Introduction
Hepatocellular carcinoma (HCC) is a primary
malignancy of the liver, and is the fifth most commonly diagnosed
cancer and the third largest cause of cancer-related mortality
worldwide (1). It is estimated
that 39,230 new HCC cases and 27,170 mortalities may occur in 2016,
based on cancer statistical data (2). A number of risk factors for HCC have
been identified, including cirrhosis, hepatitis B/hepatitis C virus
infection and nonalcoholic steatohepatitis (3). Currently, the main therapies for
patients with HCC are liver resection, transplantation,
radiofrequency ablation and adjuvant chemotherapy (4). Despite the progress in therapeutic
treatments, the five-year overall survival rate for patients with
HCC remains <5% (5). In
addition, ~85% of patients with HCC are diagnosed with locally
advanced tumor or distant metastasis, and result in poor prognosis
(1). Therefore, a full
understanding of the underlying mechanisms of the formation and
progression of HCC is urgent, and investigations on novel
therapeutic targets for the therapy of patients with this
malignancy are required.
MicroRNAs (miRNAs) are endogenous, noncoding short
RNA molecules that are 20–24 nucleotides in length (6). miRNAs function as specific gene
regulators by interacting preferentially with the 3′ untranslated
region (3′UTR) of their target genes in a base-pairing manner, and
subsequently inducing mRNA degradation or translational repression
(7). To date, various miRNAs have
been investigated that may serve important roles in diverse
biological processes, such as cell proliferation, viability,
apoptosis, cell cycle, angiogenesis, invasion, motility and
differentiation (8–10). A previous study reported that
>50% of known miRNAs are located at genomic regions or in
fragile sites closely correlated with cancer (11). Additional studies reported that
miRNAs were abnormally expressed in a variety of human cancers,
such as HCC (12), gastric cancer
(13), colorectal cancer (14), glioma (15) and bladder cancer (16). These dysregulated miRNAs may act as
oncogenes or tumor suppressors in tumorigenesis and tumor
development, which depends on tumor type and the roles of their
target genes (17,18). In addition, it has been previously
demonstrated that miRNAs may be used as diagnostic, therapeutic and
prognostic targets for certain human cancers, including HCC
(19–21).
Previous studies have shown that miR-302a was
abnormally expressed and served significant roles in several human
cancers (22–24). One such study revealed that
miR-302a overexpression improved the radiosensitivity of
radioresistant breast cancer cells to radiation therapy in
vitro and in vivo (22). The upregulation of miR-302a also
inhibited the cell invasion and metastasis of breast cancer in
vitro and in vivo (23). However, little is known about the
expression level and functions of miR-302a in HCC. In addition,
abnormal expression of VEGFA, a 35–45 kDa heparin-binding
glycoprotein and a key regulator of angiogenesis, has also been
observed in multiple human cancers, including HCC (25,26).
Thus, the aim of the present study was to investigate miR-302a
expression in HCC and its correlation with clinicopathological
significance as well as VEGFA expression. In vitro
functional experiments were performed to explore these roles in
addition to the molecular mechanisms of action of miR-302a in HCC
cell proliferation, invasion and apoptosis.
Materials and methods
Tissue samples
The present study was approved by the Ethics
Committee of the Yancheng City No. 1 People's Hospital (Yancheng,
China). Written informed consent was obtained from each patient
enrolled in this study. A total of 47 paired HCC tissues and
matched normal adjacent tissues (NATs) were obtained from patients
with HCC patients that received liver resection surgery at Yancheng
City No. 1 People's Hospital between January 2011 and December
2014. None of the patients received radiofrequency ablation or
adjuvant chemotherapy prior to surgery. All tissues samples were
immediately frozen with liquid nitrogen and subsequently stored at
−80°C until RNA extraction.
Cell lines and cell culture
The human HCC cell lines, HepG2 and Hep3B, were
purchased from American Type Culture Collection (ATCC; Manassas,
VA, USA). The human HCC cell lines, SMMC-7721 and Huh7, and the
human immortalized normal liver epithelial cell line (L-O2) were
purchased from Shanghai Institute of Biochemistry and Cell Biology
(Shanghai, China). All cells were grown in Dulbecco's modified
Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) containing 10% fetal bovine serum (FBS; Gibco;
Thermo Fisher Scientific, Inc.) and 1% penicillin/streptomycin, at
37°C in a humidified 5% CO2.
Cell transfection
miRNA (miR)-302a mimics, miRNA negative controls
(miR-NC), short interfering (si)RNA targeting vascular endothelial
growth factor A (si-VEGFA) and siRNA negative control (si-NC) were
purchased from Guangzhou Sagene Biotech Co. (Guangzhou, China).
Prior to transfection, cells were seeded into 6-well plates in DMEM
and incubated at 37°C until the cell density reached 60–70%. Cells
were transfected with miR-302a mimics (50 pmol/ml), miR-NC (50
pmol/ml), si-VEGFA (50 pmol/ml) or si-NC (50 pmol/ml) using
Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) at
room temperature for 12 h, following the manufacturer's protocol.
Following transfection, the cell culture medium was replaced with
DMEM containing 10% FBS. Following 48 h post-transfection, reverse
transcription-quantitative polymerase chain reaction (RT-qPCR) was
performed to detect miR-302a and VEGFA mRNA expression. The cell
invasion assay and apoptosis assay were also performed at 48 h
following transfection. Western blot analysis was conducted at 72 h
post-transfection, and the MTT assay was performed 24 h following
transfection.
