15.2±0.74a | 44.4±2.59 | 51.6±3.33 |
|
HAP/sh-scramble |
17.3±0.97a | 46.9±3.95 | 50.1±2.58 |
| HAP/sh-Stat3 |
39.2±0.74b |
66.7±3.99a |
24.8±3.62a |
Expression levels of Stat3-associated
genes
Previous studies have indicated that Stat3 induces
the expression of multiple genes, including the anti-apoptotic
protein Bcl-2 and Cyclin D1, which promotes cell division (22). To determine the effect of sh-Stat3
treatment on the expression of Stat3-associated genes, RT-qPCR and
western blot assays were performed. The results demonstrated that
in HAP/sh-Stat3 treated cells compared with Mock treated cells,
Bcl-2, VEGF and Cyclin D1 mRNA expression levels were significantly
decreased (P<0.01, P<0.01 and P<0.01, respectively;
Table II) and protein expression
levels also significantly decreased compared with HAP/sh-scramble
treated cells (P<0.01, P<0.01 and P<0.01, respectively;
Fig. 4). However, caspase3 and Bax
levels were significantly increased in HAP/sh-Stat3 treated cells
compared with controls (mRNA expression levels, P<0.01 and
P<0.01, respectively; Table
II; protein expression levels, P<0.01 and P<0.01,
respectively; Fig. 4). These
results indicate that treatment with HAP-delivered sh-Stat3
interferes with the transcriptional activity of Stat3 and alters
the expression of its downstream genes.
Discussion
Prostate cancer is the second most frequently
diagnosed cancer and the sixth leading cause of cancer-associated
mortality in men worldwide (23,24).
Despite advances in surgical techniques and radiotherapy, prostate
cancer continues to pose a medical challenge, with gaining an
in-depth understanding of its molecular pathogenesis and the
development of novel therapeutic options being of significant
importance to global health. As prostate cancer represents an
accumulation of genetic mutations in cells and resultant altered
function, a potential novel treatment for this cancer is gene
therapy (25).
Stat3 is a transcription factor encoded by the STAT3
gene in humans. It is a member of the STAT protein family, which
mediates multiple cellular functions including immunity,
proliferation, apoptosis and differentiation. Elevated Stat3
activity has been previously observed in primary tumor tissues and
prostate cancer cell lines, and is correlated with tumorigenesis
(26). Notably, abnormal Stat3
activation is associated with prostate cancer progression (27). Constitutive activation of Stat3
signalling represents one of the key molecular events in the
multistep process of carcinogenesis, and inhibition of Stat3
activation is a potential therapeutic strategy for anti-tumor
therapy (28).
In a previous study, DNA vector-based Stat3-specific
RNAi was demonstrated to decrease Stat3 signalling (13). However, the efficacy of
shRNA-mediated interference relies on efficient delivery of shRNA
oligonucleotides and the major weaknesses that limit most delivery
systems' wide application are the potential for oncogenesis or
mutagenesis, host immune responses, and high cost (29). Nonviral shRNA delivery does not
elicit an immune response, demonstrates improved drug target
validation and permits multiple administrations of shRNA, an
essential attribute for the therapeutic application of shRNA. Sun
et al (9) used HAP as a
novel vector for inner ear gene therapy, and previous results have
demonstrated that Ca2+ modified HAP mediates sh-Stat3
plasmid transfection into mouse prostate cancer cells in
vivo and in vitro, causing significant inhibition of
cancer growth (12). This
supported previous reports which demonstrated HAP transfection
efficiency reaches ~50–80% of liposome-mediated transfection
(30). The present study revealed
that HAP alone exerted antitumor effects: HAP carrying sh-scramble
inhibited cancer cell proliferation compared with control, although
this inhibition was much stronger when HAP was combined with
sh-Stat3. The mechanisms may be related to the intrinsic properties
of nanoparticles, and need further research. This result was
consistent with the study of Zhu et al (30), in which treatment with HAP alone
inhibited hepatocellular carcinoma cell proliferation in
vitro. Therefore, the antitumor effect of HAP may be further
improved while combined with sh-Stat3.
In the present study, the involvement of Stat3 in
promoting prostate cancer cells proliferation in vitro was
confirmed. Stat3 mRNA and protein expression levels were
down-regulated in RM1 cells following sh-Stat3 treatment, implying
that HAP transports sh-Stat3 into cancer cells, resulting in the
inhibition of Stat3 expression levels. These findings demonstrate
that the HAP that carried sh-Stat3 exerts a potent antitumor effect
in vitro through decreasing cell viability and the promotion
of apoptosis. The data displayed in Table II suggest that inhibition of
cancer cell viability was due to a combination of cell cycle arrest
and activation of apoptosis. Furthermore, the expression of
pro-apoptotic factors including caspase3 and Bax were significantly
upregulated in the sh-Stat3 group, while anti-apoptotic factors
such as Cyclin D1 and Bcl-2 were significantly downregulated.
