Introduction
Non-small cell lung cancers (NSCLCs) account for
>80% of all lung cancers and represent the leading cause of
cancer mortality worldwide, and in China alone (1,2).
Most NSCLC patients are initially diagnosed with advanced-stage
disease, which is defined as an unresectable, widely metastatic
tumor and is followed by poor prognosis.
Epidermal growth factor receptor-tyrosine kinase
inhibitors (EGFR-TKIs) are helpful for patients with
EGFR-activating mutations, especially exon 19 deletions
(19DEL) and exon 21 L858R mutations (L858R), which account for 85%
of all clinically important mutations (progression-free survival
(PFS) was 13.1 months with erlotinib compared to 4.6 months with
platinum-based chemotherapy and 8.0 months with gefitinib compared
to 6.3 months with platinum-based chemotherapy) (3,4).
However, on average, most patients undergoing EGFR-TKI treatment
develop resistance within 1 year (5,6), and
>50% of them carry the exon 20 T790M mutation (T790 M) in
EGFR (7). Recently, the
efficiency of AZD9291, a third-generation EGFR-TKI, was
demonstrated in lung cancer patients resistant to prior therapy
with EGFR-TKIs who harbored T790M (PFS was 9.6 months in
T790M-positive patients and 2.8 months in T790M-negative patients)
(8). Increasing evidence has
suggested that there may be a low-abundance, intrinsic T790M
mutation prior to EGFR-TKI therapy (9–11),
which enforces the need for exploring T790M status before
treatment. American Society of Clinical Oncology, European Society
for Medical Oncology and National Comprehensive Cancer Network
guidelines recommend clarifying EGFR mutation profiles
before TKI therapy or after the emergence of TKI resistance to
provide a precise and effective therapeutic regimen (12–14).
DNA extracted from tumor tissue remains the major
source for mutation analysis in clinical settings. Circulating
cell-free DNA (cfDNA) in plasma provides a homogeneous
representation of all tumor DNA and can help implement
non-invasive, continual monitoring of tumor mutation profiles while
obviating the reliance on invasive biopsies, tissue archives and
tumor heterogeneity. However technical challenges remain in
analyzing plasma cfDNA for EGFR mutations, such as the low
quantity and variability in the tumor-derived cfDNA fraction
between individuals.
Picoliter-droplet digital polymerase chain reaction
(ddPCR) is a promising approach with ultra-sensitive detection and
absolute quantification. This method compartmentalizes samples into
millions of picoliter droplets containing single DNA molecules and
analyses the terminal fluorescence of each droplet after parallel
amplification. Multiplex ddPCR can effectuate multiple mutation
assays simultaneously in one well with minimal DNA sample
consumption (15).
This study developed two multiplex ddPCR panels for
quantitative analysis of treatment-related EGFR mutations in
plasma cfDNA and compared the results generated by multiplex ddPCR
assays on plasma samples and by amplification refractory mutation
system (ARMS) assays on matched tumor tissue specimens from
advanced NSCLC patients.
Materials and methods
Design of the multiplex ddPCR
panels
The authors developed two independent multiplex
ddPCR panels for clinical application, a 4-plex panel for 19DEL and
T790M mutations plus the corresponding wild-type assays and a
5-plex panel to identify L858R (L858R-1 and L858R-2) and T790M
mutations.
Tables I and
II present the sequences and
concentrations of primers and TaqMan® probes (Thermo Fisher
Scientific, Inc., Waltham, MA, USA) used (16,17).
Probes for 19DEL were targeted at in-frame deletions in exon 19 of
the EGFR polypeptide. The probes recognized c. 2573T>G
for L858R-1, c. 2573T>G and c. 2574G>T for L858R-2, and c.
2369C>T for T790M. The PCR mixture for each multiplex panel
consisted of 20 µl TaqMan® Genotyping Master Mix (Thermo Fisher
Scientific, Inc.), 1.6 µl Droplet Stabilizer (RainDance
Technologies, Lexington, MA, USA), forward and reverse primers, and
probes in different concentrations and contained at least 50 ng of
target DNA template to a final reaction volume of 40 µl.
 | Table I.Primers used for the multiplex
droplet digital PCR. The total volume of the PCR mixture was 40 µl,
with 0.5 µM each of forward and reverse primers included in each
assay. |
Table I.
Primers used for the multiplex
droplet digital PCR. The total volume of the PCR mixture was 40 µl,
with 0.5 µM each of forward and reverse primers included in each
assay.
| Targeted
sequences | Forward primer
(5′-3′) | Reverse primer
(5′-3′) |
|---|
| 19DELa (c.746-c.750) |
GAAAGTTAAAATTCCCGTCGCTAT |
ACCCCCACACAGCAAAGC |
| L858R-1
(c.2573T>G) |
GCAGCATGTCAAGATCACAGATT |
CCTCCTTCTGCATGGTATTCTTTCT |
| L858R-2
(c.2573T>G, c.2574G>T) |
|
|
| T790M
(c.2369C>T) |
CCTCACCTCCACCGTGCA |
AGGCAGCCGAAGGGCA |
 | Table II.TaqMan® probes used for the multiplex
ddPCR. |
Table II.
TaqMan® probes used for the multiplex
ddPCR.
| Assay | Probes | Reporter dye | Sequence
(5′-3′) | Final concentration
(µM) |
|---|
| 4-plex ddPCR panel
assay | WT (19DEL) | VIC |
AATTAAGAGAAGCAACATC | 0.2 |
|
| Exon 19
reference-type | FAM |
ACATCGAGGATTTCCTTGT | 0.1 |
|
| WT (T790M) | TET |
T+CATC+A+C+GC/ZEN/A+GCTC | 0.2 |
|
| T790M | FAM |
T+CATC+A+T+GC/ZEN/A+GCTC | 0.2 |
| 5-plex ddPCR panel
assay | WT (L858R) | VIC |
AGTTTGGCCAGCCCAA | 0.1 |
|
| L858R-1 | FAM |
AGTTTGGCCCGCCCAA | 0.1 |
|
|
| VIC |
AGTTTGGCCCGCCCAA | 0.2 |
|
| L858R-2 | FAM |
AGTTTGGCACGCCCAA | 0.2 |
|
|
| VIC |
AGTTTGGCACGCCCAA | 0.2 |
|
| WT (T790M) | TET |
T+CATC+A+C+GC/ZEN/A+GCTC | 0.2 |
|
| T790M | FAM |
T+CATC+A+T+GC/ZEN/A+GCTC | 0.1 |
Plasmid DNA preparation and DNA
controls
The performance of each ddPCR assay was assessed
using fragmented plasmid DNA containing wild-type or mutant
EGFR sequence. Plasmids carrying wild-type (Plasmid #11011)
or 19DEL mutant (Plasmid #32062) sequences were purchased from
Addgene (http://www.addgene.org). The point
mutations L858R-1, L858R-2 and T790M were introduced using a
Phusion Site-Directed Mutagenesis kit (Thermo Fisher Scientific,
Inc.), following the manufacturer's procedure (18), and were confirmed by Sanger
sequencing. Plasmid containing mutant sequence was serially diluted
with plasmid containing wild-type sequence to yield a mixture in
which the mutant copy number was approximately 0.01–10% of the
wild-type copy number.