RT-qPCR
Total RNA was extracted from tissue (1 g) or cells
(1×106 cells) using the TRIzol Reagent (Invitrogen;
Thermo Fisher Scientific, Inc.), according to the manufacturer's
protocol. To quantify miR-302a expression, cDNA was synthesized by
reverse transcription using a TaqMan® MicroRNA Reverse
Transcription kit (Applied Biosystems; Thermo Fisher Scientific,
Inc.) following the manufacturer's protocol. The TaqMan microRNA
assay (Applied Biosystems; Thermo Fisher Scientific, Inc.) was used
to detect miR-302a expression; U6 was used as an internal control.
The cycling conditions were as follows: 50°C for 2 min, 95°C for 10
min and then 40 cycles of denaturation at 95°C for 15 sec, and
annealing/extension at 60°C for 60 sec. For VEGFA mRNA expression,
total RNA was reverse transcribed to cDNA with the PrimeScript RT
Reagent kit (Takara Bio, Inc., Otsu, Japan). qPCR was performed
with SYBR Premix Ex Taq (Takara Bio, Inc.). β-actin was served as
an internal control for VEGFA mRNA expression. The thermocycling
conditions were as follows: 5 min at 95°C, followed by 40 cycles of
95°C for 30 sec and 65°C for 45 sec. The primers were designed as
follows: miR-302a, 5′-CGTGGATGTACTTGCTTTGAA-3′ (forward) and
5′-TCACCAAAACATGGAAGCAC-3′ (reverse); U6,
5′-GCTTCGGCAGCACATATACTAAAAT-3′ (forward) and
5′-CGCTTCACGAATTTGCGTGTCAT-3′ (reverse); VEGFA,
5′-ACTTTCTGCTGTCTTGGGTG-3′ (forward) and 5′-CTGCATGGTGATGTTGGACT-3′
(reverse); and β-actin, 5′-GGGACCTGACTGACTACCTC-3′ (forward) and
5′-TCATACTCCTGCTTGCTGAT-3′ (reverse). All reactions were performed
in triplicate. The relative expression levels were calculated using
the 2−ΔΔCq method (27).
MTT cell proliferation assay
The MTT assay (Sigma-Aldrich; Merck KGaA, Darmstadt,
Germany) was performed to evaluate cell proliferation. Transfected
cells (3,000 cells/well) were seeded into 96-well plates 24 h
post-transfection. Following incubation at 37°C for 24, 36, 48, and
72 h, MTT assay was conducted according to the manufacturer's
protocol. Briefly, 20 µl MTT solution (5 mg/ml) was added into each
well and incubated for 4 h at 37°C. The medium was then replaced
with 150 µl DMSO (Sigma-Aldrich; Merck KGaA) to dissolve the purple
formazan crystals, and absorbance at 490 nm was determined using a
Tecan Infinite M200 microplate reader (Tecan Group Ltd., Männedorf,
Switzerland). This assay was repeated three times.
Cell invasion assay
The invasive ability of cells was assessed using a
Corning Costar Transwell Chamber (Corning, Inc., Corning, NY, USA)
coated with Matrigel (BD Biosciences, San Jose, CA, USA).
Transfected cells (5×104) in FBS-free DMEM were seeded
into the upper chamber at 48 h post-transfection. DMEM (500 µl)
supplemented with 20% FBS was added into the lower chamber.
Following incubation at 37°C for 48 h, cells that remained on top
surface of the membrane were removed with cotton swabs. Cells on
the bottom surface of the membrane were fixed with 100% methanol
for 10 min and stained with 0.5% crystal violet for 10 min at room
temperature. A total of five randomly selected fields of the
invading cells were captured and counted with an inverted
microscope (Olympus Corporation, Tokyo, Japan). This assay was
repeated three times.
Apoptosis assay
Apoptotic rates were determined using the
fluorescein isothiocyanate (FITC)-Annexin V Apoptosis Detection kit
I (BD Biosciences), according to the manufacturer's protocol.
Following 48 h transfection, cells in 6-well plates
(1×106 cells/well) were harvested using 0.25% trypsin
(Gibco; Thermo Fisher Scientific, Inc.), washed twice with cold
PBS, centrifuged at 500 × g for 5 min at 4°C, and resuspended in 1X
binding buffer. Subsequently, cells were transferred to a 5 ml
culture tube, and 5 µl FITC-annexin V and 10 µl propidium iodide
(PI) were added into the culture tube. Following incubation at room
temperature in the dark for 15 min, apoptosis was analyzed using a
flow cytometer (Beckman Coulter, Inc., Brea, CA, USA) within 1 h of
staining. Data were analyzed using CellQuest® software
(version 3.3; BD Biosciences) and experiments repeated three
times.
miRNA target prediction
Potential targets of miR-302a were predicted using
the algorithms provided in the online target prediction sites
TargetScan (http://www.targetscan.org) and PicTar
(http://pictar.mdc-berlin.de).
Dual-luciferase reporter assay
Wild-type and mutant 3′UTRs of VEGFA were
synthesized by Shanghai GenePharma Co., Ltd. (Shanghai, China) and
also subcloned into the pmirGLO vector; they were subsequently
named pmirGLO-Wt-VEGFA-3′UTR and pmirGLO-Mut-VEGFA-3′UTR,
respectively. For the luciferase reporter assay, HEK293T cells
(ATCC) were seeded into the 24-well plates at a density of 50–60%
confluence. Following overnight incubation at 37°C, cells were
transfected with either pmirGLO-Wt-VEGFA-3′UTR or
pmirGLO-Mut-VEGFA-3′UTR, and either miR-302a mimics or miR-NC, at
room temperature using Lipofectamine 2000. Firefly and
Renilla luciferase activities were detected at 48 h
post-transfection with Dual-Luciferase Reporter Assay System
(Promega Corporation, Madison, WI, USA). The relative luciferase
activity was normalized to Renilla luciferase activity and
experiments were repeated three times.
