Introduction
Hypertrophic scars are defined as visible and
elevated scars without spreading into surrounding tissues and with
a tendency to regress spontaneously. They are characterized by
proliferation of the dermal tissue, with excessive deposition of
fibroblast (FB)-derived extracellular matrix (ECM) proteins,
particularly collagen, over long periods, and by persistent
inflammation and fibrosis (1,2).
Numerous treatments have been described, including
surgical excision, pressure therapy, intralesional interferon,
laser therapy and silicone gel sheeting (3–7).
However, no optimal treatment method has been established,
primarily due to limited understanding of the precise underlying
mechanisms. Abnormal activation of FBs and accumulation of collagen
collaborate to induce hypertrophic scar formation (8). The ECM is primarily derived from FBs,
and its activation is considered to facilitate re-epithelialization
(9). Furthermore, reduced function
of FBs reduces ECM production and leads to cell apoptosis, leading
to maturation of the scar (10).
The balance between pro- and anti-fibrotic activity is critical to
normotrophic scar formation, and failing to regulate activated FBs
leads to pathologic scar formation, including hypertrophic scars.
Therefore, identifying molecules that strengthen or debilitate may
have therapeutic value for the treatment of hypertrophic scars.
Homeobox (HOX) genes encode a group of transcription
factors that bind to specific DNA strands via the homeodomain
(11). A total of 39 genes are
classified into four clusters: HOXA, B, C and D (12). HOXD3 and HOXA3 accelerate wound
closure in poorly healing diabetic mice, with improved angiogenesis
(13,14). In contrast to HOXA3 and HOXD3,
HOXB13 was demonstrated to impair wound healing (15,16).
These studies have associated HOX genes with wound healing, an
essential process in scar formation, indicating that HOX genes are
potentially involved in hypertrophic scar formation. However, to
the best of the authors' knowledge, no previous studies have
investigated this association.
HOXB9 is a widely understood to be involved in the
development of mammary glands, sternum and angiogenesis (17,18).
Previous studies have revealed that HOXB9 is involved in the breast
cancer, lung adenocarcinoma and gastric carcinoma, serving a role
in promoting or inhibiting the tumor process (18–24).
HOXB9 may have an effect on dermal FBs, and
facilitate or attenuate hypertrophic scar formation in vivo.
Therefore, the present study examined its expression in
hypertrophic scar tissues, and tested its effects on contraction.
This study further investigated the potential biochemical
mechanisms involved in the effects of HOXB9 on hypertrophic
formation.
Materials and methods
Ethics statement
All experimental procedures were conducted under a
protocol approved by the Ethical Committee of Xiangyang Central
Hospital (Xiangyang, China).
Cell culture and treatment
Six patients (2 males and 4 females) were enrolled
from March-May 2016 in the Plastic Surgery Department of Xiangyang
Central Hospital. The age of the patients ranged 16–40.
Hypertrophic scar tissue from the arm and adjacent healthy skin
samples were collected from the patients prior to surgical
treatment. The tissue was fixed in 4% formalin. After fixation, the
tissue was embedded with paraffin wax. Written consent was obtained
from the patients themselves or their legal guardians. Dermal FBs
were isolated and cultured as described previously (25). Briefly, tissues were trimmed to
remove excessive adipose and rinsed with PBS three times. Next,
tissues were sectioned into small pieces and incubated in
Dulbecco's Modified Eagle's medium (DMEM; Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) containing 0.1% collagenase
type I (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) at 37°C for
3 h. The isolated FBs were subsequently cultured in DMEM containing
10% fetal calf serum (Gibco; Thermo Fisher Scientific, Inc.) at
37°C in a humidified atmosphere of 5% CO2. All cells
used in this experiment were at passage 5–10. Several 60-mm dishes
of healthy skin FBs were randomly divided into different groups
(n=6).
Immunohistochemistry
Paraffin-embedded scar tissues and autologous skin
tissues were cut into 5-µm thick sections for immunohistochemical
staining. Sections were deparaffinized, dehydrated and subject to
antigen retrieval by pretreating with 7% H2O2
in distilled water, followed by 0.1 mol/l periodic acid, 0.005
mol/l NaBH4 and normal human serum (Thermo Fisher Scientific,
Inc.). The sections were separately incubated for 2 h with rabbit
anti-human fibronectin (FN; 1:1,000; catalog no. ab61214; Abcam,
Cambridge, MA, USA), rabbit anti-human collagen type I (Col1;
1:200; catalog no. ABIN2473035; Antibodies-Online, Inc., Atlanta,
GA, USA), rabbit anti-human laminin (1:300; catalog no. ab11575,
Abcam) and rabbit anti-human HOXB9 (1:500; catalog no. ABIN952780;
Antibodies-Online, Inc.) at room temperature overnight. After
washing with PBS, Envision+/HPR against rabbit (GK400305; Dako;
Agilent Technologies, Inc., Santa Clara, CA, USA) was added and
incubated for 30 min, followed by detection with
3,3′-diaminobenzidine detection (Sigma-Aldrich; Thermo Fisher
Scientific, Inc.). Slides were counterstained with hematoxylin. The
sections were washed in water, mounted and observed under a Zeiss
Axiophot microscope (Carl Zeiss AG, Oberkochen, Germany). The
images were obtained and analyzed using an Image-Pro plus software
version 6.0 system (Media Cybernetics, Inc., Rockville, MD,
USA).