Patients and sample collection
The EGFR mutation status of 33 plasma samples
was assessed from 25 NSCLC patients with stage IIIB/IV cancers that
were enrolled from Zhongshan Hospital (Shanghai, China) from
January 2015 to December 2015 and harbored EGFR mutations,
confirmed by ARMS analysis of tumor tissue. Tumor specimens in the
present study were taken for clinical diagnostic purpose, which
from the 19 patients were obtained before EGFR-TKI therapy and
re-biopsies from 3 patients were taken after acquiring resistance
to EGFR-TKIs. Contemporaneous plasma samples were subjected to the
multiplex ddPCR panels. Another 3 patients were treated with
gefitinib at the physician's discretion and were recruited for
serial plasma samples collection at 2-month intervals to monitor
the fluctuations in EGFR mutation abundance by ddPCR assays.
These patients also received the appropriate imaging diagnoses,
including computed tomography scans performed for Response
Evaluation Criteria in Solid Tumors-based treatment response
evaluation (19). The study was
approved by the Ethics Committee of Fudan University (Shanghai,
China) and informed consent was obtained from the all for use of
the blood sample and tumor tissue.
DNA extraction
Peripheral blood samples (10 ml) were collected into
EDTA-K2 containing tubes. Blood samples were centrifuged
for 10 min at 1,900 × g to obtain preliminary supernatants within 2
h following sampling. Afterwards, another centrifugation at 16,000
× g for 10 min at 4°C separated the plasma samples from the blood
cell pellet. Circulating cfDNA was extracted using a QIAamp
Circulating Nucleic Acid kit (Qiagen GmbH, Hilden, Germany)
according to the manufacturer's instructions and was eluted with 50
µl Tris-EDTA buffer. All tumor specimens were collected for
diagnostic purpose by tracheobronchoscopy, percutaneous puncture
biopsy or metastatic lymph node biopsies and reviewed by a
pathologist for tumor cell content. The tumor tissue DNA was
extracted from formalin-fixed and paraffin-embedded diagnostic
specimens using a QIAamp DNA FFPE Tissue kit (Qiagen GmbH)
following the manufacturer's instructions and was eluted with 100
µl Buffer AE (Qiagen GmbH, Hilden, Germany). The extracted cfDNA
and tumor tissue DNA concentrations were measured with a NanoDrop
ND-1000 spectrophotometer (Thermo Fisher Scientific, Inc.).
EGFR mutations in tumor tissue
specimens detected by ARMS
EGFR mutations in tumor tissue specimens were
detected using an ADx-ARMS kit (Amoy Diagnostics, Xiamen, China),
which was approved by the Chinese Food and Drug Administration for
in vitro diagnostic use. All experiments and genotype
determinations were performed according to the manufacturer's
instructions and as previously described (20).
Circulating cfDNA detected by
multiplex ddPCR panels
The multiplex ddPCR panels were performed on a
RainDrop™ system (RainDance Technologies) with two separate
microfluidic chips, each composed of eight panels for droplet
generation (Source Chip; RainDance Technologies) and fluorescence
detection (Sense Chip; RainDance Technologies). The cycling was
performed on a Veriti® Thermal Cycler with a hot lid (Applied
Biosystems; Thermo Fisher Scientific, Inc.) as follows: Initiation
at 50°C for 2 min, incubation at 95°C for 10 min, and then 45
cycles of 95°C for 15 sec and 60°C for 1 min (using a 0.5°C/min
ramp rate). These cycles were followed by a final step at 98°C for
10 min and a 12°C hold. The endpoint fluorescence signal of each
droplet was scanned and analyzed using RainDrop Analyst software II
version 1.6.0_43-b01 (RainDance Technologies), following the
manufacturer's instructions. Spectral compensation was applied to
each assay to eliminate the contamination signals between different
fluorophores. To eliminate the crosstalk fluorescence signals from
the VIC/TET and FAM fluorophores, a compensation matrix was
established based on the positive control. Positive and negative
controls were included in each run, and gates for the identified
clusters were manually selected.
Data analysis
Results were considered negative when the number of
droplets within the gated region was under the limit of blank
(LOB), otherwise positive when the gated region yielded over 20
copies. Samples with results in the grey area (droplet counts
between LOB and 20) were assayed in triplicate and the mean value
was recorded. Templates with valid droplet counts <100 were
re-extracted. Positive mutant assay results were expressed as the
percentage of target templates over the sum of both mutant and
wild-type templates.
Statistical analysis
Statistical analysis was performed using SPSS
software (version, 17.0; SPSS Inc., Chicago, IL, USA). A Poisson
model was used to define the LOB for each mutation in the multiplex
panels. The consistency between the results obtained from plasma
cfDNA by multiplex ddPCR panels and the results from ARMS assays of
matched tumor tissue specimens was calculated using a κ test. A
paired Student's t-test was applied to analyze the discrepancy in
the abundance of T790M as measured by both multiplex ddPCR panels.
The linearity range was assessed using the linear correlation
coefficient values (r2) and slope. P<0.05 was considered to
indicate a statistically significant difference.