Western blot analysis
Cell proteins were extracted using
radioimmunoprecipitation assay buffer containing a protease
inhibitor cocktail (Sigma-Aldrich; Merck KGaA). The protein
concentrations were quantified using a Bicinchoninic Acid assay kit
(Pierce; Thermo Fisher Scientific, Inc.). Equal proteins (20 µg)
were separated by 10% SDS-PAGE and subsequently transferred to
polyvinylidene difluoride membranes (EMD Millipore, Billerica, MA,
USA). Membranes were blocked with 5% skimmed in Tris-buffered
saline containing 0.1% Tween-20 (TBST) at room temperature for 1 h,
followed by incubation with primary antibodies against VEGFA
(1:1,000 dilution; cat. no. sc-65617) and GADPH (1:1,000 dilution;
cat. no. sc-32233; Santa Cruz Biotechnology, Inc., Dallas, TX, USA)
at 4°C overnight. Subsequently, the membranes were washed three
times with TBST and incubated with horseradish
peroxidase-conjugated goat anti-mouse secondary antibodies (1:5,000
dilution; cat. no. sc-2005; Santa Cruz Biotechnology, Inc.) at room
temperature for 1 h. Protein bands were visualized using
Chemiluminescence Detection Reagents (Pierce; Thermo Fisher
Scientific, Inc.). GADPH was used as a loading control and
experiments were repeated three times.
Statistical analysis
Data are expressed as the mean ± standard deviation
and calculated with SPSS version 19.0 (IBM Corp., Armonk, NY, USA).
Correlations between mRNA and miRNA expression were determined by
Spearman's correlation analysis. Student's t-tests and Pearson's
χ2 test were used to compare the differences between two
groups. P<0.05 was considered to indicate a statistically
significant difference.
Results
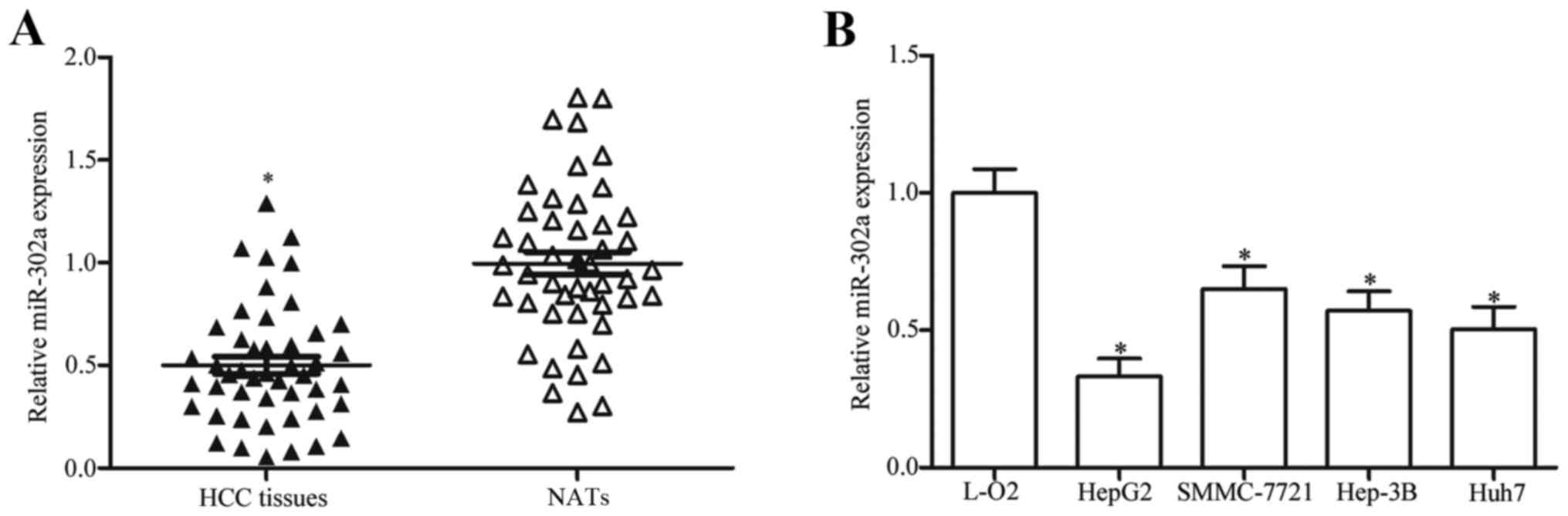
miR-302a is underexpressed in HCC
tissues and cell lines
To understand the biological roles of miR-302a in
HCC development, the expression levels were examined in HCC tissues
and compared with matched NATs. RT-qPCR analysis revealed that
miR-302a expression was significantly reduced in HCC tissues
compared with expression in the matched NATs (Fig. 1A; P<0.05). Similarly, miR-302a
expression levels were lower in the HCC cell lines HepG2,
SMMC-7721, Hep3B and Huh7 compared with the level of expression in
the L-O2 human immortalized normal liver epithelial cell (Fig. 1B; P<0.05). These results
suggested that miR-302a may serve important roles in HCC formation
and progression.
