Introduction
Early embryo growth arrest is a unique form of
spontaneous abortion, which is characterized by absent fetal
development or fetal death in the first-trimester of pregnancy
(1,2). With the environmental pollution and
increasing life stress in populations, a previous epidemiological
study demonstrated that early embryo growth arrest occurs in >4%
of pregnant women in China and the incidence is increasing rapidly
(3). Although a number of factors,
such as genetic abnormality, immune system deficits and viral
infection have been indicated in the pathogenesis of early growth
arrest, the underlying cellular and molecular mechanisms remain
poorly understood. Furthermore, although ~50% of incidence of early
embryo growth arrest can be ascribed to chromosome abnormality, the
etiology of the other clinical cases remains ambiguous (3,4).
The placenta functions as the exchange organ between
the fetal and maternal bodies, and it serves a major role in
providing nutrients and carrying over metabolic substrates via
trans-placental exchange. Fetal development and pregnancy
maintenance are dependent on normal placental growth (5). In mammals, epigenetic modification is
commonly overlaid during embryogenesis and early development of the
fetus. DNA methylation is the most important epigenetic regulation
process in genomic imprinting, transposon silencing, X-inactivation
and gene repression (6–8). DNA methylation is regulated by DNA
methyltransferases (DNMTs), including DNMT1, DNMT3A, DNMT3B and
DNMT3L. The dynamics of DNA methylation is vital for embryonic and
placental development in physiological and pathological conditions
(9–12).
The three main DNMTs (DNMT1, DNMT3A and DNMT3B) are
responsible for two special methylation processes that are required
for tissue-specific methylation patterns. DNMT1 mediates
maintenance DNA methylation, while DNMT3A and DNMT3B are
responsible for de novo methylation (7,13).
DNMT3L is highly homologous to DNMT3A and
DNMT3B in sequence, but has no catalytic activity in
vitro. Previous studies have revealed that mutation or deletion
of DNMT1, DNMT3A and DNMT3B triggered cell
death in human embryonic stem cells (12,14).
At present however, little is known about the patterns of DNA
methylation or DNMTs expression in human placenta during early
pregnancy. Some previous studies have reported low expression of
5-methyl-cytosine and relative hypo-methylation in the
developmental placenta of different pregnancy stages (15). Previous results based on gene
knockout experiments indicated that all the four DNMTs are involved
in human trans-acting imprinting defects. For instance,
DNMT3B gene mutations of specific residues in the C-terminal
catalytic domain are known to cause immunodeficiency-centromeric
instability-facial anomalies syndrome (16,17).
Aberrant expression of DNMTs may be responsible for the high rate
of abortion and abnormal embryo growth. In addition, DNMT3a
or DNMT3b knockout mice exhibited embryonic development
arrest because of the failure to initiate de novo
methylation following implantation (18,19).
In addition, wide demethylation and developmental arrest was
reported in DNMT1 knockout mice embryos at the early stage
of gestation (20).
To test the involvement of abnormal DNA
methyltransferases in early embryo growth arrest, the authors
measured the level of DNMTs expression in chorionic villi obtained
from pregnant women diagnosed with early embryo growth arrest and
control patients. A significant downregulation of the DNMT3A
protein was observed in these embryo growth arrest cases; however,
their DNMT3a mRNA expression levels were comparable to the
controls. The discordance of the mRNA and protein expression of
DNMT3A indicated that the translation process of DNMT3A was
specifically interfered during early embryo growth arrest.
Materials and methods
Clinical data and tissue samples
Chorionic villous samples were obtained from 80
pregnant women (gestational age ranging from 6 to 9 weeks) who
underwent termination of pregnancy at the Department of Obstetrics
in the Qingdao Municipal Hospital between January 2013 and June
2014. A total of 40 of the 80 cases were diagnosed with embryo
growth arrest by B-mode ultrasound; the other 40 cases were having
a normal pregnancy, but patients had voluntarily requested for the
termination of pregnancy. Patient age was between 22 and 34 years
old, and other serious medical conditions were ruled out, such as
chromosome abnormities, endocrine diseases, infection,
immunological diseases and other serious maternal complications.
Each sample (~20 g) was immediately frozen in liquid nitrogen for
30 min, and subsequently stored at −80°C for reverse
transcription-quantitative polymerase chain reaction (RT-qPCR) and
western blot analysis. The remainder of each sample was fixed with
10% buffered formalin and was paraffin-embedded for
immunohistochemistry. Written informed consent for the publication
of any associated data and accompanying images was obtained from
all patients involved. The current study was approved by the
Ethical Review Committee of Qingdao University (Qingdao, Shandong,
China).