Establishment of stable HOXB9
overexpression and knockdown in human FBs
A short hairpin (sh)RNA with high HOXB9 knockdown
efficiency was used [HOXB9 small interfering (si)RNA1:
CCCUUCAAUUUGUAGACUCUU], and a shRNA with no effect on HOXB9 levels
was used as a control (Control siRNA: UUCUCCGAACGUGUCACGU). As
described previously (26), the
lentivirus was made using a three-plasmid packaging system (catalog
no. 632455; Takara Biotechnology Co., Ltd., Dalian, China). The
lentiviral vector system had three parts before packaging,
including a pSIREN-RetroQ-ZsGreen1 vector, a pHelper 1.0 (gag/pol
element) vector, and a pHelper 2.0 (VSVG element) vector. shRNAs
targeting HOXB9 and the negative control were harbored in the
pSIREN-RetroQ-ZsGreen1 vector. DNA sequencing confirmed the
accurate insertion of the HOXB9shRNA. The pHelper 1.0 plasmid (15
µg), the pHelper 2.0 plasmid (10 µg) and the
pSIREN-RetroQ-ZsGreen1-HOXB9shRNA plasmid (Clontech Laboratories,
Inc., Mountainview, CA, USA) (20 µg) were cotransfected into
subconfluent 293T cells in serum-free medium using a cationic
liposome based transfection reagent (Lipofectamine® 2000; Thermo
Fisher Scientific, Inc.). Following 10 h of incubation, the medium
was replaced. Recombinant deficient lentiviruses with HOXB9shRNA
were harvested 72 h later. The FBs were transfected with these
recombinant deficient lentiviruses and the clones with green
fluorescence were selected under fluorescence microscopy, following
which the experimental procedures were performed. Meanwhile, a
pLKO.1-puro vector (Addgene, Inc., Cambridge, MA, USA), harboring
the HOXB9 cDNA (NCBI reference sequence ID, AH010085) lentivirus
was harvested 72 h after transfection, and 5 mg/ml polybrene was
added. Subconfluent FBs were infected with the harvested
lentivirus, and were selected in 2 mg/ml puromycin. Individual
colonies of stable overexpression of HOXB9 and control colonies
were isolated and DNA sequencing confirmed the accurate insertion
of the HOXB9 cDNA.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was isolated from FBs using TRIzol®
reagent (Bio-Rad Laboratories, Inc., Hercules, CA, USA) according
to the manufacturer's protocol. The RNA samples were treated with
RNase-free DNase (Roche Applied Science, Mannheim, Germany) at 37°C
for 30 min, followed by phenolchloroform extraction and ethanol
precipitation. Total RNA (1.2 µg) was used in the reverse
transcription reaction with oligodT primers. Samples were amplified
in an RT-PCR System (Applied Biosystems; Thermo Fisher Scientific,
Inc.) using the following conditions: Initial denaturation at 94°C
for 30 sec, followed by 30 cycles denaturation at 95°C for 30 sec,
annealing at 55°C for 30 sec, and extension at 72°C for 30 sec
followed by 6 min at 72°C. The final relative expressions of mRNA
were compared using the 2−#x0394;ΔCq method as
previously described (27). The
primers used were as follows: GAPDH forward,
5′-CTGCCGCTCCATCGTGGCTAG-3′ and reverse, 5′-GCCATGCAATGAAT-3′ for
GAP DH (107 bp band product); forward,
5′-CGCTGGTTCTAAAACTGAGTTTCTG-3′ and reverse,
5′-ATCCTGAGAAGGGCGGTGATC-3′ for laminin (117 bp band product);
forward, 5′-CCTGTCCTCTTTTCTCGTCTAATC-3′; and reverse,
5′-CTTCTCTTGGAACCAGCGTCTGG-3′ for HOXB9 (126 bp band product);
forward, 5′-GCTCCATCCAACTGGCGTGTCTGATCC-3′ and reverse,
5′-AAGACGAACCCCGTGGCATGT-3′ for HOXB9 (186 bp band product); and
forward, 5′-AGCGAAGATGGTTCACTGGGCTCCA-3′ and reverse,
5′-GACTTGTCTCTCCTCTAGGAATCC-3′ for Col1 (196 bp band product).
Western blot analysis
For western blot cell lysates were prepared using
radioimmunoprecipitation assay buffer (50 mM Tris-HCl, pH 7.4),
followed by sonication five times for 10 sec on ice. The lysate was
transferred to a microcentrifuge tube and centrifuged at 14,000 × g
for 15 min to pellet the cell debris. The supernatant containing
the protein was retained. Proteins (30 µg) were separated on a 12%
denaturing polyacrylamide gel and electro-transferred to a
polyvinylidene difluoride membrane (Merck KGaA). The membranes were
blocked with TBS with 0.1% Tween-20 containing 5% fat-free dry
milk, and subsequently incubated with a primary antibody overnight
at 4°C. The following rabbit anti-human primary antibodies were
used: FN (1:1,000; catalog no. ab61214; Abcam), Col1 (1:5,000;
catalog no. ABIN2473035; Antibodies-Online, Inc.), extracellular
signal-regulated kinase (ERK) 1/2 (1:2,000; catalog no. ab196883;
Abcam), phosphorylated (p)-ERK1/2 (1:1,000, catalog no. 9101, Cell
Signaling Technology, Inc., Danvers, MA, USA), c-Jun N-terminal
kinase (JNK; 1:1,000; catalog no. sc-572; Santa Cruz Biotechnology,
Inc., Dallas, TX, USA), p-JNK (1:1,000; catalog no. 4668; Cell
Signaling Technology, Inc.), p38 (1:200; catalog no. ab7952;
Abcam), p-p38 (1:1,000, catalog no. 9211, Cell Signaling
Technology, Inc.), HOXB9 (1:500; catalog no. ABIN952780;
Antibodies-Online, Inc.) and GAPDH (1:5,000; catalog no. 2-RGM2;
Advanced ImmunoChemical Inc., Long Beach, CA, USA). Following this,
membranes were incubated for 1 h at room temperature with
corresponding horseradish peroxidase-conjugated anti-rabbit
secondary antibodies (1:2,000; catalog no. sc-2004; Santa Cruz
Biotechnology, Inc.) and developed using enhanced chemiluminescence
(catalog no. 34077; Thermo Fisher Scientific, Inc.).
Gel contraction assay
The contraction capacity of FBs was detected using
the cell contraction assay kit (CBA-201; Cell Biolabs, Inc., San
Diego, CA, USA). FB-embedded collagen gels were prepared as
described previously (28,29). Briefly, four 24-well plates were
pre-treated with 0.4% bovine serum albumin (Sigma-Aldrich; Merck
KGaA) for 2 h. A 0.5-ml suspension containing 1×106 FBs (FB-mock),
FBs infected with an empty lentivirus (FB-negative), and
FB-HOXB9-overexpressing (FB-HOXB9over) or FB-HOXB9-silenced
(FB-HOXB9si) cells, and 2 mg/ml collagen, were added into the
wells. Following this, FB-mock, FB-negative, FB-HOXB9over and
FB-HOXB9si cells were incubated at 37°C for 48 h for
polymerization, followed by mechanical detachment from the sides of
the wells. Gels were imaged at 0, 12, 24, 36 and 48 h, and the
images were analyzed using Image-Pro Plus software, version
6.0.
Co-immunoprecipitation (co-IP)
Transfected cells were lysed in cell lysis buffer
(50 mM Tris-HCl, pH 8.0; 150 mM NaCl, 1 mM EDTA, 1% Nonidet P-40,
10% glycerol with protease inhibitor mixture) for 1 h. Whole cell
extracts were collected and precleared. The beads coated with
antibodies were incubated with the precleared whole cell extracts
at 4°C overnight. The beads were washed with cell lysis buffer four
times, following which beads were removed by low speed
centrifugation (2,000 × g) at room temperature for 1 min.
Precleared supernatants incubated with anti-HOXB9, ERK, JNK or p38
antibodies for 2 h in the presence of Protein A-agarose beads.
Immunoprecipitated material was collected on the beads, washed
extensively and purified proteins were eluted in SDS-PAGE sample
buffer. Precipitated proteins were analyzed by western blotting, as
described above.
Statistical analysis
To determine significant differences between groups,
one-way analysis of variance followed by Bonferroni's post hoc test
was performed. The statistical analyses were performed using SPSS
software version 20.0 (IBM SPSS, Armonk, NY, USA). GraphPad Prism
software version 6.0 (GraphPad Software, Inc., La Jolla, CA, USA)
was employed to produce the majority of figures. Data are presented
as the mean ± standard deviation. P<0.05 was considered to
indicate a statistically significant difference.