Association between miR-302a
expression and clinicopathological significances of patients with
HCC
The clinicopathological significance of miR-302a
expression in patients with HCC was also investigated (Table I). Low miR-302a expression was
significantly correlated with TNM stage (P=0.028) and lymph node
metastasis (P=0.037), but not with age (P=0.510), sex (P=0.919),
differentiation (P=0.595) or tumor size (P=0.440).
 | Table I.Correlations between
clinicopathological characteristics and miR-302a expression in
patients with hepatocellular carcinoma. |
Table I.
Correlations between
clinicopathological characteristics and miR-302a expression in
patients with hepatocellular carcinoma.
|
|
| miR-302a
expression |
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
significances | Cases (n) | Low | High | P-value |
|---|
| Age (years) |
|
|
| 0.510 |
|
<50 | 25 | 16 | 9 |
|
|
≥50 | 22 | 12 | 10 |
|
| Sex |
|
|
| 0.919 |
|
Male | 35 | 21 | 14 |
|
|
Female | 12 | 7 | 5 |
|
| Tumor size
(cm) |
|
|
| 0.440 |
|
<5 | 24 | 13 | 11 |
|
| ≥5 | 23 | 15 | 8 |
|
| TNM stage |
|
|
| 0.028 |
|
I–II | 23 | 10 | 13 |
|
|
III–IV | 24 | 18 | 6 |
|
|
Differentiation |
|
|
| 0.595 |
|
Well/moderate | 25 | 14 | 11 |
|
|
Poor/undifferentiated | 22 | 14 | 8 |
|
| Lymph node
metastasis |
|
|
| 0.037 |
|
Negative | 26 | 12 | 14 |
|
|
Positive | 21 | 16 | 5 |
|
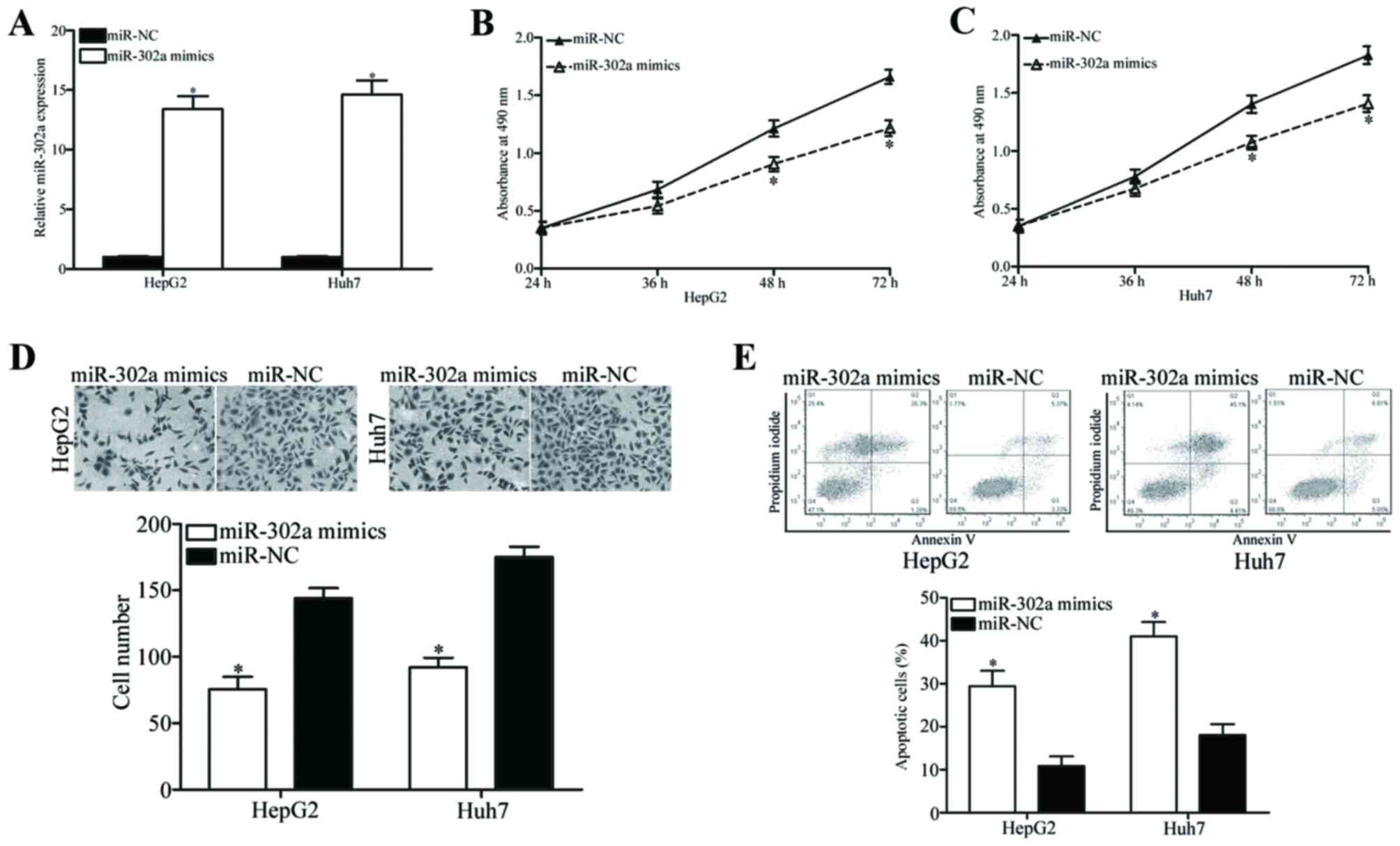
miR-302a attenuates cell
proliferation, invasion and induces apoptosis in HCC
To investigate the roles of miR-302a in HCC
progression, HepG2 and Huh7 cells were transfected with miR-302a
mimics to increase endogenous miR-302a expression level, which was
confirmed by RT-qPCR analysis (Fig.