RNA extraction, cDNA synthesis and
RT-qPCR
Total RNA was extracted from the chorionic villi
with a PureLink™ RNA Mini kit (Thermo Fisher Scientific,
Inc., Waltham, MA, USA), according to the manufacturer's
instructions. RNA quantity and quality was determined by a NanoDrop
2000 spectrophotometer (Thermo Fisher Scientific, Inc., Wilmington,
DE, USA). cDNA was then synthesized from 1 µg total RNA with
SuperScript™ III Reverse transcriptase (Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions. The
mRNA expressions of DNMTs and β-actin were measured by the Master
Cycler ep realplex PCR system (Eppendorf, Hamburg, Germany) using a
QuantiFast SYBR-Green PCR kit (Qiagen GmbH, Hilden, Germany)
according to the manufacturer's protocols. The PCR cycling
parameters were set as follows: 95°C for 5 min, followed by 40
cycles of PCR reaction at 95°C for 5 sec, and finally 60°C for 30
sec. β-actin was used as an internal control. All reactions were
run in triplicate. The threshold cycle is defined as the fractional
cycle number at which the fluorescence passes the fixed threshold.
The data were obtained by normalizing DNMT1, DNMT3a,
DNMT3b and DNMT3L genes threshold values with
corresponding β-actin threshold value, and then analyzed
with 2−ΔΔCq method (21). The primer sequences are presented
in Table I.
 | Table I.Sequences of specific gene primers
used for reverse transcription-quantitative polymerase chain
reaction. |
Table I.
Sequences of specific gene primers
used for reverse transcription-quantitative polymerase chain
reaction.
| Gene | Forward sequence
(5′-3′) | Reverse sequence
(5′-3′) |
|---|
| β-actin |
CACTCTTCCAGCCTTCCTTC |
GTACAGGTCTTTGCGGATGT |
| DNMT1 |
GACCCGTCTCTTGAAGGTGG |
CTTCTCCTGCATCAGCCCAA |
| DNMT3A |
GGTCACGCAAAACAGAACCC |
CCTTGGTGAAACCCTTTGCG |
| DNMT3B |
CTCTTCCTCAGCTGTGTGGG |
CTGTCGGCACTGTGGTTTTG |
| DNMT3L |
GATTCTTTGCCCCCATAGCCT |
TTCAAGGTTCCAGTGGTCCG |
Immunohistochemical staining and
analysis
The biopsied tissues were initially fixed in 10%
buffered formalin, embedded in paraffin and cut into 4 µm sections.
Tissue sections were then de-paraffinized and dehydrated in graded
ethanol and dimethylbenzene, respectively. Antigen retrieval was
performed by boiling sections in EDTA buffer (pH 8.0) for 2.5 min
in a pressure cooker. The endogenous peroxidase activity was
blocked using 0.3% hydrogen peroxide. After rinsing in
phosphate-buffered saline (PBS), the sections were incubated for 1
h with polyclonal rabbit anti-DNMT1 antibody (1:200, NB100-264,
Novus Biologicals, LLC, Littleton, CO, USA), polyclonal rabbit
anti-DNMT3A antibody (1:400, NB100-265, Novus Biologicals, LLC),
polyclonal rabbit anti-DNMT3B antibody (1:800, NB100-266, Novus
Biologicals, LLC) or polyclonal rabbit anti-DNMT3L antibody (1:100,
ab115522, Abcam, Cambridge, UK). After rinse in PBS for several
times, the sections were incubated with a biotinylated goat
anti-rabbit secondary antibody (UltraSensitive™ SP kit,
Fuzhou Maixin Biotech Co., Ltd., Fuzhou, China) at 37°C for 30 min.
Sections were then stained using 3, 3-diaminobenzidine chromogen
solution (Zhongshan Golden Bridge Biotechnology Co., Ltd., Beijing,
China) and counterstained with hematoxylin (Fuzhou Maixin Biotech
Co., Ltd). All slides were observed separately by two
pathologists.