Results
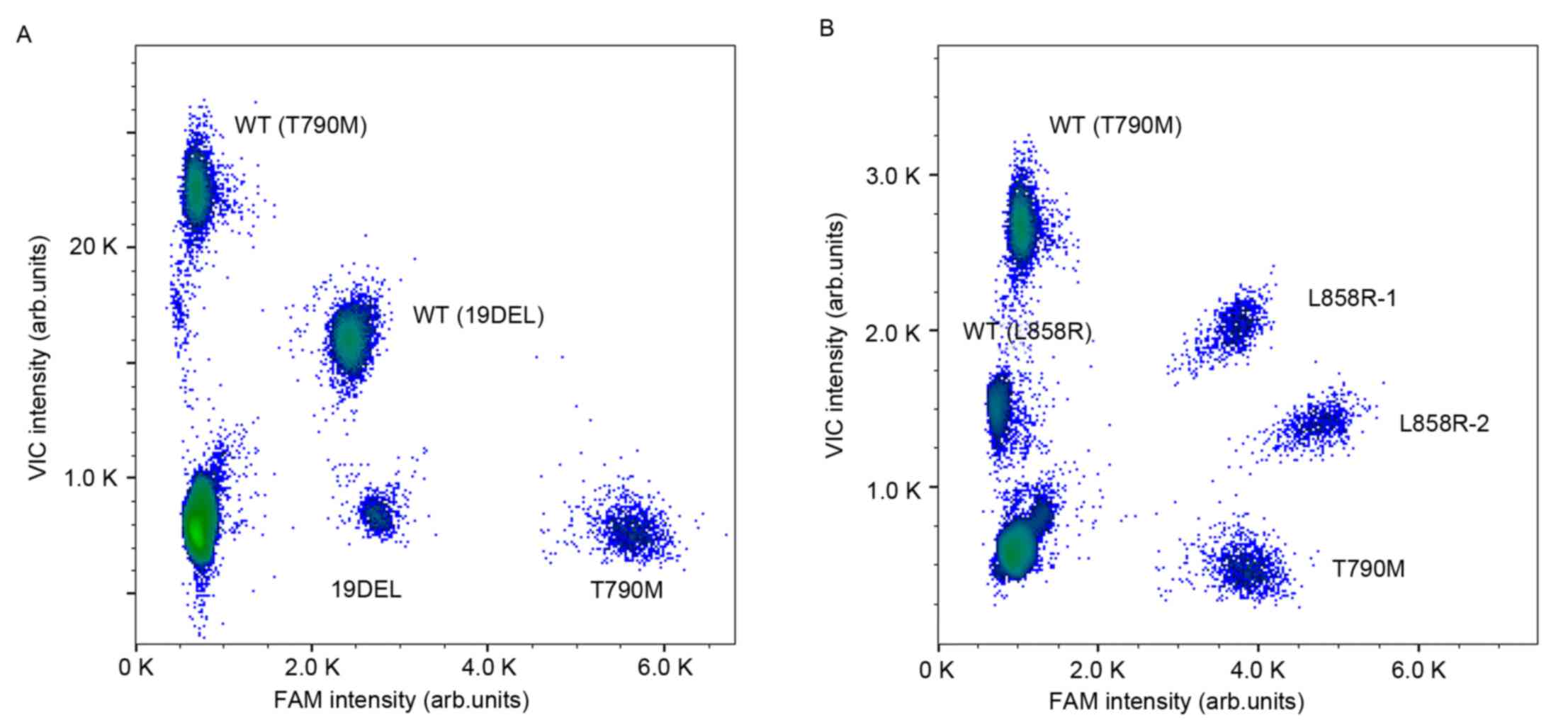
Multiplex ddPCR panels detected the
three most common EGFR mutations
Droplet digital PCR assays compartmentalize the
reaction mixture into millions of picoliter-sized droplets so that
each contains only one DNA string as the template, and an
independently PCR is then performed within the droplets in
parallel. Multiplex ddPCR panels can efficiently detect multiple
mutations simultaneously in one assay with minimal sample
consumption. The abridged general view of the workflow is described
in Fig. 1. Different fluorophores
and a diverse end point fluorescence intensity determined by the
concentration and combination use of TaqMan® probes distinguished
the droplets containing specific target templates into different
clusters in a 2-dimensional histogram. The cluster representing
19DEL or L858R is located along the diagonal line for the VIC and
FAM co-conjugated probes used (as presented in Fig. 2).
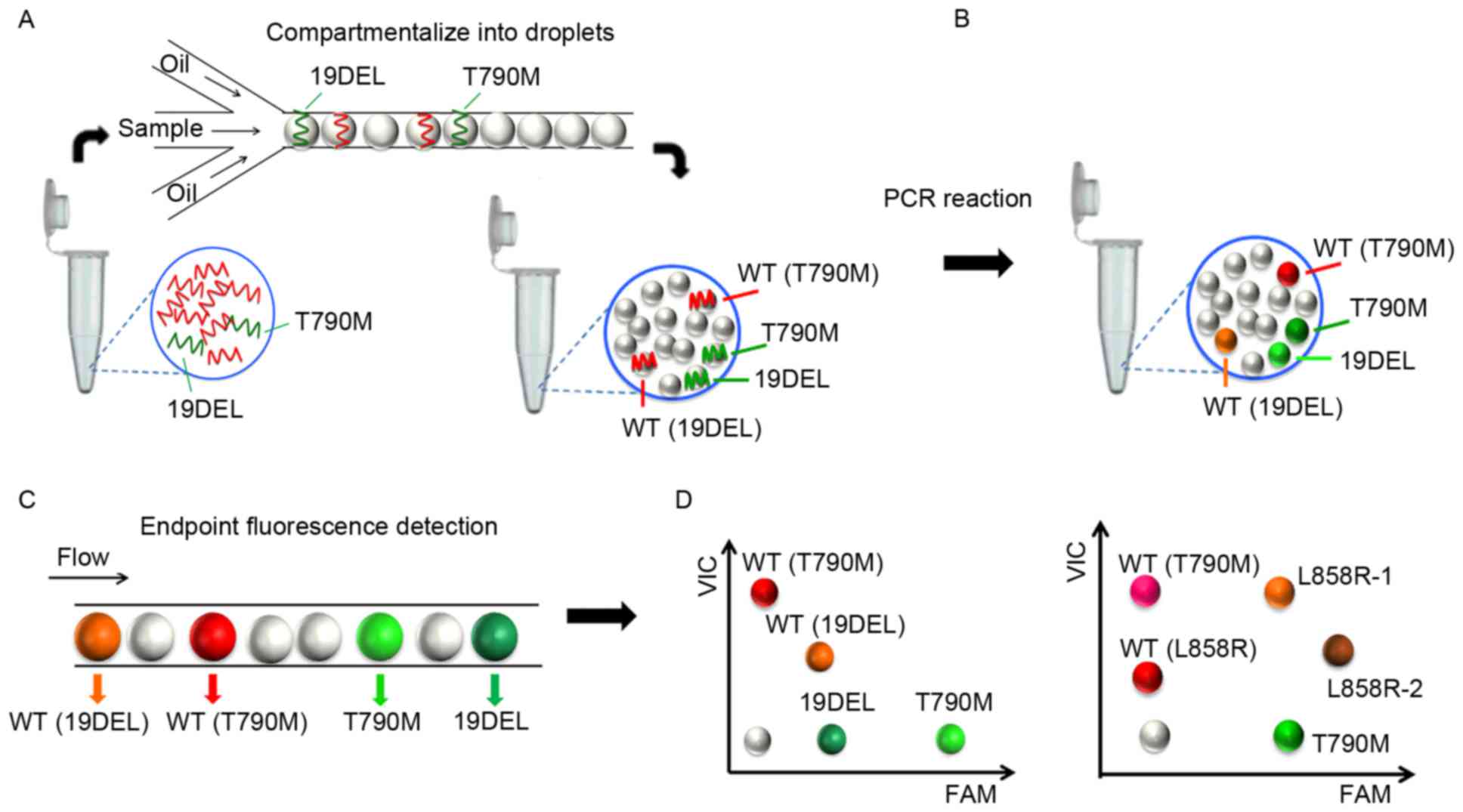 | Figure 1.Workflow of the multiplex ddPCR
panels. (A) The PCR mixture for each assay consisted of DNA
templates, primer pairs, probes, master mix and stabilizer to a
final volume of 40 µl and was (B) compartmentalized into >5
million picoliter-sized droplets for independent PCR reactions. (C)
The endpoint fluorescence signal of each droplet was scanned and
analyzed. (D) Different fluorophores and diverse end point
fluorescence intensities distinguished the droplets containing
target templates into different clusters. ddPCR, picoliter-droplet
digital polymerase chain reaction; WT, wild-type; 19DEL, exon 19
deletions; L858R-1, exon 21 L858R mutation (c.2573T>G); L858R-2,
exon 21 L858R mutation (c.2573T>G, c.2574G>T); T790M, exon 20
T790M mutation; FAM, 6-carboxyfluorescein; VIC, green fluorescent
protein. |
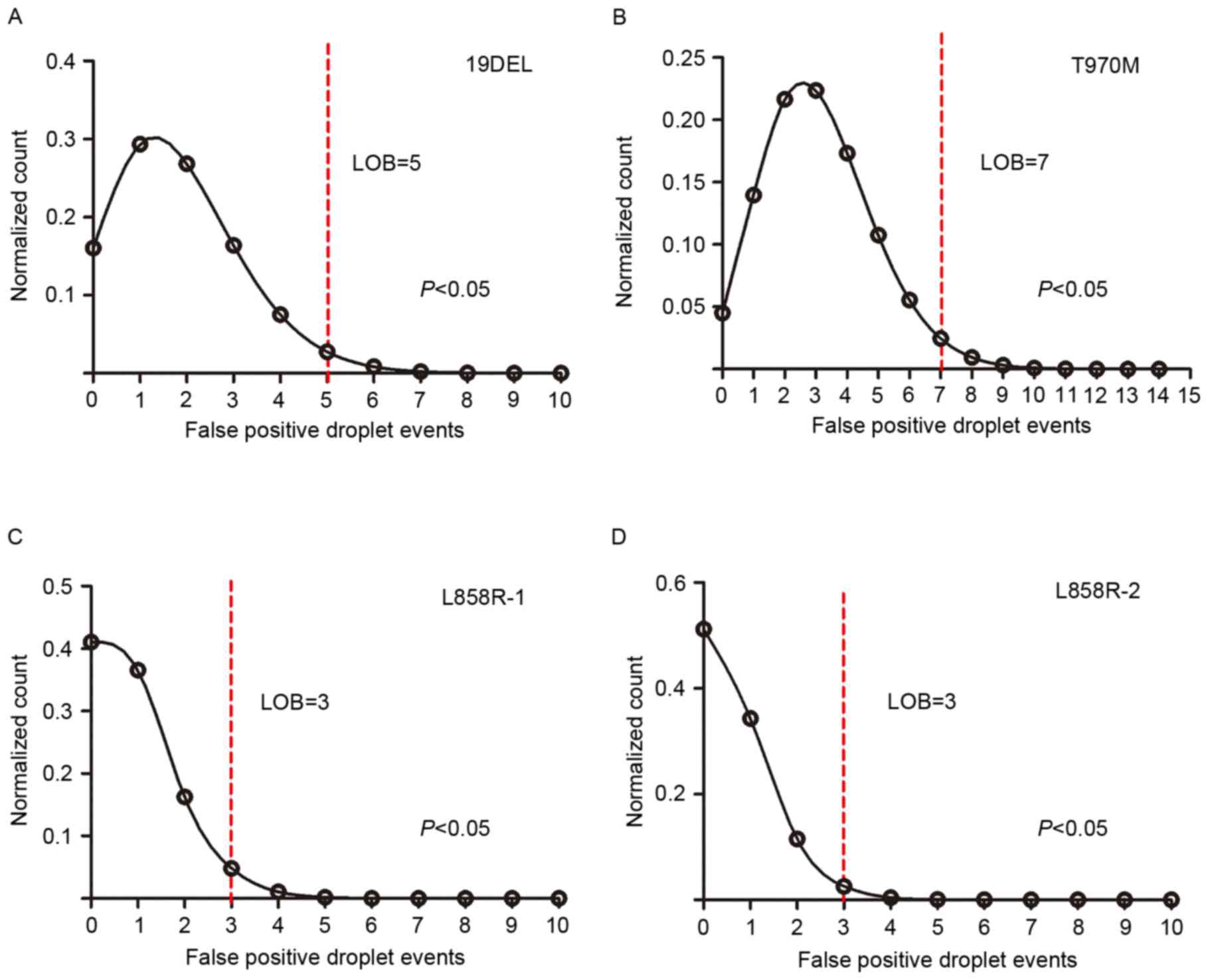
Performance assessments of the
multiplex ddPCR panels
As the defining characteristic of an assay, the LOB
determines the lower limit of detection. The authors subjected
fragmented plasmids containing only wild-type sequence to both
multiplex ddPCR panels in 24 replicates to observe the
false-positive events in each mutant assay, which allowed us to