2A; P<0.05 vs. miR-NC). The effects of miR-302a expression
on HCC cell proliferation, invasion and apoptosis were also
examined. MTT assay results demonstrated that miR-302a
overexpression inhibited the proliferation of HepG2 and Huh7 cells
compared with control miR-NC transfected cells (Fig. 2B and C, respectively; P<0.05 at
48 and 72 h). Similarly, cell invasion assays revealed that the
number of invading HepG2 and Huh7 cells was significantly reduced
following transfection with miR-302a mimics compared with cells
transfected with miR-NC (Fig. 2D;
P<0.05). In addition, overexpression of miR-302a significantly
enhanced apoptotic rates in HepG2 and Huh7 cells (Fig. 2E; P<0.05 vs. miR-NC). These
results suggested that miR-302a may serve an important role in the
occurrence and progression of HCC.
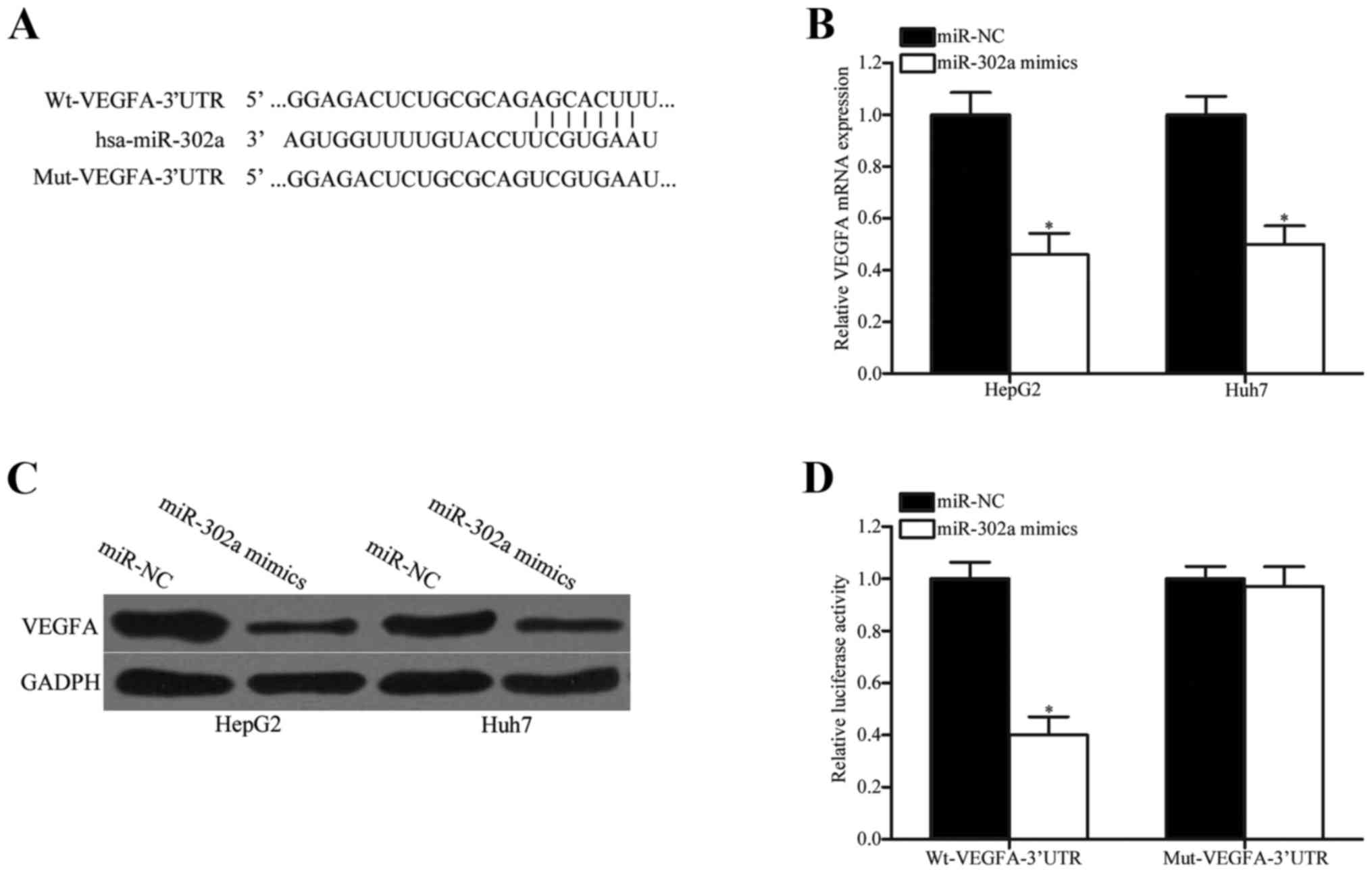
miR-302a reduces VEGFA expression by
directly binding to its 3′UTR
miR-302a was demonstrated to act as a tumor
suppressor in HCC; therefore, the mechanisms underlying the tumor
suppressive roles of miR-302a in HCC were examined. Candidate
targets of miR-302a were queried using online bioinformatics
databases; the results indicated that the 3′UTR of VEGFA contained
complementary sites for miR-302a (Fig.
3A). To verify this hypothesis, RT-qPCR and western blotting
were used to examine the endogenous levels of VEGFA mRNA and
protein expression in HepG2 and Huh7 cells transfected with
miR-302a mimics or miR-NC. Overexpression of miR-302a significantly
reduced both the mRNA (Fig. 3B;
P<0.05) and the protein (Fig.