Western blotting detection of
DNMTs
The frozen tissue was re-suspended in passive lysis
buffer (Promega Corporation, Madison, WI, USA) including freshly
added protease inhibitors and phosphatase inhibitors cocktails
(Sigma-Aldrich, Darmstadt, Germany). Protein concentrations in the
supernatant fractions were determined using a bicinchoninic acid
assay kit (Thermo Fisher Scientific, Inc.). Protein lysates were
separated with 8 or 12% SDS-acrylamide gels, and transferred onto
nitrocellulose membranes. After blocking for 60 min in a 5% skim
milk solution, membranes were incubated overnight at 4°C with
monoclonal rabbit anti-DNMT1 antibody at 1:1,000 (D59A4, Cell
Signaling Technology, Inc., Danvers, MA, USA), monoclonal rabbit
anti-DNMT3A at 1:1,000 (D23G1, Cell Signaling Technology, Inc.) and
polyclonal rabbit anti-DNMT3B at 1:1,000 (NB100-266, Novus
Biologicals, LLC), polyclonal rabbit anti-DNMT3L at 1:1,000
(ab115522, Abcam) or monoclonal mouse anti-β-actin at 1:5,000
(A5441, Sigma-Aldrich). Signals from
horseradish-peroxidase-conjugated secondary antibodies were
visualized by enhanced chemilluminescence solution (GE Healthcare
Life Sciences, Chalfont, UK) and processed with an UVP visualizer
(UVP, Inc., Upland, CA, USA). Quantification was performed using
VisionWorks LS version 7.0 from the UVP visualizer itself.
Experiments were repeated in triplicate.
Statistical analysis
The statistical analysis was performed using SPSS
version 17.0 software (SPSS Inc., Chicago, IL, USA). Student's t
test was used to compare the quantitative data between the two
groups and a one-way analysis of variance (ANOVA) followed by
Bonferroni post hoc test was used for multiple comparisons. The
results are presented as the mean ± standard error of the mean and
a two-sided P<0.05 was considered to indicate a statistically
significant difference.
Results
DNMT mRNA expression in chorionic
villi of early embryo growth arrest patients
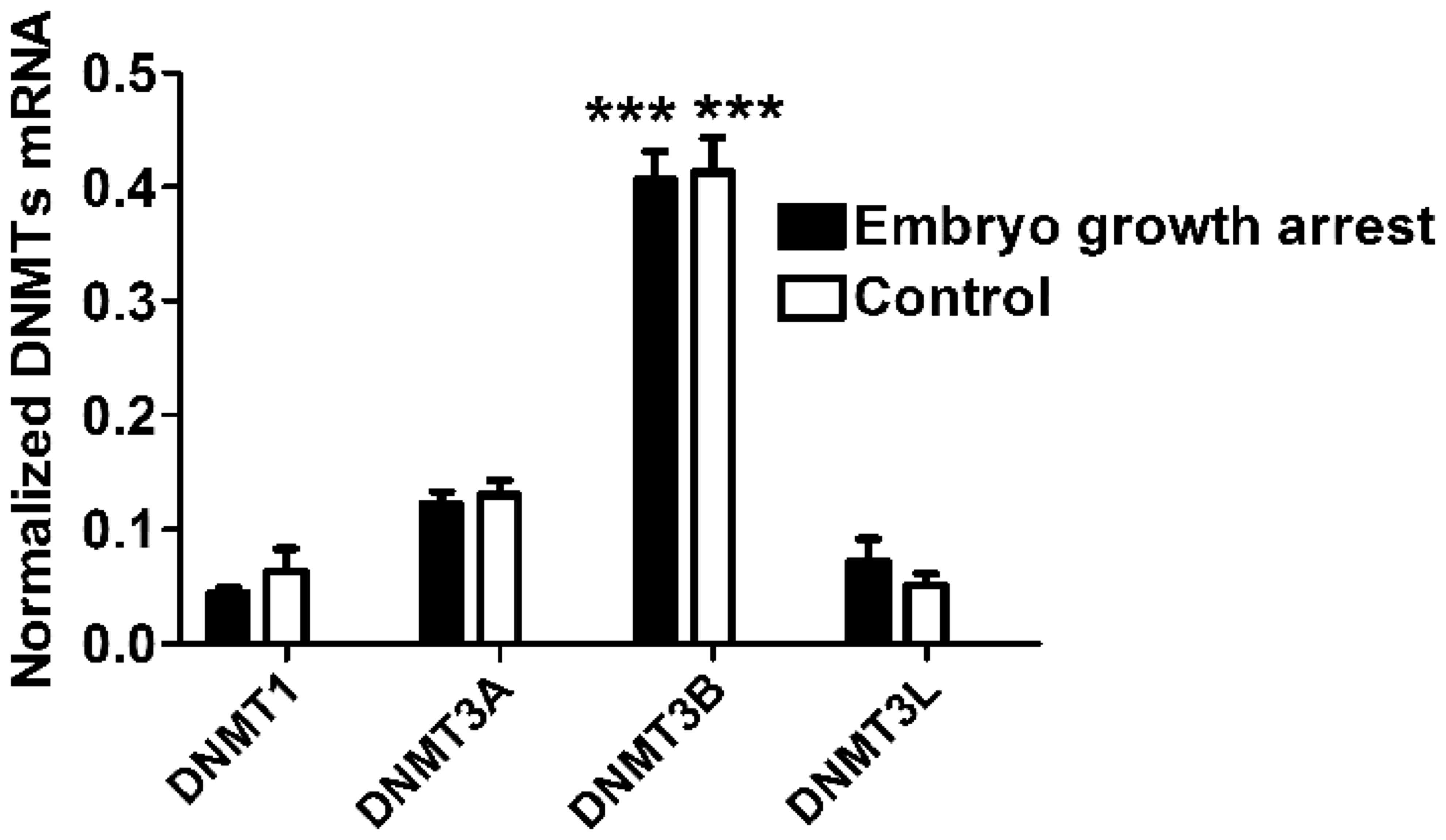
First, the authors measured the level of mRNA
expression of DNMT1, DNMT3A, DNMT3B and DNMT3L in chorionic villi
obtained from the placental tissue of both groups. DNMT3B presented
the highest level of expression compared to other three DNMTs in
both groups (One-way ANOVA, n=40 for each group, P<0.0001,
Fig. 1), indicating that DNMT3B is
the major type of DNA methyltransferase expressed in chorionic
villi. In addition, it was identified that the mRNA expression of
four DNMTs (DNMT1, DNMT3A, DNMT3B and DNMT3L) in chorionic villi of
early embryo growth arrest patients were comparable to that in the
controls, respectively (unpaired Student's t-test, n=40 for each
group, P>0.05, Fig. 1).