Previous studies have revealed that Stat3 upregulates several
anti-apoptotic proteins' expression, including Bcl-2 and B cell
lymphoma-extra large (Bcl-xL) (31), which are key components of
mitochondrial apoptotic pathways. Therefore, sh-Stat3
transfection-induced apoptosis of cancer cells may partially result
from the activation of mitochondrial apoptosis pathways (32). Additionally, western blot assays
revealed that VEGF expression levels in cancer cells were
significantly decreased following sh-Stat3 transfection. Reduced
VEGF levels resulting from Stat3 knockdown were expected because
VEGF expression has been previously demonstrated to be upregulated
by Stat3 (33).
The present study confirmed that HAP is a useful
vector for plasmid-based shRNA delivery into cancer cells in
vitro, with the proliferation of RM1 cancer cells inhibited by
the HAP-delivered sh-Stat3. These findings suggest the potential of
HAP as an effective gene delivery vehicle for shRNA-based cancer
therapy. However, further modification of HAP is required to
enhance transfection efficacy and expedite clinical applications.
In conclusion, nanoparticle-mediated sh-Stat3 delivery has
potential clinical applications for the treatment of prostate
cancers.
Acknowledgements
The present study was funded by the National Natural
Science Foundation of China (grant nos. 81201188 and 81472344).
References
|
1
|
Do TN, Lee WH, Loo CY, Zavgorodniy AV and
Rohanizadeh R: Hydroxyapatite nanoparticles as vectors for gene
delivery. Ther Deliv. 3:623–632. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bose S and Tarafder S: Calcium phosphate
ceramic systems in growth factor and drug delivery for bone tissue
engineering: A review. Acta Biomater. 8:1401–1421. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Olton D, Li J, Wilson ME, Rogers T, Close
J, Huang L, Kumta PN and Sfeir C: Nanostructured calcium phosphates
(NanoCaPs) for non-viral gene delivery: Influence of the synthesis
parameters on transfection efficiency. Biomaterials. 28:1267–1279.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ginebra MP, Canal C, Espanol M, Pastorino
D and Montufar EB: Calcium phosphate cements as drug delivery
materials. Adv Drug Deliv Rev. 64:1090–1110. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Loo SC, Moore T, Banik B and Alexis F:
Biomedical applications of hydroxyapatite nanoparticles. Curr Pharm
Biotechnol. 11:333–342. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chen L, McCrate JM, Lee JC and Li H: The
role of surface charge on the uptake and biocompatibility of
hydroxyapatite nanoparticles with osteoblast cells. Nanotechnology.
22:1057082011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zakaria SM, Zein Sharif SH, Othman MR,
Yang F and Jansen JA: Nanophase hydroxyapatite as a biomaterial in
advanced hard tissue engineering: A review. Tissue Eng Part B Rev.
19:431–441. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tan K, Cheang P, Ho IA, Lam PY and Hui KM:
Nanosized bioceramic particles could function as efficient gene
delivery vehicles with target specificity for the spleen. Gene
Ther. 14:828–835. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sun H, Jiang M and Zhu SH: In vitro and in
vivo studies on hydroxyapatite nanoparticles as a novel vector for
inner ear gene therapy. Zhonghua Er Bi Yan Hou Tou Jing Wai Ke Za
Zhi. 43:51–57. 2008.(In Chinese). PubMed/NCBI
|
|
10
|
Wu X, Ding D, Jiang H, Xing X, Huang S,
Liu H, Chen Z and Sun H: Transfection using hydroxyapatite
nanoparticles in the inner ear via an intact round window membrane
in chinchilla. J Nanopart Res. 14:7082012. View Article : Google Scholar
|
|
11
|
Yan-Zhong Z, Yan-Yan H, Jun Z, Shai-Hong
Z, Zhi-You L and Ke-Chao Z: Characteristics of functionalized
nano-hydroxyapatite and internalization by human epithelial cell.