Results
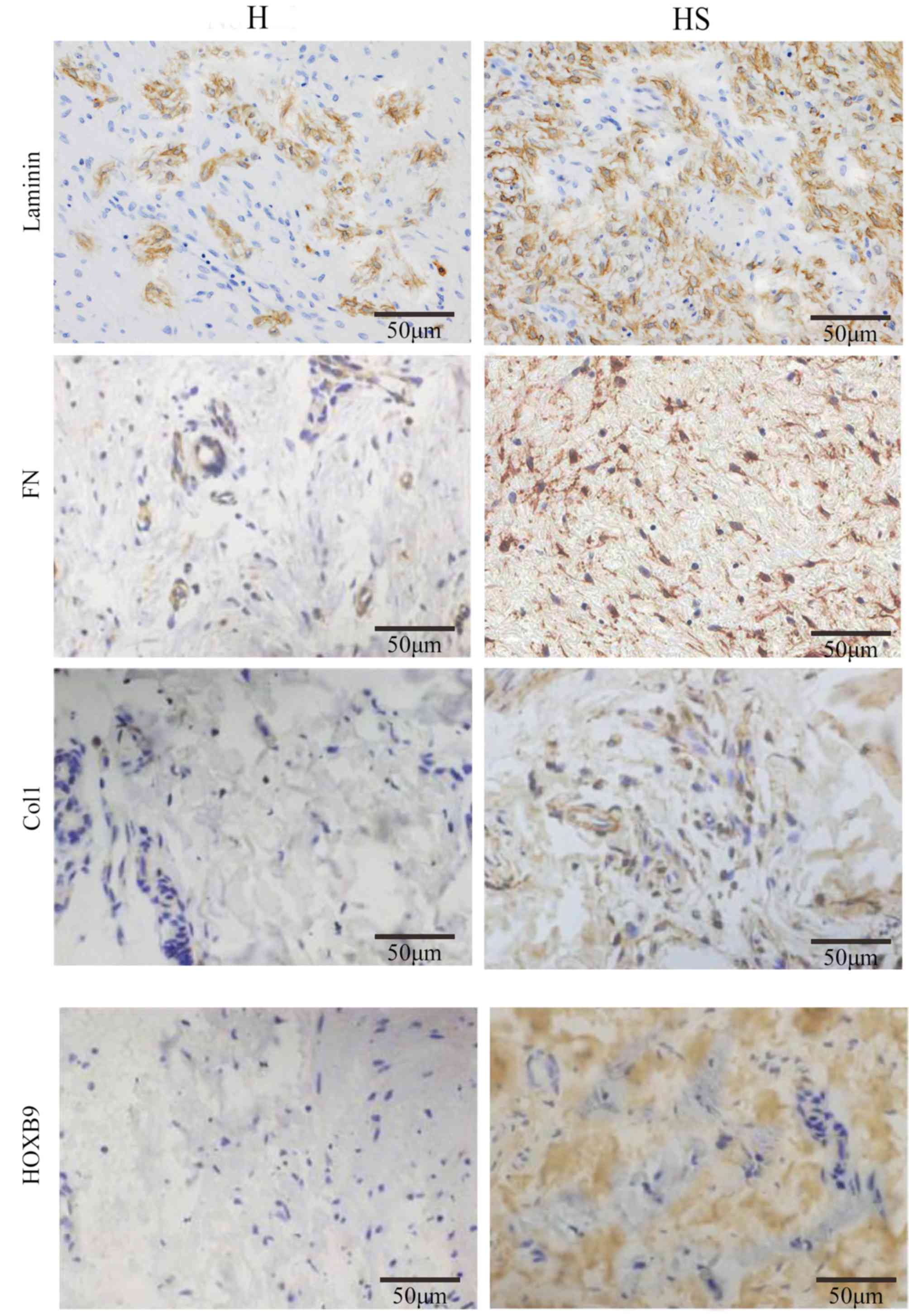
Expression levels of Col1, FN, laminin
and HOXB9 are elevated in hypertrophic scar tissues compared with
healthy skin tissues
The ECM consists of numerous components, including
collagen, FN and laminin. Previous studies have demonstrated that
the ECM is more abundant in hypertrophic scar tissue than that in
healthy skin (30–32). The present study tested the
expression of Col1, FN, laminin and HOXB9 in hypertrophic scar and
healthy skin tissues. These four proteins were elevated in the
hypertrophic scar tissues compared with corresponding healthy skin
tissues in six patients (P<0.05), suggesting that the samples
selected were in accordance with the pathological standard of
hypertrophic scar tissue (Fig.
1).
Expression of Col1, FN and laminin are
associated with HOXB9
A lentiviral system was used to overexpress or
silence HOXB9 in FBs (FB-HOXB9over and FB-HOXB9si, respectively).
Compared with the FB-mock group, mRNA and protein expression levels
of HOXB9 were downregulated in FB-HOXB9si cells; whereas they were
upregulated in FB-HOXB9over cells (Fig. 2A). Laminin protein expression
decreased by 60.33±3.25% in the FB-HOXB9si group and increased by
4.12±0.33 fold in the FB-HOXB9over group, compared with the FB-mock
group (P<0.01; Fig. 2B). FN
protein expression levels decreased by 52.27±4.33% in the
FB-HOXB9si group and increased by 1.18±0.23-fold in the
FB-HOXB9over group, compared with the FB-mock group (P<0.01;
Fig. 2C). Col1 protein expression
levels decreased by 44.53±2.85% in the FB-HOXB9si group and
increased by 2.12±0.37-fold FB-HOXB9over group, compared with the
FB-mock group (P<0.01; Fig.
2D). mRNA expression levels of laminin and Col1 were consistent
with the corresponding protein expression levels (P<0.01;
Fig. 2B and D), whereas FN mRNA
expression levels were not influenced by HOXB9 (P>0.05; Fig. 2C).