DNMT protein expression in chorionic
villi of early embryo growth arrest patients by IHC
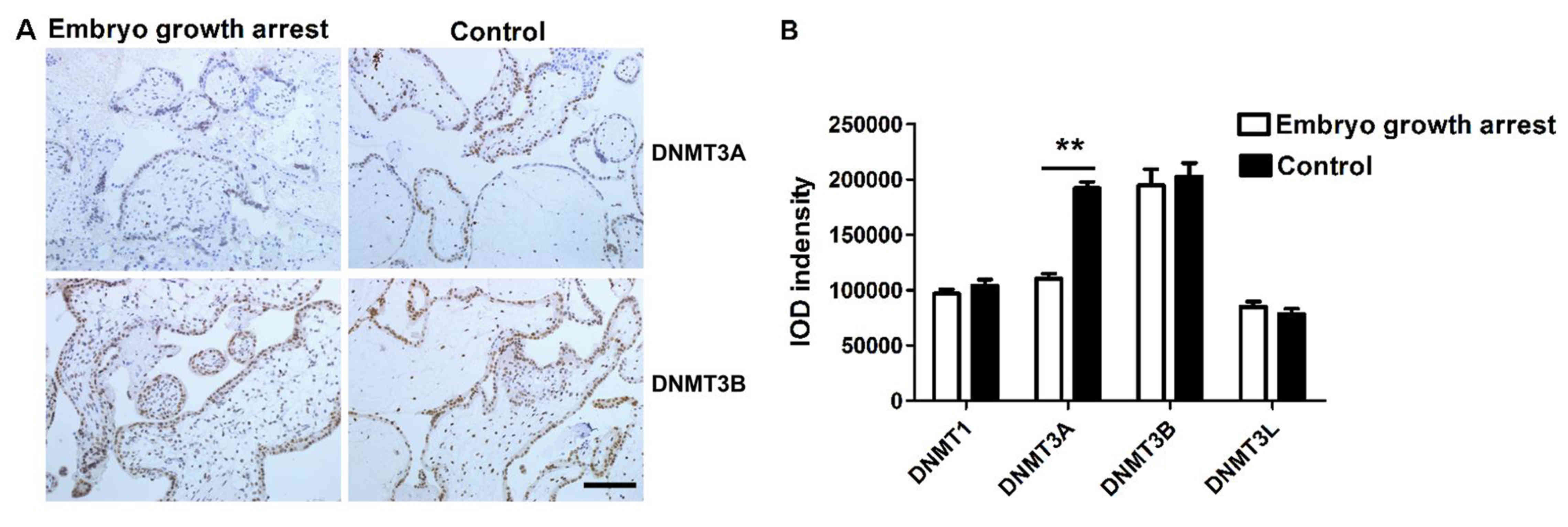
Next, IHC studies were conducted to determine
whether the protein expression of DNMTs was altered in patients
with early embryo growth arrest. IHC staining demonstrated that all
four types of DNMTs were expressed in trophoblast cells and
chorionic villi. DNMT3A and DNMT3B positive staining was localized
in the cell nucleus (Fig. 2A)
while DNMT1and DNMT3L were in the cytoplasm (data not shown).
