|
1
|
Oliveira C, Pinheiro H, Figueiredo J,
Seruca R and Carneiro F: Familial gastric cancer: Genetic
susceptibility, pathology, and implications for management. Lancet
Oncol. 16:e60–e70. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kang W, Tong JH, Lung RW, Dong Y, Zhao J,
Liang Q, Zhang L, Pan Y, Yang W, Pang JC, et al: Targeting of yap1
by microrna-15a and microrna-16-1 exerts tumor suppressor function
in gastric adenocarcinoma. Mol Cancer. 14:522015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: Globocan 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lin SJ, Gagnon-Bartsch JA, Tan IB, Earle
S, Ruff L, Pettinger K, Ylstra B, van Grieken N, Rha SY, Chung HC,
et al: Signatures of tumour immunity distinguish Asian and
non-Asian gastric adenocarcinomas. Gut. 64:1721–1731. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Stewart B and Wild CP: World cancer report
2014. World Health Organization; 2015
|
|
6
|
Cao W, Tian W, Hong J, Li D, Tavares R,
Noble L, Moss SF and Resnick MB: Expression of bile acid receptor
TGR5 in gastric adenocarcinoma. Am J Physiol Gastrointest Liver
Physiol. 304:G322–G327. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Takahashi M, Nakajima M, Ogata H, Domeki
Y, Ohtsuka K, Ihara K, Kurayama E, Yamaguchi S, Sasaki K, Miyachi K
and Kato H: Cd24 expression is associated with progression of
gastric cancer. Hepatogastroenterology. 60:653–658. 2013.PubMed/NCBI
|
|
8
|
Jiang Y, He Y, Li H, Li HN, Zhang L, Hu W,
Sun YM, Chen FL and Jin XM: Expressions of putative cancer stem
cell markers ABCB1, ABCG2, and CD133 are correlated with the degree
of differentiation of gastric cancer. Gastric cancer. 15:440–450.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yk W, Cf G, T Y, Z C, Xw Z, Xx L, Nl M and
Wz Z: Assessment of ERBB2 and EGFR gene amplification and protein
expression in gastric carcinoma by immunohistochemistry and
fluorescence in situ hybridization. Mol Cytogenet. 4:142011.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hayashi M, Inokuchi M, Takagi Y, Yamada H,
Kojima K, Kumagai J, Kawano T and Sugihara K: High expression of
HER3 is associated with a decreased survival in gastric cancer.
Clin Cancer Res. 14:7843–7849. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Suh JH, Won KY, Kim GY, Bae GE, Lim SJ,
Sung JY, Park YK, Kim YW and Lee J: Expression of tumoral FOXP3 in
gastric adenocarcinoma is associated with favorable
clinicopathological variables and related with hippo pathway. Int J
Clin Exp Pathol. 8:14608–14618. 2015.PubMed/NCBI
|
|
12
|
Zhao J, Bai Z, Feng F, Song E, Du F, Zhao
J, Shen G, Ji F, Li G and Ma X: Cross-talk between EPAS-1/HIF-2α
and PXR signaling pathway regulates multi-drug resistance of
stomach cancer cell. Int J Biochem Cell Biol. 72:73–88. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kitagawa M, Lee SH and McCormick F: Skp2
suppresses p53-dependent apoptosis by inhibiting p300. Mol Cell.