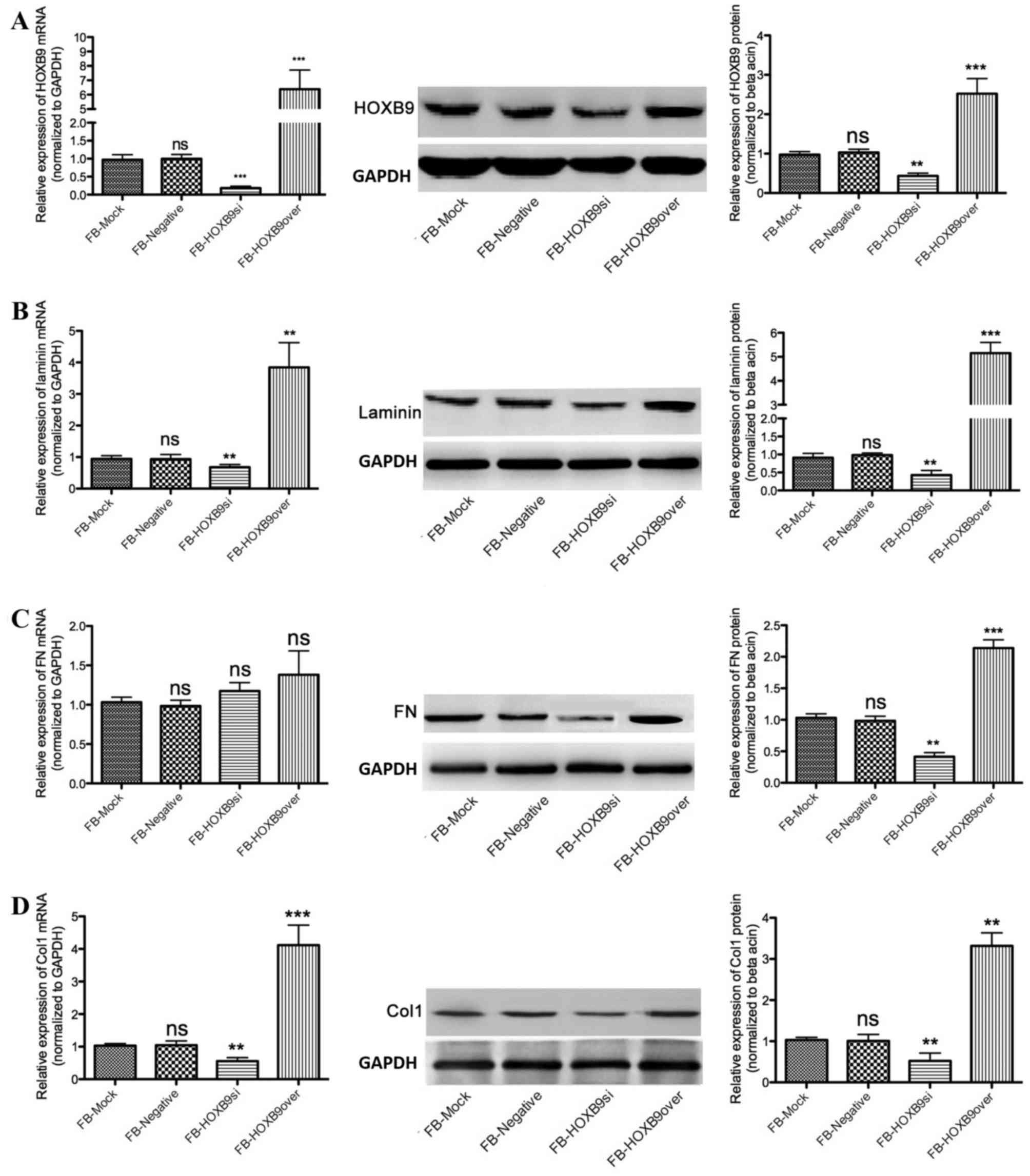 | Figure 2.HOXB9 regulates the expression of
laminin, FN and Col1 in FBs. Reverse transcription-quantitative
polymerase chain reaction and western blotting analyses were
performed to assess differential mRNA and protein expression
levels, respectively, in FB mock, FB-negative, FB-HOXB9si and
FB-HOXB9over cells. (A) FBs were constructed to stably overexpress
(FB-HOXB9over) or silence (FB-HOXB9si) HOXB9 using lentiviruses
(P<0.001). (B) mRNA and protein expression levels of laminin
were decreased in FB-HOXB9si cells and increased in FB-HOXB9over
cells, compared with the FB-mock and FB-negative groups
(P<0.01). (C) FN and protein expression levels were decreased in
FB-HOXB9si cells and increased in FB-HOXB9over cells, compared with
the FB-mock and FB-negative groups (P<0.01); however, no
significant differences were observed in mRNA expression levels
(P>0.05). (D) Col1 mRNA and protein expression levels were
decreased in FB-HOXB9si cells and increased in FB-HOXB9over cells,
compared with FB-mock and FB-negative cells (P<0.01). Data are
presented as the mean ± standard deviation of three independent
experiments. **P<0.01 and ***P<0.001 vs. FB-mock and
FB-negative. FB, fibroblast; FN, fibronectin; HOXB9, homeobox B9;
si, silenced; over, overexpressing; Col1; collagen type I; ns,
non-significant. |
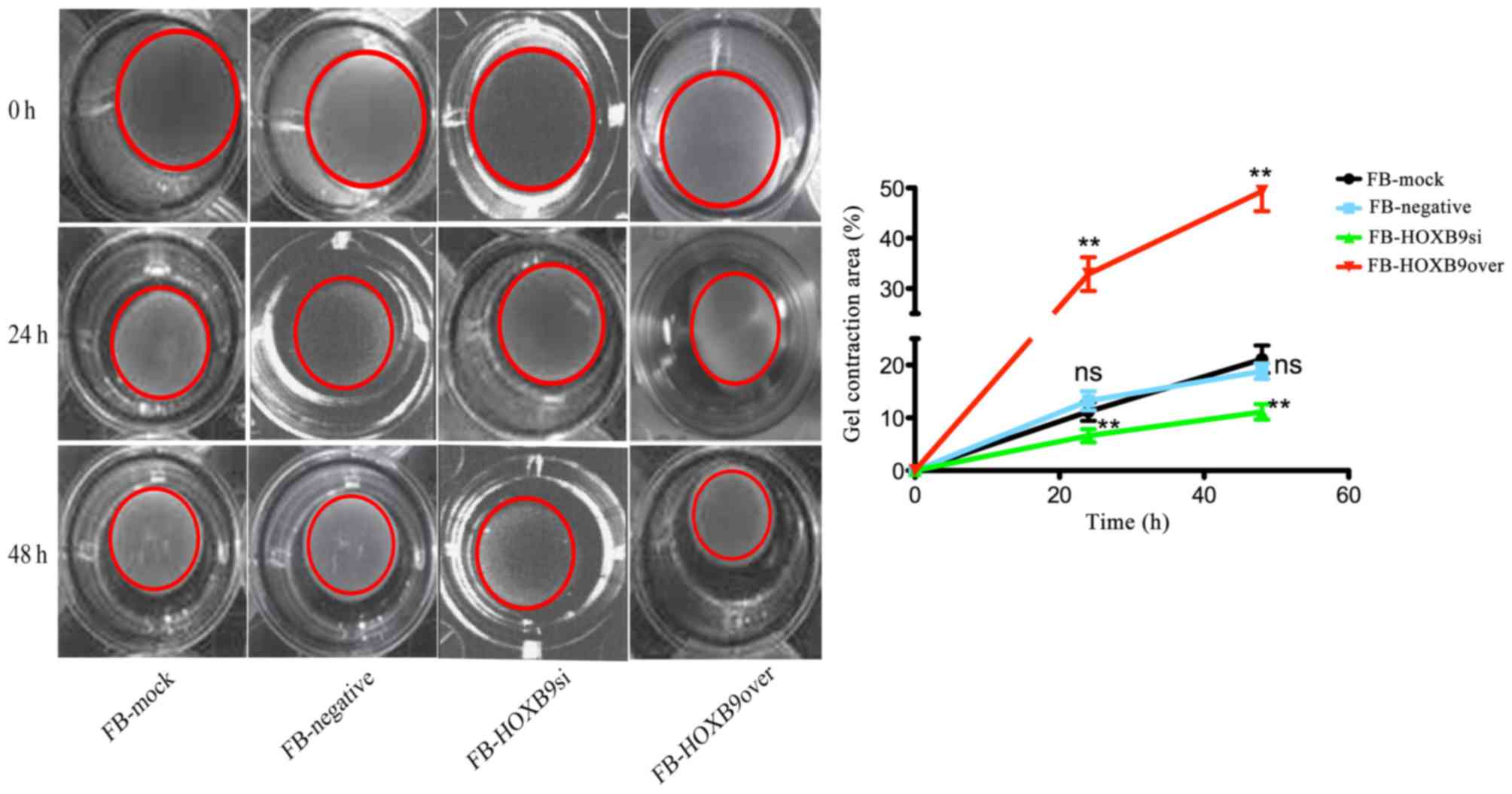
HOXB9 contributes to gel contraction
caused by FBs
FBs contract the surrounding matrix, thereby
contracting the gel. Compared with healthy skin, hypertrophic scar
tissues are characterized by stronger contraction ability. Given
that HOXB9 is highly expressed in hypertrophic scar tissues,
investigating the potential roles of HOXB9 in the formation of
hypertrophic scars may be significant. To demonstrate whether HOXB9
contributes to gel contraction, FB-mock, FB-negative, FB-HOXB9si
and FB-HOXB9over cells were seeded into gels and the gel size was
measured 0, 24 and 48 h after seeding. The results clearly
demonstrated that HOXOB9 contributes to the gel contraction caused
by FBs; the FB-HOXB9over group had a significantly increased gel
contraction area at 24 and 28 h compared with the FB-HOXB9si group
(P<0.01, Fig. 3). Knocking down
HOXB9 may attenuate the contraction ability, whereas overexpressing
HOXB9 enhances this ability, which is consistent with the results
that HOXB9 is highly expressed in hypertrophic scar tissues.
HOXB9 mediates activation of the
mitogen-activated protein kinase (MAPK) pathway
MAPK cascades have been demonstrated to serve
crucial roles in transmitting extracellular signals to cellular
response, leading to an appropriate physiological response
including cellular proliferation, differentiation, development,
inflammatory responses and apoptosis in mammalian cells. A total of
three MAPK families have been characterized, namely classical MAPK
(additionally known as ERK), c-Jun JNK and p38 kinase. Given that
the production of laminin, FN and Col1 are initiated by their
corresponding genes activated predominantly by the MAPK signaling
pathway, whether HOXB9 mediates MAPK activation was detected. The