Immunostaining intensity was calculated by Image Pro plus 6.0
analysis software (Media Cybernetics Inc., Rockville, MD, USA) and
represented with semi-quantitative integrated optical density
(IOD). The total IOD value of each sample was calculated by summing
up the staining intensity from six randomly selected visions per
section and across six representative sections. The current results
revealed that the expression of DNMT3A protein in chorionic villi
of early embryo growth arrest patients was significantly lower than
that in chorionic villi of the controls (unpaired Student's t-test,
n=40 for each group, P<0.01, Fig.
2B). The protein levels of DNMT1, DNMT3B and DNMT3L were
similar between two groups (unpaired Student's t test, n=40 per
group, P>0.05, Fig. 2B).
DNMTs protein expression in chorionic
villi of early embryo growth arrest patients by western
blotting
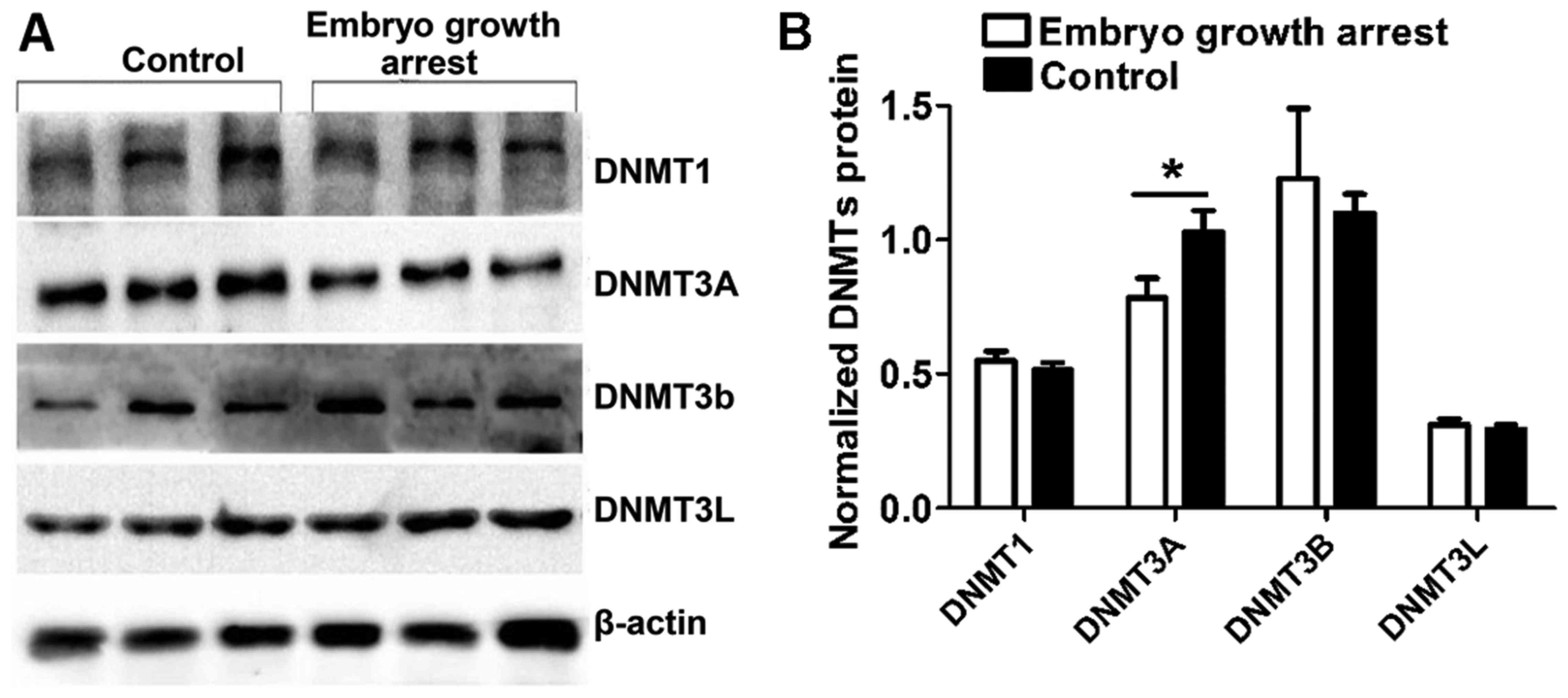
To confirm the above results, western blot analysis
was conducted to compare the DNMT expression in chorionic villi
from patients with early embryo growth arrest with healthy
controls. Data were analyzed with the UVP gel imaging system.
Consistently, it was identified that, in comparison to the
controls, DNMT3A protein expression was specifically decreased in
chorionic villi obtained from embryo growth arrest patients
(unpaired Student's t -test, n=40 per group, P<0.05, Fig. 3B). No significant difference was
presented between groups as for DNMT1, DNMT3B and DNMT3L expression
(unpaired Student's t-test, n=40 per group, P>0.05, Fig. 3).