3C; P<0.05) level of VEGFA expression in HepG2 and Huh7
cells compared with cells transfected with miR-NC. In addition, the
dual-luciferase reporter assay was performed to explore whether
miR-302a was able to directly target the 3′UTR of VEGFA. The
results demonstrated that miR-302a overexpression led to decreased
luciferase activity in HEK293T cells co-transfected with
pmirGLO-Wt-VEGFA-3′UTR (Fig. 3D;
P<0.05 vs. miR-NC). However, no significant differences were
identified in the luciferase activity of cells co-transfected with
miR-302a mimics and pmirGLO-Mut-VEGFA-3′UTR compared with miR-NC.
These results suggested that miR-302a may target the 3′UTR of VEGFA
and reduce its expression in HCC.
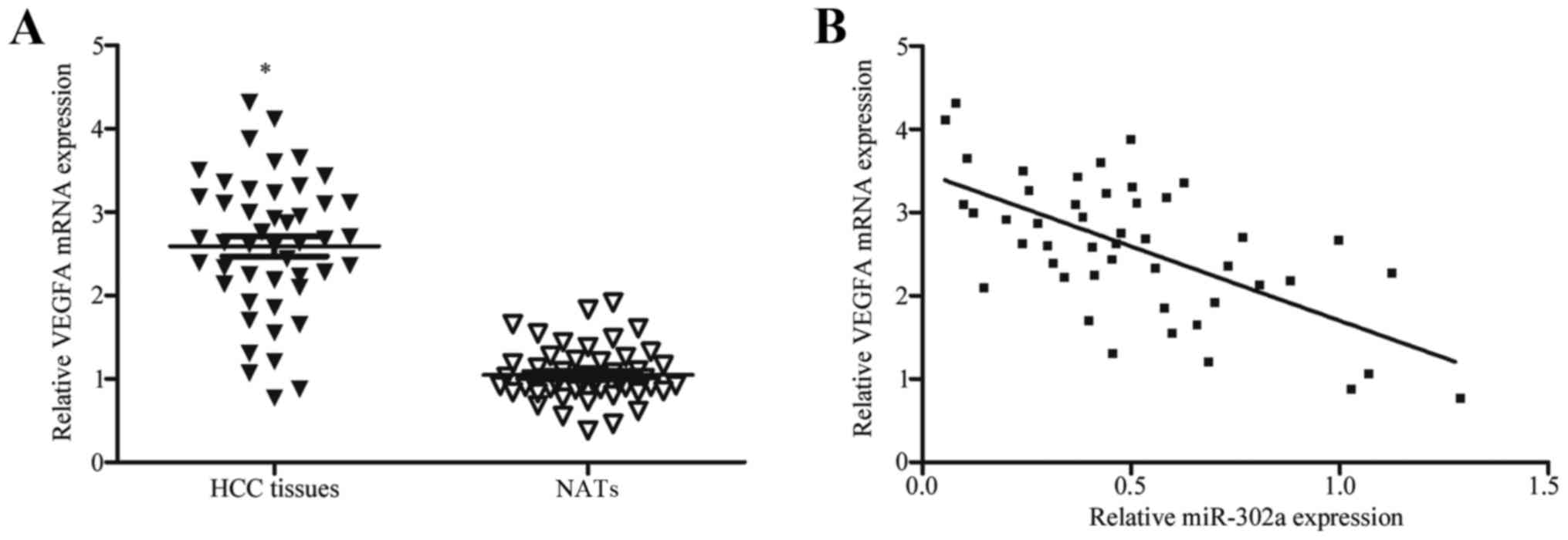
VEGFA expression is negatively
correlated with miR-302a expression in HCC tissues
VEGFA has previously been reported to be upregulated
in HCC (25,26). As VEGFA has been identified as a
direct target gene of miR-302a, the present study hypothesized that
reduced expression of miR-302a may be involved in the high mRNA
expression level of VEGFA in HCC. To confirm this hypothesis,
RT-qPCR was conducted to measure VEGFA mRNA expression in HCC
tissues and NATs. VEGFA mRNA expression was significantly higher in
HCC tissues compared with expression in NATs (Fig. 4A; P<0.05). In addition, the
correlation between miR-302a and VEGFA expression in HCC tissues
was analyzed using Spearman's correlation analysis. The results
indicated that VEGFA mRNA expression levels were negatively
correlated with miR-302a expression in HCC tissues (Fig. 4B; r=−0.6261; P<0.001). These
results further demonstrated that VEGFA may be a direct target of
miR-302a, and that decreased miR302a expression may contribute to
the upregulation of VEGFA in HCC.
miR-302a inhibits cell proliferation
and invasion, and induces apoptosis in HCC by targeting VEGFA
To determine whether VEGFA expression was involved
in the tumor suppressive roles induced by miR-302a overexpression
in HCC, its biological roles in HCC progression were evaluated.