results clearly demonstrated that knocking down HOXB9 decreased
p-ERK, p-JNK and p-p38 production by 40.12±5.79 (Fig. 4A), 68.37±8.72 (Fig. 4B) and 51.25±4.76% (Fig. 4C), respectively, compared with the
FB-mock group (P<0.05). In contrast, overexpressing HOXB9
increased p-ERK, p-JNK and p-p38 production by 70.33±9.21%
(Fig. 4A), 69.32±4.93% (Fig. 4B) and 1.8±0.42 fold (Fig. 4C), respectively, compared with the
FB-mock group (P<0.01).
 | Figure 4.HOXB9 activates the mitogen-activated
protein kinase signaling pathway via phosphorylating ERK, JNK and
p38. Representative western blot images and quantification of
protein expression levels of (A) ERK and p-ERK, (B) JNK and p-JNK,
and (C) p38 and P-38. Data are presented as the mean ± standard
deviation of three independent experiments. *P<0.05 and
**P<0.01; ***P<0.001 vs. FB-mock and FB-negative. FB,
fibroblast; HOXB9, homeobox B9; si, silenced; over, overexpressing;
p, phosphorylated; ERK, extracellular signal-regulated kinase; JNK,
c-Jun N-terminal kinase; ns, non-significant. |
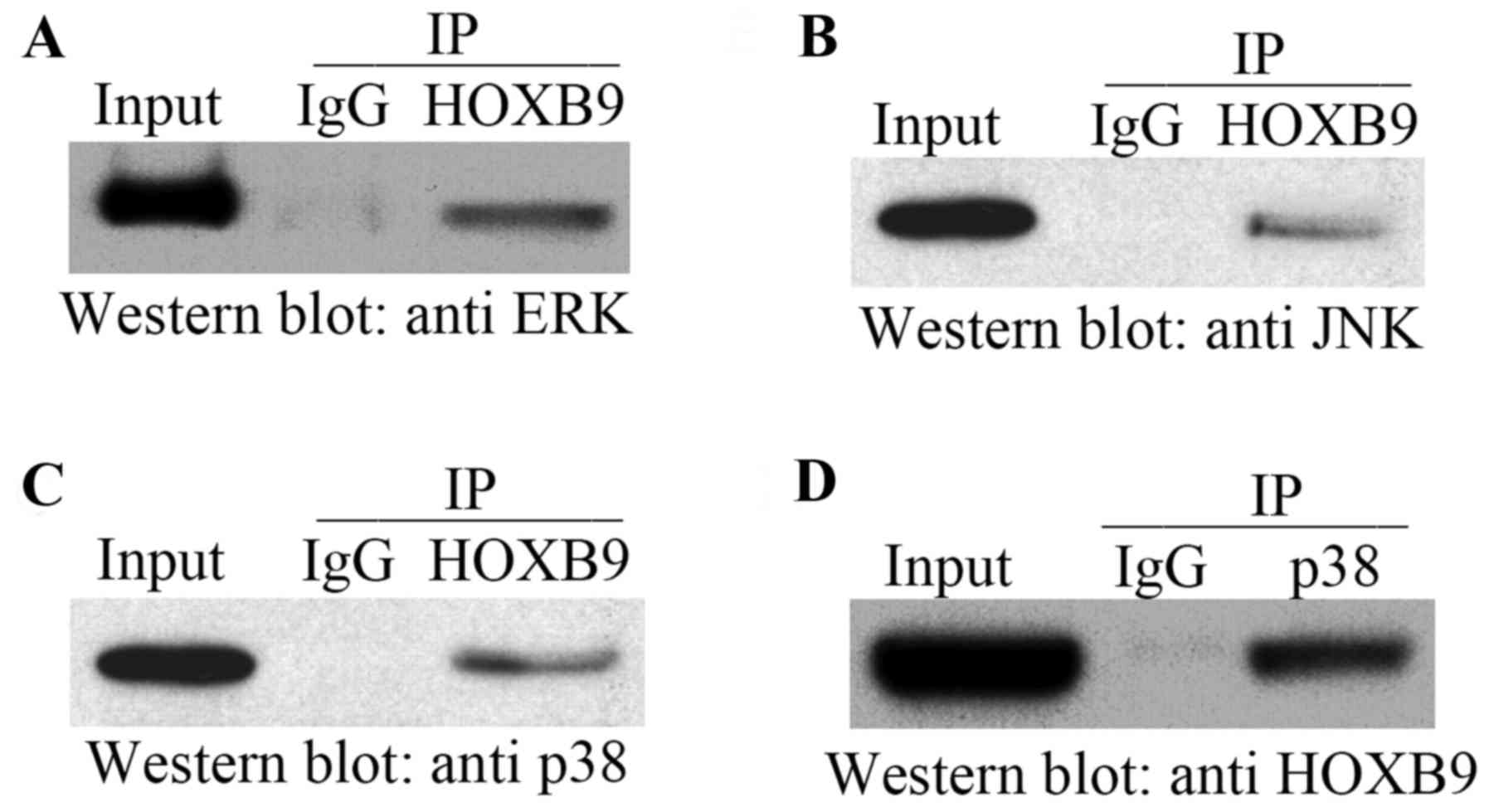
HOXB9 interacts directly with p38
To further understand the underlying mechanisms by
which HOXB9 mediates the MAPK signaling pathway, co-IP experiments
were performed using FB extracts to confirm whether HOXB9 interacts
with ERK (Fig. 5A), JNK (Fig. 5B) and p38 (Fig. 5C). All three molecules were
demonstrated to co-precipitate with HOXB9. However, the reciprocal
co-IP experiment revealed that only p38 was present in the HOXB9
precipitate (Fig. 5D). These
results confirmed that HOXB9 interacts with p38 in FB cells. It is
unclear why the interaction between HOXB9, and ERK and JNK, was
inefficient; it may be influenced by temperature and buffer
conditions, posttranslational modifications, additional cellular
factors and/or the presence of the linker region in ERK and
JNK.
Discussion
The current study aimed to investigate the role of
HOXB9 in hypertropic scar formation and the underlying mechanisms.
HOXB9 was demonstrated as highly expressed in hypertrophic scar
tissues, accompanied by high expression levels of laminin, FN and
Col1. To further examine the potential roles of HOXB9 in
hypertrophic scar formation, FB-HOXB9si and FB-HOXB9over vectors
were constructed to silence or overexpress the HOXB9 gene,
respectively. Western blotting demonstrated that overexpressing
HOXB9 elevated laminin, FN and Col1 protein expression, while
knocking down HOXB9 decreased their expression. Gel contraction
assay supported these findings; increased and reduced HOXB9
expression resulted in enhanced and reduced contraction ability of
FBs, respectively. HOXB9 elevated the levels of phosphorylated ERK,
JNK and p38, and co-IP revealed an interaction between HOXB9 and
p38. To the best of the authors' knowledge, this is the first study
to investigate the association between HOXB9 and hypertrophic scar
formation, and the underlying mechanisms.
Hypertrophic scars develop from deep dermis injury,
including burn injury, surgery and piercings. The annual incidence
is ~100 million patients in the developed world alone each year
(33). Hypertrophic scars may
greatly decrease the quality of life of the patient, both