Discussion
In recent years, serious pregnancy complications,
such as early embryo growth arrest, recurrent spontaneous
miscarriage, intrauterine growth restriction and preeclampsia, have
globally been on the increase despite improved medical environment
(1,22). These complications are the leading
causes of mortality and they put huge emotional and financial
burdens on the caregivers and patients themselves. Early embryo
growth arrest is a stop in intrauterine embryonic development in
the early period of pregnancy (1).
Distinct from spontaneous abortion, the majority of the pregnant
women with early embryo growth arrest present no obvious symptoms,
such as bleeding or abdominal pain (1,2).
Early diagnosis of embryo growth arrest primarily depends on B-mode
and Doppler ultrasound examination (23). Although previous etiology studies
have focused on chromosome abnormality (1,24–26),
sperm deformity, immune factors, environment factors and hormonal
regulation, the molecular mechanisms of embryo growth arrest remain
uncertain.
DNA methylation is crucial during the entire
embryonic development. Specifically, it is reported to be important
in various cellular processes such as cell differentiation, gene
transcription and genomic imprinting (10,11).
DNA methylation is established in epiblast cells within embryonic
days 4.5 and 6.5 (12). Previous
studies indicated that genomic imprinting, regulated by DNA
methylation, may serve a critical role in placenta growth (27,28).
Methylation levels of human placenta increase in a gestational
stage-dependent manner (26). In
addition, recent studies demonstrated the association between
changes of placental DNA methylation patterns and pregnancy
disorders (29). Furthermore,
dynamic regulation of DNA methylation by DNMTs exists in placenta
(11,12). Therefore, understanding the role
and underlying mechanisms of DNA methylation in placental
development may offer a new strategy to prevent serious pregnancy
complications.
In the present study, the authors investigated the
expression patterns of four DNMTs (DNMT1, DNMT3A, DNMT3B and
DNMT3L) in chorionic villi of pregnant women and demonstrated that
all four DNMTs were expressed in placental villi and trophoblast
cells. The expression levels of DNMT3B mRNA and protein were the
highest both in patients with early embryo growth arrest and in
individuals with normal pregnancy, suggesting that DNMT3B may be
one of the prominent DNA methyltransferases involved in placenta
development during early pregnancy. More importantly, specific
downregulation of the DNMT3A protein was observed with both
immunostaining and western blot analysis in the chorionic villi of
pregnant women with embryo growth arrest, while none of other three
DNMTs (DNMT1, DNMT3B or DNMT3L) was changed at both mRNA and
protein levels. To the best of the authors' knowledge, the
maintenance DNA methylation is a crucial and indispensable element
in embryo implantation and development process (10,11).
Previous studies suggested that both DNMT3A and DNMT3B are
essential for de novo methylation in the embryonic stem
cells and early embryos (30). In
a mouse model, the DNMT3B protein is more enriched in the embryonic
stem cells than DNMT3A (9,19). However, the DNMT3A protein begins
to be expressed again in the middle embryonic development
(E10.5-E14.5), while DNMT3B is not (9). These studies support the theory that
epigenetic alterations in early embryos may be carried forward to
subsequent developmental stages (31). Moreover, DNMT3A
transcription is presumably the vital point for embryonic
development process. Therefore, the finding that an abnormal
decrease of chorionic villous DNMT3A protein in the placenta of
early embryo growth arrest patients may be important for the
pathogenesis of early embryo growth arrest. Decreased protein
expression accompanied by normal mRNA expression of DNMT3A also
indicated that DNMT3A protein translation or post-translation
processing was specifically interfered in early embryo growth
arrest.
The placenta serves an important role in managing
intrauterine embryonic growth and development through nutrients and
waste transfer (32). Previous
studies have indicated that placental epigenetic profiles may
affect fetal growth through the interaction between intra-uterine
and extra-uterine environments (33–35).
Administration of DNA methyltransferase inhibitors to pregnant rats
resulted in smaller placentas with severe histological damage at
different gestational ages (36).
Another previous study reported that DNMT3B mRNA expression
decreases following the increase in DNMT3A and DNMT1 expression in
early pregnancy placenta (37).
Other studies indicated that the DNMT1 protein level was
significantly decreased and the global DNA methylation level was
significantly downregulated in chorionic villi of women with early
pregnancy loss (27,29). Although different subtypes of DNMTs
were reported to be downregulated in the present study compared to
the previous reports, the downregulation trends of DNMTs expression
was the same. Our findings indicated that the downregulation of
DNMT3A expression in chorionic villi is specific in early embryo
growth arrest. Insufficient maintenance methylation due to the
lower level of DNMT3A protein expression may be associated with
abnormal embryonic development in human early pregnancy loss.
