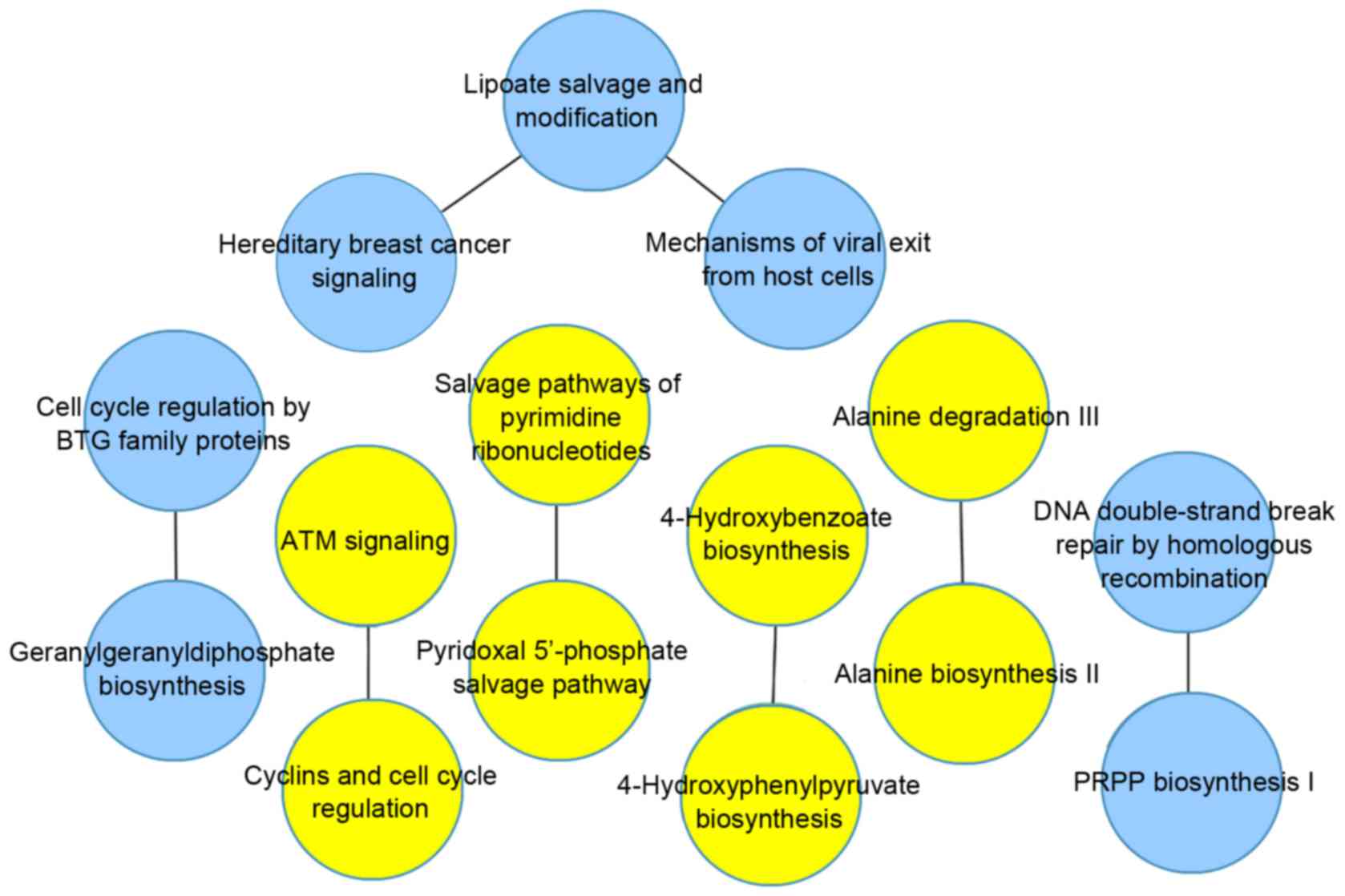
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kumar R, Saraswat MK, Sharma BC, Sakhuja P
and Sarin SK: Characteristics of hepatocellular carcinoma in India:
A retrospective analysis of 191 cases. QJM. 101:479–485. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wong PY, Xia V, Imagawa DK, Hoefs J and Hu
KQ: Clinical presentation of hepatocellular carcinoma (HCC) in
Asian-Americans versus non-Asian-Americans. J Immigr Minor Health.