physically and psychologically, by causing pruritus, pain and
contractures. Elaborate efforts have been made to understand the
underlying mechanisms of hypertrophic scar formation; however, no
satisfactory therapeutic approaches are currently available for
this type of lesion.
The ECM consists of structural proteins, collagens,
laminins, elastins and FNs to provide flexibility. Abnormal
reconstruction of the ECM during wound healing contributes to the
formation of hypertrophic scars. The present study collected
hypertrophic scar samples from six adults and demonstrated that
laminin, FN and Col1 are highly expressed in hypertrophic scar
tissues, which is consistent with previous studies (34).
Previous studies have revealed an association
between the HOX genes and wound healing; however, to the best of
the authors' knowledge, there are no studies demonstrating the
potential association between HOX genes with hypertrophic scar
formation. The present study revealed that HOXB9 is highly
expressed in hypertrophic scar tissues, implying a potential role
in facilitating the hypertrophic scar. To investigate the
underlying mechanisms by which HOXB9 may benefit hypertrophic scar
formation, FN-HOXB9si and FN-HOXB9over stable cells were
constructed. Knocking down HOXB9 markedly decreased laminin, FN and
Col1 protein expression levels, while overexpressing HOXB9 elevated
their expression. This effect was consistent with the results of
the clinical samples, indicating that HOXB9 is involved in the
remodeling of the ECM, which is crucial for hypertrophic scar
formation. In order to further validate these results, a gel
contraction assay was performed to investigate HOXB9 function in
hypertrophic scar formation. Gel contraction in the FN-HOXBsi group
was significantly decreased, whereas overexpressing HOXB9 in FBs
enhanced the contraction ability and greatly increased gel
contraction, supporting the hypothesis that HOXB9 facilitates
hypertrophic scar formation.
The underlying mechanisms by which HOXB9 upregulates
the expression of laminin, FN and Col1 were investigated.
Increasing evidence has indicated that the MAPK signaling pathway
serves an important role in reconstruction of the ECM (35–37).
These results demonstrated that knocking down HOXB9 in FBs
decreased levels of p-ERK, p-JNK and p-p38, which belong to the
MAPK family; whereas overexpressing HOXB9 increased their
expression levels, implying that HOXB9 activates the MAPK signaling
pathway (38). Co-IP assays
demonstrated that HOXB9 interacts directly with ERK, JNK and p38;
however, reciprocal co-IP indicated that only p38 interacts with
HOXB9. This interaction may lead to accumulation of p-p38, p-ERK
and p-JNK, activating MAPK and subsequently elevating the
expression levels of laminin, FN and Col1, which results in
reconstruction of the ECM and facilitates hypertrophic scar
formation. As the MAPK signaling pathways relay, amplify and
integrate signals from a wide range of stimuli from the ECM
including cytokines, collagen and fibronectin (39), it is possible that these elevated
ECM components in return strengthened the activated MAPK signaling
pathways (Fig. 6).