29:217–231. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang Z, Fukushima H, Inuzuka H, Wan L, Liu
P, Gao D, Sarkar FH and Wei W: Skp2 is a promising therapeutic
target in breast cancer. Front Oncol. 1:pii: 187022012. View Article : Google Scholar
|
|
15
|
Wei Z, Jiang X, Qiao H, Zhai B, Zhang L,
Zhang Q, Wu Y, Jiang H and Sun X: STAT3 interacts with skp2/p27/p21
pathway to regulate the motility and invasion of gastric cancer
cells. Cell Signal. 25:931–938. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cheadle C, Vawter MP, Freed WJ and Becker
KG: Analysis of microarray data using z score transformation. J Mol
Diagn. 5:73–81. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Smyth GK: Limma: Linear models for
microarray dataBioinformatics and Computational Biology Solutions
using R and Bioconductor. Gentleman. Carey V..Dudoit S..Irizarry
R..Huber W.: Springer; New York: pp. 397–420. 2005, View Article : Google Scholar
|
|
18
|
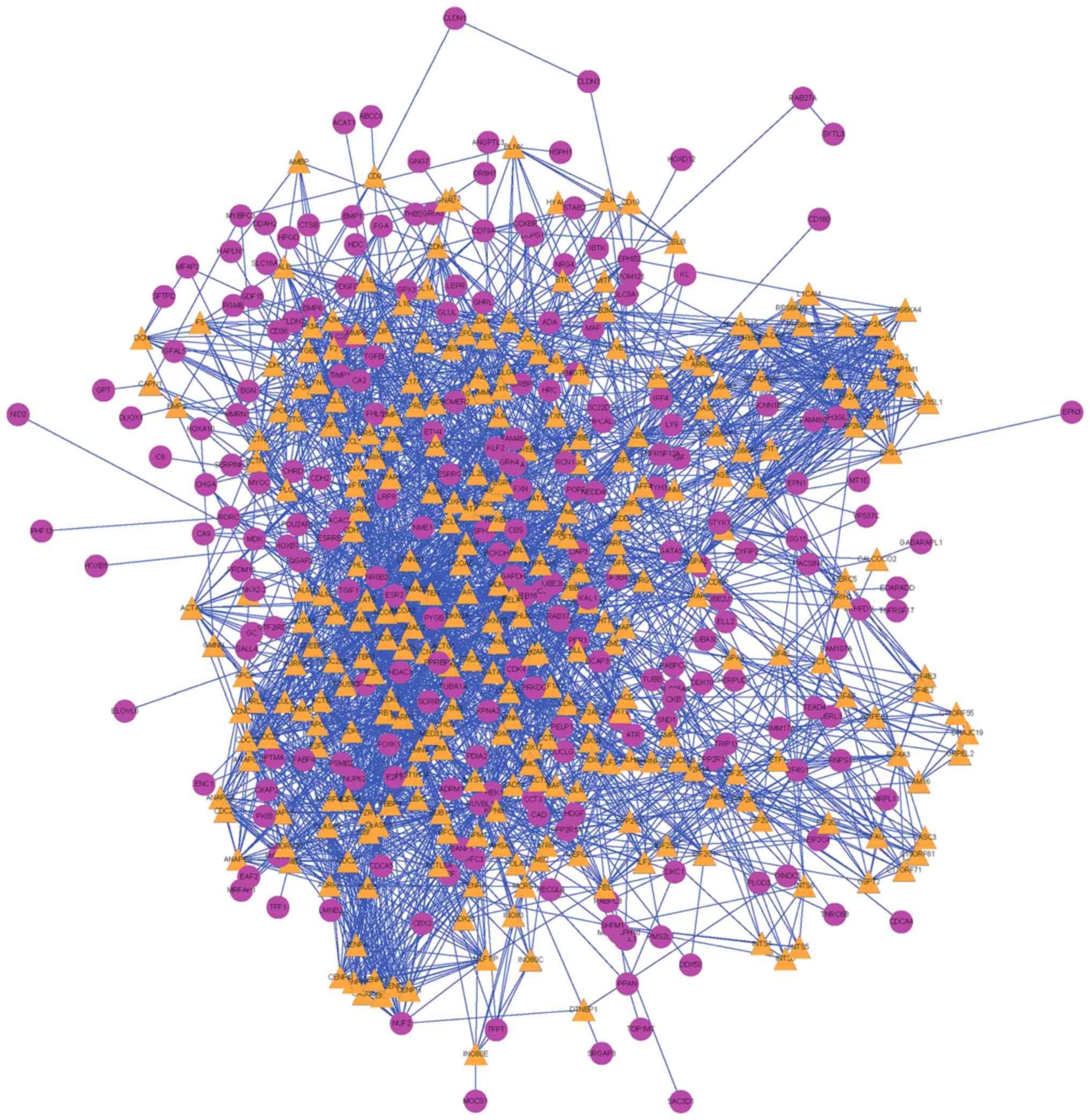
Doncheva NT, Assenov Y, Domingues FS and
Albrecht M: Topological analysis and interactive visualization of
biological networks and protein structures. Nat Protoc. 7:670–685.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
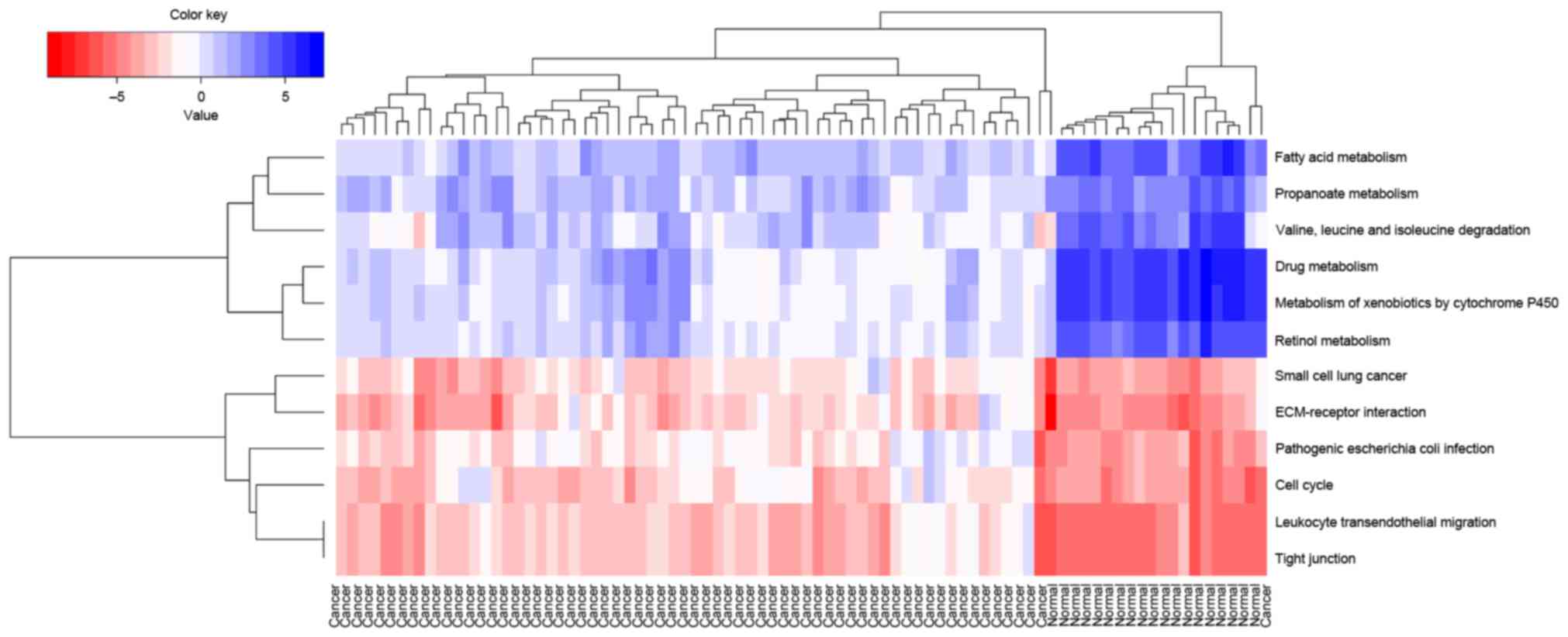
Dennis G Jr, Sherman BT, Hosack DA, Yang
J, Gao W, Lane HC and Lempicki RA: David: Database for annotation,
visualization, and integrated discovery. Genome biol. 4:P32003.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mao X, Cai T, Olyarchuk JG and Wei L:
Automated genome annotation and pathway identification using the
KEGG orthology (KO) as a controlled vocabulary. Bioinformatics.