In summary, DNA methylation has a critical role in
placenta development, and changes in methylation pattern can lead
to adverse birth outcome. Placental epigenome may serve as a
modulator of disease pathogenesis. The current findings confirmed
that downregulation of DNMT3A protein expression and the ensuing
disturbance of the maintenance DNA methylation may serve an
important role in the pathogenesis of early embryo growth arrest.
Further studies are required to disclose the specific target genes
regulated by the changes of DNA methylation in placental
pathogenesis. In addition, studies are required to elucidate the
distinct nature of DNA methylation in the maternal-fetal
interaction during pregnancy.
Acknowledgements
The current work was supported by the Municipal
Science and Technology Foundation of Qingdao (grant no.
2012-WSZD009).
Glossary
Abbreviations
Abbreviations:
|
DNMT
|
DNA methyltransferase
|
|
IUGR
|
intrauterine growth restriction
|
|
ICF syndrome
|
immunodeficiency-centromeric
instability-facial anomalies syndrome
|
References
|
1
|
Yan J, Fan L, Zhao Y, You L, Wang L, Zhao
H, Li Y and Chen ZJ: DYZ1 copy number variation, Y chromosome
polymorphism and early recurrent spontaneous abortion/early embryo
growth arrest. Eur J Obstet Gynecol Reprod Biol. 159:371–374. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Luan L, Zhu X and Ma Y: Study on
expression of TGF-β1, MMP-9 and TIMP-1 in the chorionic villi of
early embryo growth arrest. Pro Obstetrics Gynecol. 2012.
|
|
3
|
Xiao FZ and Hu ML: The prevent and reason
analysis of the patients of embryo damage. China Practical
Medicine. 8:27–29. 2013.(In Chinese).
|
|
4
|
Wang Y, Han YF, Wang ZY and Wen-Yuan LI:
Clinical study on intervention measures of re-pregnancy in people
with embryo damage history. Guide of China Medicine. 19:56–57.
2014.(In Chinese).
|
|
5
|
Bauer MK, Harding JE, Bassett NS, Breier
BH, Oliver MH, Gallaher BH, Evans PC, Woodall SM and Gluckman PD:
Fetal growth and placental function. Mol Cell Endocrinol.
140:115–120. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Beck S and Rakyan VK: The methylome:
Approaches for global DNA methylation profiling. Trends Genet.
24:231–237. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Oda M, Yamagiwa A, Yamamoto S, Nakayama T,
Tsumura A, Sasaki H, Nakao K, Li E and Okano M: DNA methylation
regulates long-range gene silencing of an X-linked homeobox gene
cluster in a lineage-specific manner. Genes Dev. 20:3382–3394.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
MacDonald WA and Mann MR: Epigenetic
regulation of genomic imprinting from germ line to preimplantation.
Mol Reprod Dev. 81:126–140. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Watanabe D, Suetake I, Tada T and Tajima
S: Stage- and cell-specific expression of DNMT3a and DNMT3b during
embryogenesis. Mech Dev. 118:187–190. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Koukoura O, Sifakis S and Spandidos DA:
DNA methylation in the human placenta and fetal growth (Review).
Mol Med Rep. 5:883–889. 2012.PubMed/NCBI
|
|
11
|
O'Doherty AM, Magee DA, O'Shea LC, Forde
N, Beltman ME, Mamo S and Fair T: DNA methylation dynamics at
imprinted genes during bovine pre-implantation embryo development.
BMC Dev Biol. 15:132015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Uysal F, Akkoyunlu G and Ozturk S: Dynamic
expression of DNA methyltransferases (DNMTs) in oocytes and early
embryos. Biochimie. 116:103–113. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen T and Li E: Structure and function of
eukaryotic DNA methyltransferases. Curr Top Dev Biol. 60:55–89.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yoder MC, Hiatt K, Dutt P, Mukherjee P,
Bodine DM and Orlic D: Characterization of definitive
lymphohematopoietic stem cells in the day 9 murine yolk sac.
Immunity. 7:335–344. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Katari S, Turan N, Bibikova M, Erinle O,
Chalian R, Foster M, Gaughan JP, Coutifaris C and Sapienza C: DNA
methylation and gene expression differences in children conceived
in vitro or in vivo. Hum Mol Genet. 18:3769–3778. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Huang K, Wu Z, Liu Z, Hu G, Yu J, Chang
KH, Kim KP, Le T, Faull KF, Rao N, et al: Selective demethylation
and altered gene expression are associated with ICF syndrome in
human-induced pluripotent stem cells and mesenchymal stem cells.