13:842–848. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lo CM, Ngan H, Tso WK, Liu CL, Lam CM,
Poon RT, Fan ST and Wong J: Randomized controlled trial of
transarterial lipiodol chemoembolization for unresectable
hepatocellular carcinoma. Hepatology. 35:1164–1171. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ma S, Jiao B and Liu X, Yi H, Kong D, Gao
L, Zhao G, Yang Y and Liu X: Approach to radiation therapy in
hepatocellular carcinoma. Cancer Treat Rev. 36:157–163. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Abou-Alfa GK: Hepatocellular carcinoma:
Molecular biology and therapy. Semin Oncol. 33:(6 Suppl 11).
S79–S83. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sherman M: Hepatocellular carcinoma:
Epidemiology, risk factors, and screening. Semin Liver Dis.
25:143–154. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Attwa MH and El-Etreby SA: Guide for
diagnosis and treatment of hepatocellular carcinoma. World J
Hepatol. 7:1632–1651. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Di Bisceglie AM, Sterling RK, Chung RT,
Everhart JE, Dienstag JL, Bonkovsky HL, Wright EC, Everson GT,
Lindsay KL, Lok AS, et al: Serum alpha-fetoprotein levels in
patients with advanced hepatitis C: Results from the HALT-C Trial.
J Hepatol. 43:434–441. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Woo HG, Park ES, Cheon JH, Kim JH, Lee JS,
Park BJ, Kim W, Park SC, Chung YJ, Kim BG, et al: Gene
expression-based recurrence prediction of hepatitis B virus-related
human hepatocellular carcinoma. Clin Cancer Res. 14:2056–2064.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tackels-Horne D, Goodman MD, Williams AJ,
Wilson DJ, Eskandari T, Vogt LM, Boland JF, Scherf U and Vockley
JG: Identification of differentially expressed genes in
hepatocellular carcinoma and metastatic liver tumors by
oligonucleotide expression profiling. Cancer. 92:395–405. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Thomas MB, Jaffe D, Choti MM, Belghiti J,
Curley S, Fong Y, Gores G, Kerlan R, Merle P, O'Neil B, et al:
Hepatocellular carcinoma: Consensus recommendations of the National
Cancer Institute Clinical Trials Planning Meeting. J Clin Oncol.
28:3994–4005. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Papatheodoridis GV, Chan HL, Hansen BE,
Janssen HL and Lampertico P: Risk of hepatocellular carcinoma in
chronic hepatitis B: Assessment and modification with current
antiviral therapy. J Hepatol. 62:956–967. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yao M, Yao DF, Bian YZ, Wu W, Yan XD, Yu
DD, Qiu LW, Yang JL, Zhang HJ, Sai WL and Chen J: Values of
circulating GPC-3 mRNA and alpha-fetoprotein in detecting patients
with hepatocellular carcinoma. Hepatobiliary Pancreat Dis Int.
12:171–179. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lee SY, Song KH, Koo I, Lee KH, Suh KS and
Kim BY: Comparison of pathways associated with hepatitis B- and
C-infected hepatocellular carcinoma using pathway-based class
discrimination method. Genomics. 99:347–354. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ahn T, Lee E, Huh N and Park T:
Personalized identification of altered pathways in cancer using
accumulated normal tissue data. Bioinformatics. 30:i422–i429. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kim BY, Choi DW, Woo SR, Park ER, Lee JG,
Kim SH, Koo I, Park SH, Han CJ, Kim SB, et al:
Recurrence-associated pathways in hepatitis B virus-positive
hepatocellular carcinoma. BMC Genomics. 16:2792015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yoon SY, Kim JM, Oh JH, Jeon YJ, Lee DS,
Kim JH, Choi JY, Ahn BM, Kim S, Yoo HS, et al: Gene expression
profiling of human HBV- and/or HCV-associated hepatocellular
carcinoma cells using expressed sequence tags. Int J Oncol.