generate the LOB (the count distribution is presented in Fig. 3). A Poisson model was applied to
fit the false-positive distribution, and the LOB was determined
using the 95% confidence interval. The LOBs were 5 for 19DEL, 3 for
L858R-1, 3 for L858R-2, and 7 for T790M (the LOB for T790M
generated from the two panels was the same, data not shown). To
assess the specificity of the panel assays, plasmids containing
either the L858R-1 or the L858R-2 sequence, which have only one
base difference between them, were subjected to the 5-plex panel in
triplicate; no cross-reactivity was observed (all false-positive
events were less than the LOB, data not shown).
The quantitative performance of multiplex ddPCR
panels was assessed using a series of dilutions from fragmented
plasmid sequences (Fig. 4). Each
dilution was analyzed in triplicate, and the averaged results were
plotted on 2-dimensional histograms (as shown in Fig. 4A and C for each panel). The mutant
allele expected copy number was plotted against the actual copy
number (Fig. 4B and D), and
regression analysis revealed a strong correlation (the slopes were
1.008 for 19DEL, 1.007 for T790M, 0.996 for L858R-1, and 0.992 for
L858R-2; all slopes had r2 values greater than 0.990).
Detectable abundance ranged from 0.01 to 10%. There was no
significant difference between the T790M abundance in the same
sample by either panel (t=1.386, P>0.05). Therefore, it follows
that the quantitative analysis of 19DEL, L858R-1, L858R-2 and T790M
using these two multiplex ddPCR panels is reliable and has
ultra-sensitivity, which allows the detection and quantification of
mutant alleles, even at ultra-low ratios (0.01%).
Multiplex ddPCR analysis of EGFR
mutations in circulating cfDNA from patients with advanced
NSCLC
To demonstrate the feasibility of applying these two
multiplex panels to clinical practice, analysis was performed on
plasma samples collected from 22 patients with advanced NSCLC whose
EGFR mutation profiles had been obtained by ARMS in
contemporaneously collected tumor tissues (Table III). The inter-individual
concentrations of extracted plasma cfDNA ranged from 16.4 to 102.5
ng/µl, and concentration did not correlate with the content of
particular mutant abundance (15).