Nanoscale Res Lett. 6:6002011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liang ZW, Guo BF, Li Y, Li XJ, Li X, Zhao
LJ, Gao LF, Yu H, Zhao XJ, Zhang L and Yang BX: Plasmid-based Stat3
siRNA delivered by hydroxyapatite nanoparticles suppresses mouse
prostate tumour growth in vivo. Asian J Androl. 13:481–486. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gao L, Zhang L, Hu J, Li F, Shao Y, Zhao
D, Kalvakolanu DV, Kopecko DJ, Zhao X and Xu DQ: Down-regulation of
signal transducer and activator of transcription 3 expression using
vector-based small interfering RNAs suppresses growth of human
prostate tumor in vivo. Clin Cancer Res. 11:6333–6341. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yamada M, Ueno T, Tsukimura N, Ikeda T,
Nakagawa K, Hori N, Suzuki T and Ogawa T: Bone integration
capability of nanopolymorphic crystalline hydroxyapatite coated on
titanium implants. Int J Nanomedicine. 7:859–873. 2012.PubMed/NCBI
|
|
15
|
Fernandez JM, Molinuevo MS, Cortizo MS and
Cortizo AM: Development of an osteoconductive
PCL-PDIPF-hydroxyapatite composite scaffold for bone tissue
engineering. J Tissue Eng Regen Med. 5:e126–e135. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ye F, Guo H, Zhang H and He X: Polymeric
micelle-templated synthesis of hydroxyapatite hollow nanoparticles
for a drug delivery system. Acta Biomater. 6:2212–2218. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu H, Xi P, Xie G, Chen F, Li Z, Bai D
and Zeng Z: Biocompatible hydroxyapatite nanoparticles as a redox
luminescence switch. J Biol Inorg Chem. 16:1135–1140. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Barghi L, Asgari D, Barar J, Nakhlband A
and Valizadeh H: Synthesis, characterization and in vitro
anti-tumoral evaluation of Erlotinib-PCEC nanoparticles. Asian Pac
J Cancer Prev. 15:10281–10287. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xu J, Xu P, Li Z, Huang J and Yang Z:
Oxidative stress and apoptosis induced by hydroxyapatite
nanoparticles in C6 cells. J Biomed Mater Res A. 100:738–745. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang L, Gao L, Li Y, Lin G, Shao Y, Ji K,
Yu H, Hu J, Kalvakolanu DV, Kopecko DJ, et al: Effects of
plasmid-based Stat3-specific short hairpin RNA and GRIM-19 on PC-3M
tumor cell growth. Clin Cancer Res. 14:559–568. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Barton BE, Karras JG, Murphy TF, Barton A
and Huang HF: Signal transducer and activator of transcription 3
(STAT3) activation in prostate cancer: Direct STAT3 inhibition
induces apoptosis in prostate cancer lines. Mol Cancer Ther.
3:11–20. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Da Silveira RA, Hermes CL, Almeida TC,
Bochi GV, De Bona KS, Moretto MB and Moresco RN: Ischemia-modified
albumin and inflammatory biomarkers in patients with prostate
cancer. Clin Lab. 60:1703–1708. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Dumache R, Puiu M, Motoc M, Vernic C and
Dumitrascu V: Prostate cancer molecular detection in plasma samples
by glutathione S-transferase P1 (GSTP1) methylation analysis. Clin
Lab. 60:847–852. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Aneknan P, Kukongviriyapan V, Prawan A,
Kongpetch S, Sripa B and Senggunprai L: Luteolin arrests cell
cycling, induces apoptosis and inhibits the JAK/STAT3 pathway in
human cholangiocarcinoma cells. Asian Pac J Cancer Prev.
15:5071–5076. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Siveen KS, Sikka S, Surana R, Dai X, Zhang
J, Kumar AP, Tan BK, Sethi G and Bishayee A: Targeting the STAT3
signaling pathway in cancer: Role of synthetic and natural
inhibitors. Biochim Biophys Acta. 1845:136–154. 2014.PubMed/NCBI
|
|
28
|
Han Z, Wang X, Ma L, Chen L, Xiao M, Huang
L, Cao Y, Bai J, Ma D, Zhou J and Hong Z: Inhibition of STAT3
signaling targets both tumor-initiating and differentiated cell
populations in prostate cancer. Oncotarget. 5:8416–8428. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kim WJ and Kim SW: Efficient siRNA
delivery with non-viral polymeric vehicles. Pharm Res. 26:657–666.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhu SH, Huang BY, Zhou KC, Huang SP, Liu
F, Li YM, Xue ZG and Long ZG: Hydroxyapatite nanoparticles as a
novel gene carrier. J Nanopart Res. 6:307–311. 2004. View Article : Google Scholar
|
|
31
|
Lim SL, Park SY, Kang S, Park D, Kim SH,
Um JY, Jang HJ, Lee JH, Jeong CH, Jang JH, et al: Morusin induces
cell death through inactivating STAT3 signaling in prostate cancer
cells. Am J Cancer Res. 5:289–299. 2014.PubMed/NCBI
|
|
32
|
Zhou Y, Tian L, Zhang YC, Guo BF and Zhou
QW: Apoptotic effects of psiRNA-STAT3 on 4T1 breast cancer cells in
vitro. Asian Pac J Cancer Prev. 15:6977–6982. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chen Z and Han ZC: STAT3: A critical
transcription activator in angiogenesis. Med Res Rev. 28:185–200.
2008. View Article : Google Scholar : PubMed/NCBI
|