29:315–327. 2006.PubMed/NCBI
|
|
19
|
Parkinson H, Kapushesky M, Shojatalab M,
Abeygunawardena N, Coulson R, Farne A, Holloway E, Kolesnykov N,
Lilja P, Lukk M, et al: ArrayExpress-a public database of
microarray experiments and gene expression profiles. Nucleic Acids
Res. 35:(Database Issue). D747–D750. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bolstad BM, Irizarry RA, Astrand M and
Speed TP: A comparison of normalization methods for high density
oligonucleotide array data based on variance and bias.
Bioinformatics. 19:185–193. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Benjamini Y and Yekutieli D: Quantitative
trait Loci analysis using the false discovery rate. Genetics.
171:783–790. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Golub TR, Slonim DK, Tamayo P, Huard C,
Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri
MA, et al: Molecular classification of cancer: Class discovery and
class prediction by gene expression monitoring. Science.
286:531–537. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Orsetti B, Nugoli M, Cervera N, Lasorsa L,
Chuchana P, Rougé C, Ursule L, Nguyen C, Bibeau F, Rodriguez C and
Theillet C: Genetic profiling of chromosome 1 in breast cancer:
Mapping of regions of gains and losses and identification of
candidate genes on 1q. Br J Cancer. 95:1439–1447. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Burgette LF and Reiter JP: Multiple
imputation for missing data via sequential regression trees. Am J
Epidemiol. 172:1070–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tsuji S, Midorikawa Y, Takahashi T, Yagi
K, Takayama T, Yoshida K, Sugiyama Y and Aburatani H: Potential
responders to FOLFOX therapy for colorectal cancer by Random
Forests analysis. Br J Cancer. 106:126–132. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Colaprico A, Cava C, Bertoli G, Bontempi G
and Castiglioni I: Integrative analysis with Monte Carlo
Cross-Validation reveals miRNAs regulating pathways Cross-talk in
aggressive breast cancer. Biomed Res Int. 2015:8313142015.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Saito K, Shirasago Y, Suzuki T, Aizaki H,
Hanada K, Wakita T, Nishijima M and Fukasawa M: Targeting cellular
squalene synthase, an enzyme essential for cholesterol
biosynthesis, is a potential antiviral strategy against hepatitis C
virus. J Virol. 89:2220–2232. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Xu Z, Choi J, Yen TS, Lu W, Strohecker A,
Govindarajan S, Chien D, Selby MJ and Ou J: Synthesis of a novel
hepatitis C virus protein by ribosomal frameshift. EMBO J.
20:3840–3848. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ratinier M, Boulant S, Combet C,
Targett-Adams P, McLauchlan J and Lavergne JP: Transcriptional
slippage prompts recoding in alternate reading frames in the
hepatitis C virus (HCV) core sequence from strain HCV-1. J Gen
Virol. 89:1569–1578. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Carbajo-Pescador S, Ordoñez R, Benet M,
Jover R, García-Palomo A, Mauriz JL and González-Gallego J:
Inhibition of VEGF expression through blockade of Hif1α and STAT3
signalling mediates the anti-angiogenic effect of melatonin in
HepG2 liver cancer cells. Br J Cancer. 109:83–91. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Brustolim D, Ribeiro-dos-Santos R, Kast
RE, Altschuler EL and Soares MB: A new chapter opens in
anti-inflammatory treatments: The antidepressant bupropion lowers
production of tumor necrosis factor-alpha and interferon-gamma in
mice. Int Immunopharmacol. 6:903–907. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chen WC, Lai HC, Su WP, Palani M and Su
KP: Bupropion for interferon-alpha-induced depression in patients
with hepatitis C viral infection: An open-label study. Psychiatry
Investig. 12:142–145. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Packer L, Tritschler HJ and Wessel K:
Neuroprotection by the metabolic antioxidant alpha-lipoic acid.
Free Radic Biol Med. 22:359–378. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Na MH, Seo EY and Kim WK: Effects of
alpha-lipoic acid on cell proliferation and apoptosis in MDA-MB-231
human breast cells. Nutr Res Pract. 3:265–271. 2009. View Article : Google Scholar : PubMed/NCBI
|