Adequate numbers of valid droplets with >100 counts ensured the
validity of each assay; cases 9 and 17, which had no allotment for
re-extraction, were excluded because they did not have enough
droplets that detected wild-type templates. The overall concordance
rate between EGFR mutation profiles obtained from plasma or
tumor specimens was 80% (16/20). The concordance rates of the
19DEL, L858R and T790M assays were 90% (k=0.798, P<0.05), 95%
(k=0.894, P<0.05), and 95% (k=0.857; P<0.05), respectively,
with sensitivities and specificities of 90.9 and 88.9% for 19DEL,
87.5 and 100% for L858R, and 100 and 93.8% for T790M, respectively.
The ARMS results from tumor tissue specimens were used as a
reference. Plasma cfDNA assays for cases 21 and 22 revealed
additional mutant alleles that were not detected in tumor tissue
specimens by ARMS. Therefore, the authors reanalyzed the DNA
extracted from these tumor specimens in multiplex ddPCR panels and
verified the mutations with ultra-low abundance (0.29% for T790M in
case 21 and 0.57% for 19DEL in the metastatic tumor biopsied from
the lymph node in case 22; data not shown). However, two patients
were negative for the expected mutations in plasma cfDNA, which had
been detected by ARMS in tumor tissue specimens (19DEL in case 8
and L858R in case 16).
 | Table III.Results generated from plasma cfDNA
by multiplex ddPCR panels and tumor tissue DNA by ARMS in advanced
NSCLC patients. |
Table III.
Results generated from plasma cfDNA
by multiplex ddPCR panels and tumor tissue DNA by ARMS in advanced
NSCLC patients.
|
| Multiplex ddPCR
panels on plasma cfDNA |
|---|
|
|
|
|---|
| Case | Sex | Age (years) | Stage | EGFR-TKI | Site | ARMS on tumor
tissue DNA | Mutant | Abundance (%) |
|---|
| 1 | M | 53 | IV | No | Lung | 19DEL | 19DELa |
2.00 |
| 2 | M | 53 | IV | No | Lung | 19DEL | 19DEL | 15.64 |
| 3 | F | 65 | IV | No | Lung | 19DEL | 19DEL |
4.81 |
| 4 | M | 62 | IV | No | Lung | 19DEL | 19DEL | 45.68 |
| 5 | F | 41 | IV | No | Lung | 19DEL | 19DEL |
5.22 |
| 6 | F | 51 | IIIb | No | Lung | 19DEL | 19DEL | 19.62 |
| 7 | M | 46 | IV | No | Lung | 19DEL | 19DEL | 12.93 |
| 8 | F | 64 | IV | No | Lung | 19DEL | Na |
|
| 9 | M | 61 | IV | No | Lung | 19DEL | NE |
|
| 10 | M | 47 | IV | No | Lung | L858R | L858R |
7.47 |
| 11 | M | 71 | IIIb | No | Lung | L858R | L858Ra | 17.74 |
| 12 | F | 63 | IV | No | Lung | L858R | L858Ra |
1.32 |
| 13 | F | 58 | IV | No | Lung | L858R | L858R |
5.78 |
| 14 | F | 69 | IV | No | Lung | L858R | L858Ra |
5.63 |
| 15 | M | 35 | IIIb | No | Lung | L858R | L858Ra |
3.15 |
| 16 | F | 66 | IIIb | No | Lung | L858R | Na |
|
| 17 | F | 74 | IV | No | Lung | L858R | NE |
|
| 18 | M | 61 | IV | Gefitinib | Lung | T790M | T790Ma |
2.91 |
| 19 | F | 48 | IV | Gefitinib | Lung | 19DEL | 19DEL | 27.54 |
|
|
|
|
|
|
| T790M | T790M | 23.49 |
| 20 | F | 62 | IV | Gefitinib | Lung | 19DEL | 19DEL |
8.64 |
|
|
|
|
|
|
| T790M | T790M |
1.69 |
| 21 | F | 72 | IV | No | Lung | 19DEL | 19DEL | 64.02 |
|
|
|
|
|
|
|
| T790Ma |
0.91 |
| 22 | M | 68 | IV | No | Lung/lymph
node | L858R | 19DEL |
8.17 |
|
|
|
|
|
|
| T790M | L858R |
4.07 |
|
|
|
|
|
|
|
| T790Ma |
3.97 |
Therapeutic monitoring via the
multiplex ddPCR panels
To investigate whether quantitative analysis of
cfDNA EGFR mutations by either multiplex ddPCR panel could
help monitor treatment response and reveal therapeutic resistance,
3 patients were analyzed using 11 serially collected plasma samples
during gefitinib administration. Fig.
5 demonstrated that the EGFR mutant abundance in plasma
cfDNA fluctuated according to the change in tumor size as revealed
by imaging scans.
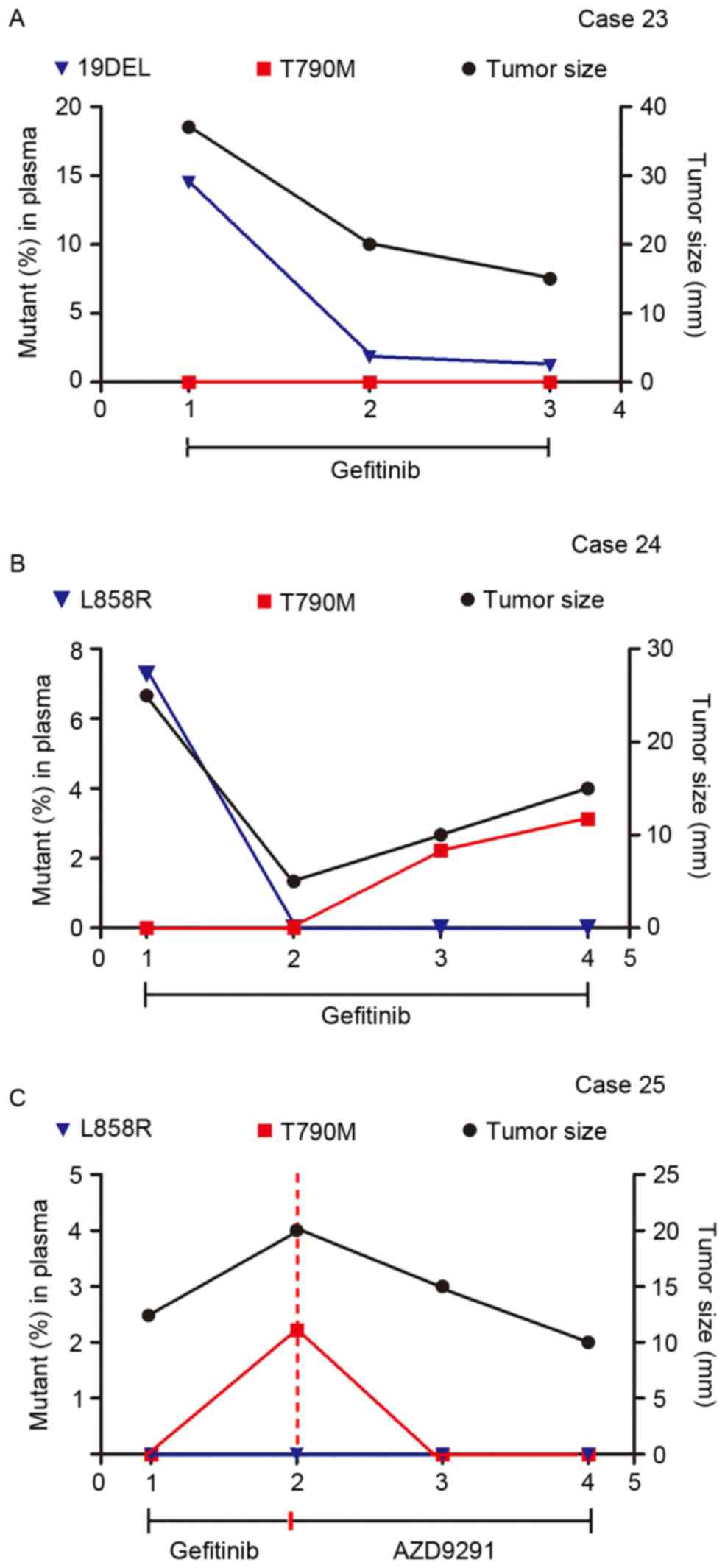 | Figure 5.Therapeutic monitoring by multiplex
picoliter-droplet digital polymerase chain reaction assays on
plasma cfDNA EGFR mutations and lung mass imaging scans.