21:3787–3793. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
de Hoon MJ, Imoto S, Nolan J and Miyano S:
Open source clustering software. Bioinformatics. 20:1453–1454.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Singh MS and Michael M: Role of xenobiotic
metabolic enzymes in cancer epidemiology. Methods Mol Biol.
472:243–264. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ding X and Kaminsky LS: Human extrahepatic
cytochromes p450: Function in xenobiotic metabolism and
tissue-selective chemical toxicity in the respiratory and
gastrointestinal tracts. Annu Rev Pharmacol Toxicol. 43:149–173.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
González CA, Sala N and Capellá G: Genetic
susceptibility and gastric cancer risk. Int J Cancer. 100:249–260.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Shah MA, Khanin R, Tang L, Janjigian YY,
Klimstra DS, Gerdes H and Kelsen DP: Molecular classification of
gastric cancer: A new paradigm. Clin Cancer Res. 17:2693–2701.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
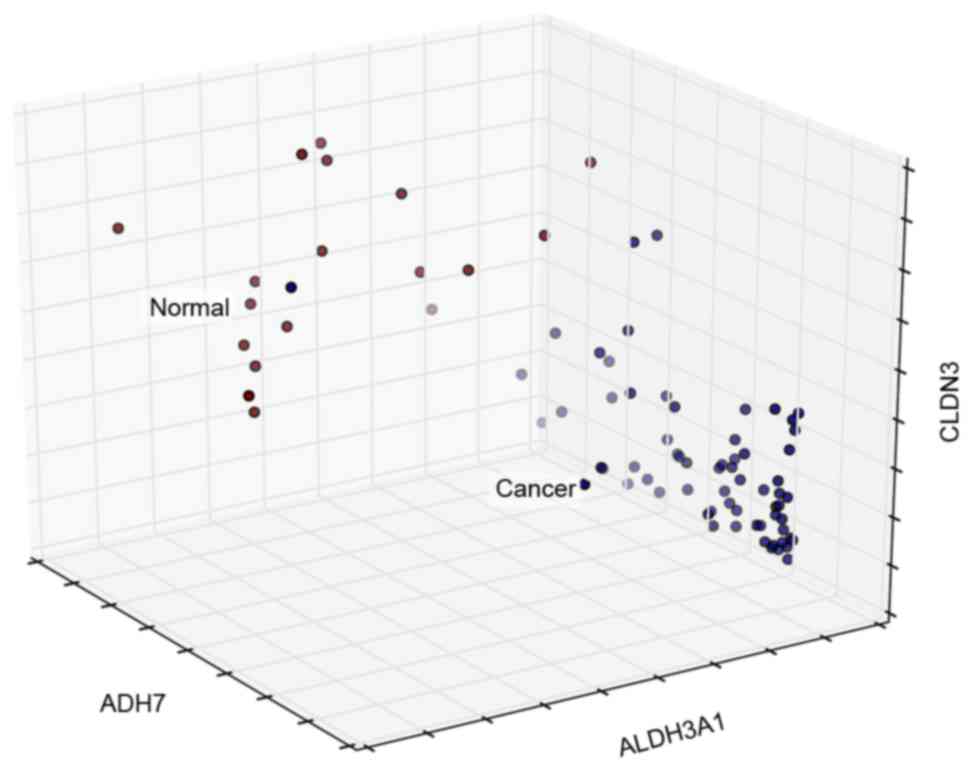
Jelski W and Szmitkowski M: Alcohol
dehydrogenase (ADH) and aldehyde dehydrogenase (ALDH) in the cancer
diseases. Clin Chim Acta. 395:1–5. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Duell EJ, Sala N, Travier N, Muñoz X,
Boutron-Ruault MC, Clavel-Chapelon F, Barricarte A, Arriola L,
Navarro C, Sánchez-Cantalejo E, et al: Genetic variation in alcohol
dehydrogenase (ADH1A, ADH1B, ADH1C, ADH7) and aldehyde
dehydrogenase (ALDH2), alcohol consumption and gastric cancer risk
in the European prospective investigation into cancer and nutrition
(EPIC) cohort. Carcinogenesis. 33:361–367. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Saadat M: Genetic polymorphisms of
glutathione s-transferase T1 (GSTT1) and susceptibility to gastric
cancer: A meta-analysis. Cancer Sci. 97:505–509. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Babhadiashar N, Sotoudeh M, Azizi E,
Bashiri J, Didevar R, Malekzadeh R and Ghahremani MH: Correlation
between cigarette smoking and urine cotinine level in Gastric
cancer patients. Iran J Pharm Res. 13:313–318. 2014.PubMed/NCBI
|
|
30
|
Rodriguez-Antona C and Ingelman-Sundberg
M: Cytochrome P450 pharmacogenetics and cancer. Oncogene.