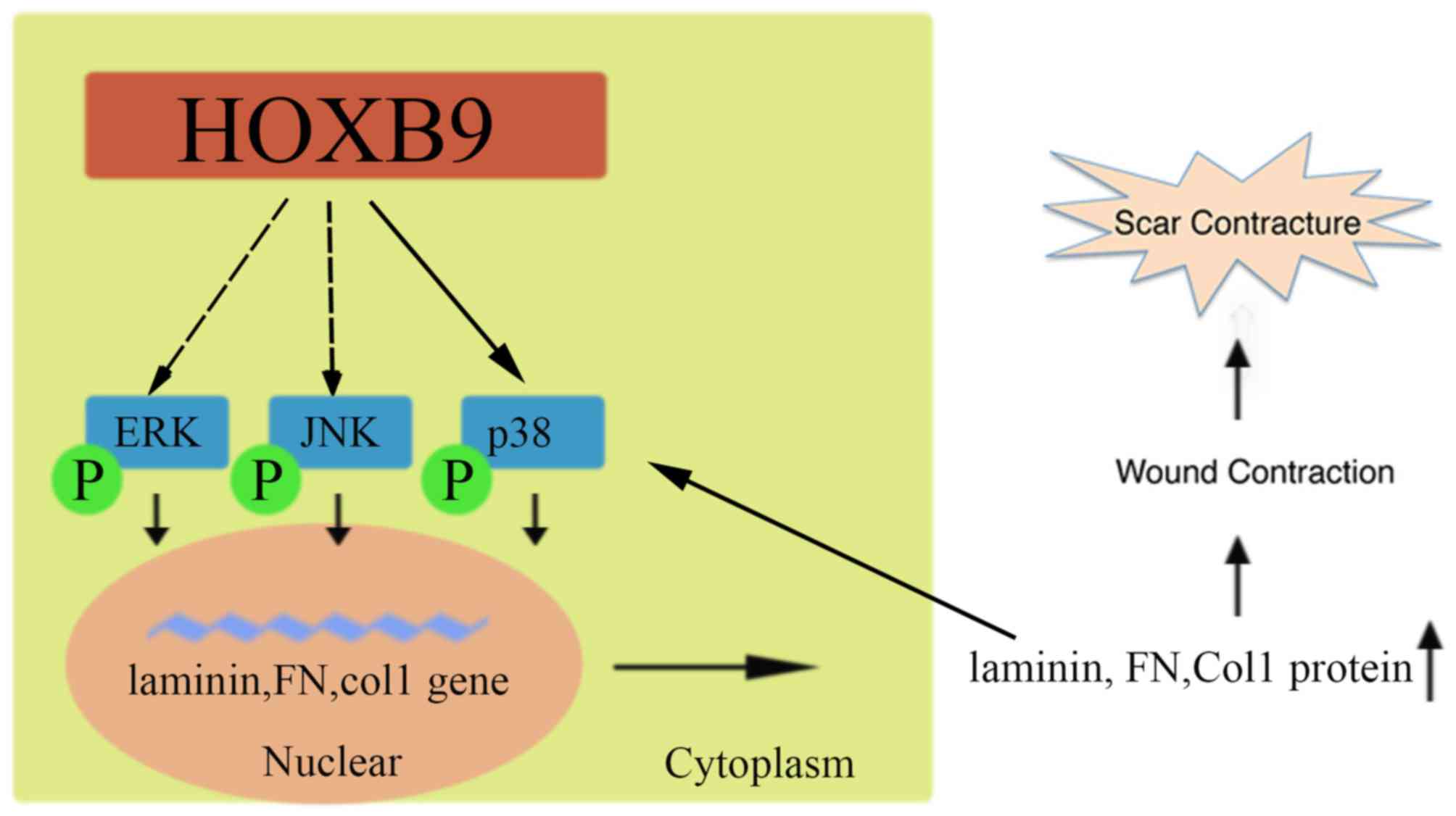 | Figure 6.Schematic diagram of HOXB9
facilitating the formation of a hypertrophic scar. HOXB9 activates
the MAPK pathway by phosphorylating ERK, JNK and p38, potentially
by interacting directly with p38. Activated MAPK leads to the
transcription of laminin, FN and Col1 mRNA, increasing their
protein expression levels, which reconstructs the ECM and
contributes to hypertrophic scar formation. Increased laminin, FN
and Col1 may in return increase p-ERK, p-JNK and p-p38 levels. FN,
fibronectin; Col1, collagen type I; HOXB9, homeobox B9; ERK,
extracellular signal-regulated kinase; JNK, c-Jun N-terminal
kinase; p, phosphorylated; MAPK, mitogen-activated protein
kinase. |
Notably, no reciprocal interactions were observed
between HOXB9, ERK and JNK, although HOXB9 upregulated p-ERK and
p-JNK levels, suggesting that additionally underlying mechanisms by
which HOXB9 may induces the phosphorylation of ERK and JNK should
be investigated. Animal models used for scar research should
additionally be examined to further verify these findings.
In conclusion, the present study demonstrated that
HOXB9 effectively facilitated hypertrophic scar formation and
strengthened the contractile ability of FBs via activating the MAPK
signaling pathway in vivo. HOXB9 may serve as a potential
therapeutic target for the treatment of hypertrophic scars or other
fibroproliferative disorders, and requires further
investigation.
Acknowledgements
The authors would like to thank Professor Shengguo
Shan (Renmin Hospital of Wuhan University, Wuhan, China) for his
assistance in preparing the manuscript.
References
|
1
|
Rabello FB, Souza CD and Júnior JA Farina:
Update on hypertrophic scar treatment. Clinics (Sao Paulo).
69:565–573. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Branski LK, Rennekampff HO and Vogt PM:
Keloid and hypertrophic scar treatment modalities. An update.
Chirurg. 83:831–846. 2012.(In German). View Article : Google Scholar
|
|
3
|
Hayashi T, Furukawa H, Oyama A, Funayama
E, Saito A, Murao N and Yamamoto Y: A new uniform protocol of
combined corticosteroid injections and ointment application reduces
recurrence rates after surgical keloid/hypertrophic scar excision.
Dermatol Surg. 38:893–897. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Huang D, Shen KH and Wang HG: Pressure
therapy upregulates matrix metalloproteinase expression and
downregulates collagen expression in hypertrophic scar tissue. Chin
Med J (Engl). 126:3321–3324. 2013.PubMed/NCBI
|
|
5
|
Chesnut C, Mednik S and Lask G:
Hypertrophic scar treatment with intralesional triamcinolone
acetonide and pulsed dye laser results in necrosis. Cutis.
94:E12–E13. 2014.PubMed/NCBI
|
|
6
|
On HR, Lee SH, Lee YS, Chang HS, Park C
and Roh MR: Evaluating hypertrophic thyroidectomy scar outcomes
after treatment with triamcinolone injections and copper bromide
laser therapy. Lasers Surg Med. 47:479–484. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Al-Mohamady Ael-S, Ibrahim SM and Muhammad
MM: Pulsed dye laser versus long pulsed Nd:YAG laser in the
treatment of hypertrophic scars and keloid: A comparative
randomized split-scar trial. J Cosmet Laser Ther. 18:208–212. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhu Z, Ding J, Shankowsky HA and Tredget
EE: The molecular mechanism of hypertrophic scar. J Cell Commun
Signal. 7:239–252. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Werner S, Krieg T and Smola H:
Keratinocyte-fibroblast interactions in wound healing. J Invest
Dermatol. 127:998–1008. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li B and Wang JH: Fibroblasts and
myofibroblasts in wound healing: Force generation and measurement.