Abundance of EGFR mutations in plasma cfDNA fluctuated with
tumor size, as revealed by imaging scans. (A) 19DEL abundance
persistently decreased when tumors shrank in Case 23. (B) T790M
rose with the tumor size in Case 24, which indicated an acquired
therapeutic resistance to gefitinib. (C) Case 25 presented a change
in T790M abundance after receiving AZD9291 treatment. cfDNA,
circulating cell-free DNA; EGFR, epidermal growth factor receptor;
Mut, mutant; FAM, 6-carboxyfluorescein; VIC, green fluorescent
protein; 19DEL, exon 19 deletions; L858R, exon 21 L858R mutation
(either L858R-1 or L858R-2); T790M, exon 20 T790M mutation. |
Case 23 (Fig. 5A)
is a 46-year-old male with no history of smoking with lung
adenocarcinoma and brain metastasis harboring a 19DEL mutation,
which was confirmed by ARMS in a tumor biopsy. The 4-plex panel
(for assaying 19DEL and T790M) was applied to monitor the
therapeutic response to gefitinib and noted a persistent decrease
in 19DEL mutant abundance that was coincident with the change in
the maximum tumor diameter. Case 24 (Fig. 5B) is a 56-year-old female
non-smoker with stage IV lung adenocarcinoma who had taken
gefitinib for 6 months before the serial plasma collection started.
Considering the L858R mutation detected by ARMS in the biopsied
tumor specimens at the time of onset, the 5-plex panel (for
assaying L858R and T790M) was applied. A decrease in L858R mutant
abundance was observed in the first follow-up period, and T790M
mutant abundance then rose, which is in accordance with the tumor
size and indicated an acquired therapeutic resistance to gefitinib.
Case 25 (Fig. 5C) is a 61-year-old
male smoker with lung adenocarcinoma and bone metastases who had
been treated with gefitinib for 10 months and developed resistance.
This patient was assessed using the 5-plex panel assay for an
EGFR activating-mutant profile, which was previously
demonstrated by ARMS assays in tumor tissue. Applying the
third-generation EGFR-TKI AZD9291 caused a changeover in the T790M
mutant abundance and disease progress.
Discussion
In the present study, two multiplex ddPCR panels
were established and quantitatively analyzed EGFR mutations
in plasma cfDNA isolated from advanced NSCLC patients to assess
their usefulness in EGFR-TKI decision-making. A previous study
demonstrated the high carrying rate of EGFR mutations in
Asian patients with advanced NSCLC; however, the coexistence of
19DEL and L858R mutants increased in only 0.8% patients (21). Thus, either of the multiplex ddPCR
panels (assaying 19DEL and L858R as TKI-sensitive mutants and T790M
as a TKI-resistant mutant) is capable of evaluating EGFR
mutations for current clinical requirements.
Circulating cfDNA assays on tumor genetic
information could serve as a ‘liquid biopsy’ to facilitate dynamic
monitoring of EGFR mutations during treatment and could
allow for constant optimization of personalized therapy (22). Moreover, circulating cfDNA mutant
assays can help generate genetic profiles from patients incapable
of having or reluctant to accept tumor biopsies. The amount of
circulating cfDNA is higher in cancer patients than in healthy
individuals due to the release of nucleic acids into the
bloodstream by tumor cell apoptosis, necrosis and secretion
(23). However, detecting
EGFR mutations in circulating cfDNA remains a challenge due
to its low abundance, which is attenuated in the overall complex
genetic background. High-sensitivity methods have been explored in
the past year. Scorpion-ARMS, denaturing high-performance liquid
chromatography detection, peptide nucleic acid mediated polymerase
chain reaction clamping and next-generation sequencing have been
evaluated as EGFR mutant assays with plasma cfDNA from NSCLC
patients and have detection sensitivities ranging from 20.7 to
65.79% of the sensitivities achieved using tumor tissue examination
methods (24–27). The detectable abundance of the
multiplex ddPCR panels in the current study ranged from 0.01 to
10%. Applying plasma cfDNA assays to a ddPCR platform has greater
detection sensitivity than the existing technologies for seeking
specific mutations with ultra-low abundance.
The overall concordance rate in the present study
between results generated from plasma samples by ddPCR and tumor
tissues by ARMS was 80%, which is similar to other studies, which
have ranged from 80.0 to 94.19% concordance (19,28–30).
Interestingly, the ddPCR assays performed on plasma cfDNA have
noted additional EGFR mutants that were absent from the
tumor tissue in two cases. Despite detection sensitivity issues,
the contradiction in results could be due to the inter- or
intra-tumor heterogeneity in tumor biopsy specimens (31,32).
The EGFR mutation status between primary tumors and
metastatic tumors is reportedly different (33,34).
ddPCR assays were performed on a metastatic tumor collected from
the lymph node of one patient and detected an additional 19DEL
mutation. Plasma cfDNA containing genetic information from both
primary and metastatic tumors can help overcome the boundedness due
to tumor heterogeneity and selection bias in biopsy and can acquire
more genetic profiling that is absent from a single snapshot.
Plasma cfDNA may degrade without timely separation when collected
in EDTA-K2-containing tubes without nuclease inhibitors
(35); the authors suspected this
limitation was the reason for the positive mutations detected in
tumor tissues with negative results in plasma cfDNA in the
study.
Several studies have demonstrated that changes in
mutant abundance in plasma cfDNA are associated with the tumor
burden and malignant progression (30,36).