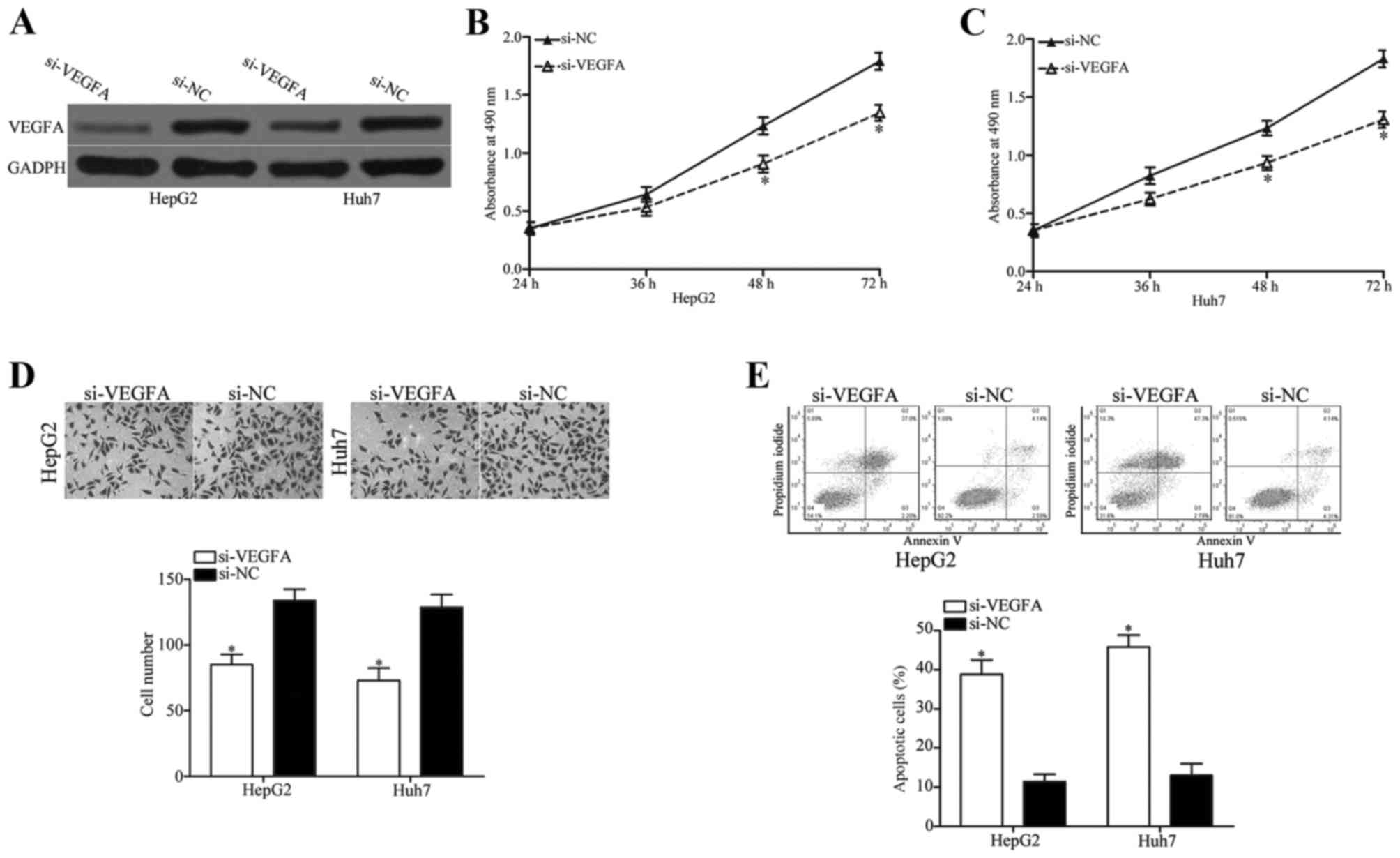
si-VEGFA was used to knockdown VEGFA expression in HepG2 and Huh7
cells (Fig. 5A). MTT assay, cell
invasion assay and apoptosis assay results revealed that knockdown
of VEGFA significantly inhibited cell proliferation (Fig. 5B and C; P<0.05 at 48 and 72 h)
and invasion (Fig. 5D; P<0.05),
and significantly induced apoptosis (Fig. 5E; P<0.05) in HepG2 and Huh7
cells compared with cells transfected with si-NC. The results
induced by VEGFA knockdown in HCC cell proliferation, invasion and
apoptosis were similar to those in cells overexpressing miR-302a,
which suggested that miR-302a may act as a tumor suppressor in HCC
progression by targeting the expression of VEGFA.
Discussion
A number of previous studies have indicated that
abnormally expressed miRNAs may be involved in the occurrence and
progression of HCC by the negative regulation of their target genes
(28,29). Therefore, further investigation of
potential miRNAs that may contribute to the initiation and
progression of HCC may provide novel targets for the diagnosis and
treatments of patients with this malignancy (30). In the present study miR-302a was
demonstrated to be significantly downregulated both in HCC tissues
and in HCC cell lines. Low miR-302a expression was significantly
correlated with TNM stage and lymph node metastasis in patients
with HCC. miR-302a overexpression reduced cell proliferation and
invasion, and induced apoptosis in HCC cells. In addition, VEGFA
was validated as a direct target gene of miR-302a in HCC cells.
These findings suggested that miR-302a underexpression may serve
important roles in HCC formation and progression.
miR-302a has been previously reported to act as a
tumor suppressor in different cancer types (22–24).
For example, miR-302a expression was low in irradiated breast
cancer cells. Upregulation of miR-302a expression enhanced
radiosensitivity of radioresistant breast cancer cells to radiation
therapy in vitro and in vivo (22). Another study reported that
expression levels of miR-302a were decreased in metastatic breast
cancer cells and tumor tissues (23). Overexpression of miR-302a
suppressed breast cancer cell invasion and metastasis in
vitro and in vivo (23). In testicular embryonal carcinoma,
miR-302a overexpression was demonstrated to improve the sensitivity
of NT2 cells to cisplatin by inducing cell apoptosis (24). In ovarian cancer, miR-302a was
revealed to be downregulated in tumor tissues and cell lines
(31). Low miR-302a expression was
correlated with tumor stage of ovarian cancer. Ectopic expression
of miR-302a inhibited ovarian cancer cell proliferation and
enhanced the cell cycle progress. In prostate cancer, miR-302a
expression was reported to be lower in tumor tissues and cell lines
(32). Expression levels of
miR-302a were significantly associated with Gleason score.