Hum Mol Genet. 23:6448–6457. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ehrlich M: The ICF syndrome, a DNA
methyltransferase 3B deficiency and immunodeficiency disease. Clin
Immunol. 109:17–28. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Arand J, Wossidlo M, Lepikhov K, Peat JR,
Reik W and Walter J: Selective impairment of methylation
maintenance is the major cause of DNA methylation reprogramming in
the early embryo. Epigenetics Chromatin. 8:12015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hirasawa R and Sasaki H: Dynamic
transition of Dnmt3b expression in mouse pre- and early
post-implantation embryos. Gene Expr Patterns. 9:27–30. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hirasawa R, Chiba H, Kaneda M, Tajima S,
Li E, Jaenisch R and Sasaki H: Maternal and zygotic Dnmt1 are
necessary and sufficient for the maintenance of DNA methylation
imprints during preimplantation development. Genes Dev.
22:1607–1616. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Spradley FT: Metabolic abnormalities and
obesity's impact on the risk for developing preeclampsia. Am J
Physiol Regul Integr Comp Physiol. 312:R5–R12. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hou SX and Dept U: Application of
ultrasound in the diagnosis of intrauterine embryo growth arrest.
World Latest Medicine Information. 15:29–30. 2015.(In Chinese).
|
|
24
|
Sharma S: Natural killer cells and
regulatory T cells in early pregnancy loss. Int J Dev Biol.
58:219–229. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zobeiri F, Sadrkhanlou RA, Salami S,
Mardani K and Ahmadi A: The effect of ciprofloxacin on sperm DNA
damage, fertility potential and early embryonic development in NMRI
mice. Vet Res Forum. 3:131–135. 2012.PubMed/NCBI
|
|
26
|
Fuke C, Shimabukuro M, Petronis A,
Sugimoto J, Oda T, Miura K, Miyazaki T, Ogura C, Okazaki Y and
Jinno Y: Age related changes in 5-methylcytosine content in human
peripheral leukocytes and placentas: An HPLC-based study. Ann Hum
Genet. 68:196–204. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Maccani MA and Marsit CJ: Epigenetics in
the placenta. Am J Reprod Immunol. 62:78–89. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hata K, Kusumi M, Yokomine T, Li E and
Sasaki H: Meiotic and epigenetic aberrations in Dnmt3L-deficient
male germ cells. Mol Reprod Dev. 73:116–122. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yin LJ, Zhang Y, Lv PP, He WH, Wu YT, Liu
AX, Ding GL, Dong MY, Qu F, Xu CM, et al: Insufficient maintenance
DNA methylation is associated with abnormal embryonic development.
BMC Med. 10:262012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lucifero D, La Salle S, Bourc'his D,
Martel J, Bestor TH and Trasler JM: Coordinate regulation of DNA
methyltransferase expression during oogenesis. BMC Dev Biol.
7:362007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Waterland RA and Jirtle RL: Early
nutrition, epigenetic changes at transposons and imprinted genes,
and enhanced susceptibility to adult chronic diseases. Nutrition.
20:63–68. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Robins JC, Marsit CJ, Padbury JF and
Sharma SS: Endocrine disruptors, environmental oxygen, epigenetics
and pregnancy. Front Biosci (Elite Ed). 3:690–700. 2011.PubMed/NCBI
|
|
33
|
Sood R, Zehnder JL, Druzin ML and Brown
PO: Gene expression patterns in human placenta. Proc Natl Acad Sci
USA. 103:5478–5483. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Filiberto AC, Maccani MA, Koestler D,
Wilhelm-Benartzi C, Avissar-Whiting M, Banister CE, Gagne LA and
Marsit CJ: Birthweight is associated with DNA promoter methylation
of the glucocorticoid receptor in human placenta. Epigenetics.
6:566–572. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Nelissen EC, van Montfoort AP, Dumoulin JC
and Evers JL: Epigenetics and the placenta. Hum Reprod Update.
17:397–417. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Vlahović M, Bulić-Jakus F, Jurić-Lekić G,
Fucić A, Marić S and Serman D: Changes in the placenta and in the
rat embryo caused by the demethylating agent 5-azacytidine. Int J
Dev Biol. 43:843–846. 1999.PubMed/NCBI
|
|
37
|
Grazul-Bilska AT, Johnson ML, Borowicz PP,
Minten M, Bilski JJ, Wroblewski R, Velimirovich M, Coupe LR, Redmer
DA and Reynolds LP: Placental development during early pregnancy in
sheep: Cell proliferation, global methylation, and angiogenesis in
the fetal placenta. Reproduction. 141:529–540. 2011. View Article : Google Scholar : PubMed/NCBI
|