Similarly, the results of our study inferred that EGFR
mutant abundance in plasma cfDNA fluctuated according to changes in
tumor size, which certified the practicality of our multiplex ddPCR
panels in therapeutic evaluation and resistance monitoring. Either
panel can be chosen flexibly, according to the mutant alleles
generated by routine assays on tumor tissue specimens in a clinical
setting, and can be applied to patients receiving TKIs to
continually inspect EGFR mutation abundance in plasma cfDNA.
With its very short retention period in plasma, circulating cfDNA
can reflect immediate tumor-associated changes (23). Thus, it can be monitored at short
intervals, even less than the 2 months described in the present
study, and can serve as the evidence for therapeutic regimen
adjustments before tumor progression imaging is performed. By
monitoring the changes in EGFR mutant abundance in plasma
cfDNA, our multiplex ddPCR panels can provide an early indication
of EGFR-TKI resistance. Patients in whom the post-treatment T790M
mutation arose would then benefit from a new treatment plan using
the new TKI, AZD9291 (37,38).
In this study, two multiplex ddPCR panels were put
forward for analysis of multiple EGFR mutant alleles with
therapeutic significance that are integrated into a single assay
consuming minimal plasma sample from NSCLC patients. The
application of these two panels in clinical settings will be
practical, flexible, and time- and cost-saving. However, it should
be noted that an LOB series was determined for each mutant based on
our multiplex ddPCR panels developed in-house; these results should
not be used to determine cutoffs for clinical decisions. The
utility, feasibility and robustness of the multiplex ddPCR panels
should be further examined in a larger-scale clinical trial.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant no. 81572064) and the
Key Developing Disciplines of Shanghai Municipal Commission of
Health and Family Planning (grant no. 2015ZB0201).
Glossary
Abbreviations
Abbreviations:
|
ddPCR
|
picoliter-droplet digital polymerase
chain reaction
|
|
EGFR
|
epidermal growth factor receptor
|
|
cfDNA
|
circulating cell-free DNA
|
|
NSCLC
|
non-small cell lung cancer
|
|
EGFR-TKIs
|
epidermal growth factor
receptor-tyrosine kinase inhibitors
|
|
19DEL
|
exon 19 deletions
|
|
L858R
|
exon 21 L858R mutations
|
|
T790M
|
exon 20 T790M mutation
|
|
LOB
|
limit of blank
|
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhou C, Wu YL, Chen G, Feng J, Liu XQ,
Wang C, Zhang S, Wang J, Zhou S, Ren S, et al: Erlotinib versus
chemotherapy as first-line treatment for patients with advanced
EGFR mutation-positive non-small-cell lung cancer (OPTIMAL,
CTONG-0802): A multicentre, open-label, randomised, phase 3 study.
Lancet Oncol. 12:735–742. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Han JY, Park K, Kim SW, Lee DH, Kim HY,
Kim HT, Ahn MJ, Yun T, Ahn JS, Suh C, et al: First-SIGNAL:
First-line single-agent iressa versus gemcitabine and cisplatin
trial in never-smokers with adenocarcinoma of the lung. J Clin
Oncol. 30:1122–1128. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mitsudomi T and Yatabe Y: Mutations of the
epidermal growth factor receptor gene and related genes as
determinants of epidermal growth factor receptor tyrosine kinase
inhibitors sensitivity in lung cancer. Cancer Sci. 98:1817–1824.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sequist LV, Waltman BA, Dias-Santagata D,
Digumarthy S, Turke AB, Fidias P, Bergethon K, Shaw AT, Gettinger
S, Cosper AK, et al: Genotypic and histological evolution of lung
cancers acquiring resistance to EGFR inhibitors. Sci Transl Med.
3:75ra262011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yu HA, Arcila ME, Rekhtman N, Sima CS,
Zakowski MF, Pao W, Kris MG, Miller VA, Ladanyi M and Riely GJ:
Analysis of tumor specimens at the time of acquired resistance to
EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers.
Clin Cancer Res. 19:2240–2247. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jänne PA, Yang JC, Kim DW, Planchard D,
Ohe Y, Ramalingam SS, Ahn MJ, Kim SW, Su WC, Horn L, et al: AZD9291
in EGFR inhibitor-resistant non-small-cell lung cancer. N Engl J
Med. 372:1689–1699. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Su KY, Chen HY, Li KC, Kuo ML, Yang JC,
Chan WK, Ho BC, Chang GC, Shih JY, Yu SL and Yang PC: Pretreatment
epidermal growth factor receptor (EGFR) T790M mutation predicts
shorter EGFR tyrosine kinase inhibitor response duration in
patients with non-small-cell lung cancer. J Clin Oncol. 30:433–440.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hata A, Katakami N, Yoshioka H, Takeshita
J, Tanaka K, Nanjo S, Fujita S, Kaji R, Imai Y, Monden K, et al:
Rebiopsy of non-small cell lung cancer patients with acquired
resistance to epidermal growth factor receptor-tyrosine kinase
inhibitor: Comparison between T790M mutation-positive and
mutation-negative populations. Cancer. 119:4325–4332. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang Z, Chen R, Wang S, Zhong J, Wu M,
Zhao J, Duan J, Zhuo M, An T, Wang Y, et al: Quantification and
dynamic monitoring of EGFR T790M in plasma cell-free DNA by digital
PCR for prognosis of EGFR-TKI treatment in advanced NSCLC. PLoS
One. 9:e1107802014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Reck M, Popat S, Reinmuth N, de Ruysscher
D, Kerr KM and Peters S: ESMO Guidelines Working Group: Metastatic
non-small-cell lung cancer (NSCLC): ESMO clinical practice
guidelines for diagnosis, treatment and follow-up. Ann Oncol. 25
Suppl 3:iii27–39. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Leighl NB, Rekhtman N, Biermann WA, Huang
J, Mino-Kenudson M, Ramalingam SS, West H, Whitlock S and
Somerfield MR: Molecular testing for selection of patients with
lung cancer for epidermal growth factor receptor and anaplastic
lymphoma kinase tyrosine kinase inhibitors: American society of
clinical oncology endorsement of the College of American
Pathologists/International Association for the study of lung
cancer/association for molecular pathology guideline. J Clin Oncol.
32:3673–3679. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
National Comprehensive Cancer Network:
NCCN guidelines for patients. Version 1. 2015, Non-Small Cell Lung
Cancer (S/OL); November 6–2015
|
|
15
|
Taly V, Pekin D, Benhaim L, Kotsopoulos
SK, Le Corre D, Li X, Atochin I, Link DR, Griffiths AD, Pallier K,
et al: Multiplex picodroplet digital PCR to detect KRAS mutations
in circulating DNA from the plasma of colorectal cancer patients.