25:1679–1691. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hurley TD, Edenberg HJ and Li TK: The
pharmacogenomics of alcoholismPharmacogenomics: The Search for
Individualized Therapies. Wiley-VCH; Weinheim: pp. 417–441. 2002,
View Article : Google Scholar
|
|
32
|
Vaglenova J, Martínez SE, Portí S, Duester
G, Farrés J and Parés X: Expression, localization and potential
physiological significance of alcohol dehydrogenase in the
gastrointestinal tract. Eur J Biochem. 270:2652–2662. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jairam S and Edenberg HJ:
Single-nucleotide polymorphisms interact to affect adh7
transcription. Alcohol Clin Exp Res. 38:921–929. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Jelski W, Kutylowska E, Laniewska-Dunaj M
and Szmitkowski M: Alcohol dehydrogenase (ADH) and aldehyde
dehydrogenase (ALDH) as candidates for tumor markers in patients
with pancreatic cancer. J Gastrointestin Liver Dis. 20:255–259.
2011.PubMed/NCBI
|
|
35
|
Yokoyama A and Omori T: Genetic
polymorphisms of alcohol and aldehyde dehydrogenases and risk for
esophageal and head and neck cancers. Jpn J Clin Oncol. 33:111–121.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Tsukita S and Furuse M: Pores in the wall:
Claudins constitute tight junction strands containing aqueous
pores. J Cell Biol. 149:13–16. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hoevel T, Macek R, Swisshelm K and Kubbies
M: Reexpression of the TJ protein CLDN1 induces apoptosis in breast
tumor spheroids. Int J Cancer. 108:374–383. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ueda J, Semba S, Chiba H, Sawada N, Seo Y,
Kasuga M and Yokozaki H: Heterogeneous expression of claudin-4 in
human colorectal cancer: Decreased claudin-4 expression at the
invasive front correlates cancer invasion and metastasis.
Pathobiology. 74:32–41. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Satake S, Semba S, Matsuda Y, Usami Y,
Chiba H, Sawada N, Kasuga M and Yokozaki H: Cdx2 transcription
factor regulates claudin-3 and claudin-4 expression during
intestinal differentiation of gastric carcinoma. Pathol Int.
58:156–163. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hewitt KJ, Agarwal R and Morin PJ: The
claudin gene family: Expression in normal and neoplastic tissues.
BMC Cancer. 6:1862006. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Matsuda Y, Semba S, Ueda J, Fuku T, Hasuo
T, Chiba H, Sawada N, Kuroda Y and Yokozaki H: Gastric and
intestinal claudin expression at the invasive front of gastric
carcinoma. Cancer Sci. 98:1014–1019. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Choi YL, Kim J, Kwon MJ, Choi JS, Kim TJ,
Bae DS, Koh SS, In YH, Park YW, Kim SH, et al: Expression profile
of tight junction protein claudin 3 and claudin 4 in ovarian serous
adenocarcinoma with prognostic correlation. 22:1185–1195. 2007.
|
|
43
|
Jung H, Jun KH, Jung JH, Chin HM and Park
WB: The expression of claudin-1, claudin-2, claudin-3, and
claudin-4 in gastric cancer tissue. J Surg Res. 167:e185–e191.
2011. View Article : Google Scholar : PubMed/NCBI
|