J Tissue Viability. 20:108–120. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gehring WJ, Affolter M and Bürglin T:
Homeodomain proteins. Annu Rev Biochem. 63:487–526. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Apiou F, Flagiello D, Cillo C, Malfoy B,
Poupon MF and Dutrillaux B: Fine mapping of human HOX gene
clusters. Cytogenet Cell Genet. 73:114–115. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mace KA, Hansen SL, Myers C, Young DM and
Boudreau N: HOXA3 induces cell migration in endothelial and
epithelial cells promoting angiogenesis and wound repair. J Cell
Sci. 118:2567–2577. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hansen SL, Myers CA, Charboneau A, Young
DM and Boudreau N: HoxD3 accelerates wound healing in diabetic
mice. Am J Pathol. 163:2421–2431. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Mack JA and Maytin EV: Persistent
inflammation and angiogenesis during wound healing in K14-directed
Hoxb13 transgenic mice. J Invest Dermatol. 130:856–865. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mack JA, Abramson SR, Ben Y, Coffin JC,
Rothrock JK, Maytin EV, Hascall VC, Largman C and Stelnicki EJ:
Hoxb13 knockout adult skin exhibits high levels of hyaluronan and
enhanced wound healing. FASEB J. 17:1352–1354. 2003.PubMed/NCBI
|
|
17
|
Chen F and Capecchi MR: Paralogous mouse
Hox genes, Hoxa9, Hoxb9 and Hoxd9, function together to control
development of the mammary gland in response to pregnancy. Proc
Natl Acad Sci USA. 96:541–546. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Seki H, Hayashida T, Jinno H, Hirose S,
Sakata M, Takahashi M, Maheswaran S, Mukai M and Kitagawa Y: HOXB9
expression promoting tumor cell proliferation and angiogenesis is
associated with clinical outcomes in breast cancer patients. Ann
Surg Oncol. 19:1831–1840. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kwon OS, Oh E, Park JR, Lee JS, Bae GY,
Koo JH, Kim H, Choi YL, Choi YS, Kim J and Cha HJ: GalNAc-T14
promotes metastasis through Wnt dependent HOXB9 expression in lung
adenocarcinoma. Oncotarget. 6:41916–41928. 2015.PubMed/NCBI
|
|
20
|
Zhan J, Wang P, Niu M, Wang Y, Zhu X, Guo
Y and Zhang H: High expression of transcriptional factor HoxB9
predicts poor prognosis in patients with lung adenocarcinoma.
Histopathology. 66:955–965. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hayashida T, Takahashi F, Chiba N,
Brachtel E, Takahashi M, Godin-Heymann N, Gross KW, Vivanco Md,
Wijendran V, Shioda T, et al: HOXB9, a gene overexpressed in breast
cancer, promotes tumorigenicity and lung metastasis. Proc Natl Acad
Sci USA. 107:1100–1105. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Nguyen DX, Chiang AC, Zhang XH, Kim JY,
Kris MG, Ladanyi M, Gerald WL and Massagué J: WNT/TCF signaling
through LEF1 and HOXB9 mediates lung adenocarcinoma metastasis.
Cell. 138:51–62. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sha S, Gu Y, Xu B, Hu H, Yang Y, Kong X
and Wu K: Decreased expression of HOXB9 is related to poor overall
survival in patients with gastric carcinoma. Dig Liver Dis.
45:422–429. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chang Q, Zhang L, He C, Zhang B, Zhang J,
Liu B, Zeng N and Zhu Z: HOXB9 induction of
mesenchymal-to-epithelial transition in gastric carcinoma is
negatively regulated by its hexapeptide motif. Oncotarget.
6:42838–42853. 2015.PubMed/NCBI
|
|
25
|
He T, Bai X, Yang L, Fan L, Li Y, Su L,
Gao J, Han S and Hu D: Loureirin B inhibits hypertrophic scar
formation via inhibition of the TGF-β1-ERK/JNK Pathway. Cell
Physiol Biochem. 37:666–676. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Root DE, Hacohen N, Hahn WC, Lander ES and
Sabatini DM: Genome-scale loss-of-function screening with a
lentiviral RNAi library. Nat Methods. 3:715–719. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ngo P, Ramalingam P, Phillips JA and
Furuta GT: Collagen gel contraction assay. Methods Mol Biol.
341:103–109. 2006.PubMed/NCBI
|
|
29
|
Vernon RB and Gooden MD: An improved
method for the collagen gel contraction assay. In Vitro Cell Dev
Biol Anim. 38:97–101. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Viera MH, Amini S, Valins W and Berman B:
Innovative therapies in the treatment of keloids and hypertrophic
scars. J Clin Aesthet Dermatol. 3:20–26. 2010.
|
|
31
|
Shaarawy E, Hegazy RA and Hay RM Abdel:
Intralesional botulinum toxin type A equally effective and better
tolerated than intralesional steroid in the treatment of keloids: A
randomized controlled trial. J Cosmet Dermatol. 14:161–166. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gauglitz GG, Bureik D, Dombrowski Y,
Pavicic T, Ruzicka T and Schauber J: Botulinum toxin A for the
treatment of keloids. Skin Pharmacol Physiol. 25:313–318. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gauglitz GG, Korting HC, Pavicic T,
Ruzicka T and Jeschke MG: Hypertrophic scarring and keloids:
Pathomechanisms and current and emerging treatment strategies. Mol
Med. 17:113–125. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Xue M and Jackson CJ: Extracellular matrix
reorganization during wound healing and its impact on abnormal
scarring. Adv Wound Care (New Rochelle). 4:119–136. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Crean JK, Finlay D, Murphy M, Moss C,
Godson C, Martin F and Brady HR: The role of p42/44 MAPK and
protein kinase B in connective tissue growth factor induced
extracellular matrix protein production, cell migration, and actin
cytoskeletal rearrangement in human mesangial cells. J Biol Chem.
277:44187–44194. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Xiao G, Gopalakrishnan R, Jiang D, Reith
E, Benson MD and Franceschi RT: Bone morphogenetic proteins,
extracellular matrix, and mitogen-activated protein kinase
signaling pathways are required for osteoblast-specific gene
expression and differentiation in MC3T3-E1 cells. J Bone Miner Res.
17:101–110. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Qin H, Ishiwata T, Wang R, Kudo M,
Yokoyama M, Naito Z and Asano G: Effects of extracellular matrix on
phenotype modulation and MAPK transduction of rat aortic smooth
muscle cells in vitro. Exp Mol Pathol. 69:79–90. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kyriakis JM and Avruch J: Mammalian MAPK
signal transduction pathways activated by stress and inflammation:
A 10-year update. Physiol Rev. 92:689–737. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang W and Liu HT: MAPK signal pathways
in the regulation of cell proliferation in mammalian cells. Cell
Res. 12:9–18. 2002. View Article : Google Scholar : PubMed/NCBI
|