Clin Chem. 59:1722–1731. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yung TK, Chan KC, Mok TS, Tong J, To KF
and Lo YM: Single-molecule detection of epidermal growth factor
receptor mutations in plasma by microfluidics digital PCR in
non-small cell lung cancer patients. Clin Cancer Res. 15:2076–2084.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Watanabe M, Kawaguchi T, Isa S, Ando M,
Tamiya A, Kubo A, Saka H, Takeo S, Adachi H, Tagawa T, et al:
Ultra-sensitive detection of the pretreatment EGFR T790M mutation
in non-small cell lung cancer patients with an EGFR-activating
mutation using droplet digital PCR. Clin Cancer Res. 21:3552–3560.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Thermo Scientific. User Guide: Phusion
site-directed mutagenesis kit. January 4–2015
|
|
19
|
Nishino M, Jackman DM, Hatabu H, Jänne PA,
Johnson BE and Van den Abbeele AD: Imaging of lung cancer in the
era of molecular medicine. Acad Radiol. 18:424–436. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhu G, Ye X, Dong Z, Lu YC, Sun Y, Liu Y,
McCormack R, Gu Y and Liu X: Highly sensitive droplet digital PCR
method for detection of EGFR-activating mutations in plasma
cell-Free DNA from patients with advanced non-small cell lung
cancer. J Mol Diagn. 17:265–272. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shi Y, Au JS, Thongprasert S, Srinivasan
S, Tsai CM, Khoa MT, Heeroma K, Itoh Y, Cornelio G and Yang PC: A
prospective, molecular epidemiology study of EGFR mutations in
Asian patients with advanced non-small-cell lung cancer of
adenocarcinoma histology (PIONEER). J Thorac Oncol. 9:154–162.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Heitzer E, Ulz P and Geigl JB: Circulating
tumor DNA as a liquid biopsy for cancer. Clin Chem. 61:112–123.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Diaz LA Jr and Bardelli A: Liquid
biopsies: Genotyping circulating tumor DNA. J Clin Oncol.
32:579–586. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bai H, Mao L, Wang HS, Zhao J, Yang L, An
TT, Wang X, Duan CJ, Wu NM, Guo ZQ, et al: Epidermal growth factor
receptor mutations in plasma DNA samples predict tumor response in
Chinese patients with stages IIIB to IV non-small-cell lung cancer.
J Clin Oncol. 27:2653–2659. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Douillard JY, Ostoros G, Cobo M, Ciuleanu
T, Cole R, McWalter G, Walker J, Dearden S, Webster A, Milenkova T
and McCormack R: Gefitinib treatment in EGFR mutated caucasian
NSCLC: Circulating-free tumor DNA as a surrogate for determination
of EGFR status. J Thorac Oncol. 9:1345–1353. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kim HR, Lee SY, Hyun DS, Lee MK, Lee HK,
Choi CM, Yang SH, Kim YC, Lee YC, Kim SY, et al: Detection of EGFR
mutations in circulating free DNA by PNA-mediated PCR clamping. J
Exp Clin Cancer Res. 32:502013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Couraud S, Vaca-Paniagua F, Villar S,
Oliver J, Schuster T, Blanché H, Girard N, Trédaniel J,
Guilleminault L, Gervais R, et al: Noninvasive diagnosis of
actionable mutations by deep sequencing of circulating free DNA in
lung cancer from never-smokers: A proof-of-concept study from
BioCAST/IFCT-1002. Clin Cancer Res. 20:4613–4624. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ishii H, Azuma K, Sakai K, Kawahara A,
Yamada K, Tokito T, Okamoto I, Nishio K and Hoshino T: Digital PCR
analysis of plasma cell-free DNA for non-invasive detection of drug
resistance mechanisms in EGFR mutant NSCLC: Correlation with paired
tumor samples. Oncotarget. 6:30850–30858. 2015.PubMed/NCBI
|
|
29
|
Seki Y, Fujiwara Y, Kohno T, Takai E,
Sunami K, Goto Y, Horinouchi H, Kanda S, Nokihara H, Watanabe S, et
al: Picoliter-droplet digital polymerase Chain reaction-based
analysis of cell-free plasma DNA to assess EGFR mutations in lung
adenocarcinoma that confer resistance to tyrosine-kinase
inhibitors. Oncologist. 21:156–164. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lee JY, Qing X, Xiumin W, Yali B, Chi S,
Bak SH, Lee HY, Sun JM, Lee SH, Ahn JS, et al: Longitudinal
monitoring of EGFR mutations in plasma predicts outcomes of NSCLC
patients treated with EGFR TKIs: Korean lung cancer consortium
(KLCC-12-02). Oncotarget. 7:6984–6993. 2016.PubMed/NCBI
|
|
31
|
Taniguchi K, Okami J, Kodama K,
Higashiyama M and Kato K: Intratumor heterogeneity of epidermal
growth factor receptor mutations in lung cancer and its correlation
to the response to gefitinib. Cancer Sci. 99:929–935. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Bai H, Wang Z, Wang Y, Zhuo M, Zhou Q,
Duan J, Yang L, Wu M, An T, Zhao J and Wang J: Detection and
clinical significance of intratumoral EGFR mutational heterogeneity
in Chinese patients with advanced non-small cell lung cancer. PLoS
One. 8:e541702013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chen ZY, Zhong WZ, Zhang XC, Su J, Yang
XN, Chen ZH, Yang JJ, Zhou Q, Yan HH, An SJ, et al: EGFR mutation
heterogeneity and the mixed response to EGFR tyrosine kinase
inhibitors of lung adenocarcinomas. Oncologist. 17:978–985. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Shimizu K, Yukawa T, Hirami Y, Okita R,
Saisho S, Maeda A, Yasuda K and Nakata M: Heterogeneity of the EGFR
mutation status between the primary tumor and metastatic lymph node
and the sensitivity to EGFR tyrosine kinase inhibitor in non-small
cell lung cancer. Target Oncol. 8:237–242. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Sillence KA, Roberts LA, Hollands HJ,
Thompson HP, Kiernan M, Madgett TE, Welch CR and Avent ND: Fetal
sex and RHD genotyping with digital PCR demonstrates greater
sensitivity than real-time PCR. Clin Chem. 61:1399–1407. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Reinert T, Schøler LV, Thomsen R, Tobiasen
H, Vang S, Nordentoft I, Lamy P, Kannerup AS, Mortensen FV,
Stribolt K, et al: Analysis of circulating tumor DNA to monitor
disease burden following colorectal cancer surgery. Gut.
65:625–634. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Cross DA, Ashton SE, Ghiorghiu S, Eberlein
C, Nebhan CA, Spitzler PJ, Orme JP, Finlay MR, Ward RA, Mellor MJ,
et al: AZD9291, an irreversible EGFR TKI, overcomes T790M-mediated
resistance to EGFR inhibitors in lung cancer. Cancer Discov.
4:1046–1061. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Jiang T and Zhou C: Clinical activity of
the mutant-selective EGFR inhibitor AZD9291 in patients with EGFR
inhibitor-resistant non-small cell lung cancer. Transl Lung Cancer
Res. 3:370–372. 2014.PubMed/NCBI
|