Upregulation of miR-302a in prostate cancer cells enhanced G1/S
cell cycle arrest and repressed cell proliferation in vitro
and in vivo. A previous study demonstrated that miR-302a was
reduced in colorectal cancer cells (33). Resumption expression of miR-302a
decreased cell proliferation and invasion in colorectal cancer.
These findings suggested that miR-302a may be a novel therapeutic
strategy for patients with these cancer types.
miRNAs participate in various physiological or
pathological biological processes by negatively regulating the
expression of their target genes; therefore, it is essential to
identify the direct target genes of miR-302a in HCC. Previous
studies have indicated that miR-302a targets many important genes,
such as AKT serine/threonine kinase 1 (AKT1), RAD52 homolog DNA
repair protein and C-X-C motif chemokine receptor 4 (22,23)
in breast cancer, syndecan 1 in ovarian cancer (31), AKT in prostate cancer (32), and mitogen-activated protein kinase
in colorectal cancer (33). The
present study used bioinformatics analysis and the luciferase
reporter assay to demonstrate that the 3′UTR of VEGFA is a direct
target gene of miR-302a in HCC. In addition, miR-302a was
demonstrated to negatively regulate VEGFA expression at both the
mRNA and protein level in HCC. VEGFA mRNA was significantly
expressed in HCC tissues and was inversely correlated with miR-302a
expression. In addition, similar effects were observed when VEGFA
expression was knocked down in HCC as when miR-302a was
overexpressed.
VEGFA is a 35–45 kDa heparin-binding glycoprotein
and a key regulator of angiogenesis (34); angiogenesis is known to be a
crucial factor in local tumor growth tumors and metastasis
progression. An increasing number of studies have indicated that
VEGFA was abnormally expressed in multiple human cancers, including
HCC (25,26), gastric cancer (35), bladder cancer (36), retinoblastoma (37) and colorectal cancer (38). Additional studies have demonstrated
that VEGFA serves important roles in vasculogenesis, angiogenesis,
cell proliferation, migration, invasion, apoptosis and tumor
angiogenesis (39–42). Therefore, targeting VEGFA may be an
effective therapeutic strategy for patients with certain tumors.
Several agents that target VEGFA are in development, some of which
are in clinical trials for cancer treatment (34). The present study demonstrated that
miR-302a targeted VEGFA to inhibit HCC cell growth and metastasis,
and enhanced apoptosis. These results suggested that the
miR-302a/VEGFA pathway may provide novel therapeutic targets for
patients with HCC.
In conclusion, miR-302a may act as a tumor
suppressor in HCC, at least in part, via the inhibition of VEGFA
expression, which suggested that the miR-302a/VEGFA axis may
provide novel therapeutic targets for the treatment of HCC. Future
studies should investigate the upstream regulatory mechanism
underlying miR-302a expression.
References
|
1
|
El-Serag HB and Rudolph KL: Hepatocellular
carcinoma: Epidemiology and molecular carcinogenesis.
Gastroenterology. 132:2557–2576. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dhanasekaran R, Limaye A and Cabrera R:
Hepatocellular carcinoma: Current trends in worldwide epidemiology,
risk factors, diagnosism, and therapeutics. Hepat Med. 4:19–37.
2012.PubMed/NCBI
|
|
4
|
Xu L, Zhang M, Zheng X, Yi P, Lan C and Xu
M: The circular RNA ciRS-7 (Cdr1as) acts as a risk factor of
hepatic microvascular invasion in hepatocellular carcinoma. J
Cancer Res Clin Oncol. 143:17–27. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hou B, Jian Z, Chen S, Ou Y, Li S and Ou
J: Expression of miR-216a in pancreatic cancer and its clinical
significance. Nan Fang Yi Ke Da Xue Xue Bao. 32:1628–1631. 2012.(In
Chinese). PubMed/NCBI
|
|
6
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Krol J, Loedige I and Filipowicz W: The
widespread regulation of microRNA biogenesis, function and decay.
Nat Rev Genet. 11:597–610. 2010.PubMed/NCBI
|
|
8
|
Choi E, Choi E and Hwang KC: MicroRNAs as
novel regulators of stem cell fate. World J Stem Cells. 5:172–187.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhou W, Zou B, Liu L, Cui K, Gao J, Yuan S
and Cong N: MicroRNA-98 acts as a tumor suppressor in
hepatocellular carcinoma via targeting SALL4. Oncotarget.
7:74059–74073. 2016.PubMed/NCBI
|
|
10
|
Wang CY, Zhang JJ, Hua L, Yao KH, Chen JT
and Ren XQ: MicroRNA-98 suppresses cell proliferation, migration
and invasion by targeting collagen triple helix repeat containing 1
in hepatocellular carcinoma. Mol Med Rep. 13:2639–2644. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Calin GA, Sevignani C, Dumitru CD, Hyslop
T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M
and Croce CM: Human microRNA genes are frequently located at
fragile sites and genomic regions involved in cancers. Proc Natl
Acad Sci USA. 101:2999–3004. 2004; View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu S, Liu K, Zhang W, Wang Y, Jin Z, Jia
B and Liu Y: miR-449a inhibits proliferation and invasion by
regulating ADAM10 in hepatocellular carcinoma. Am J Transl Res.
8:2609–2619. 2016.PubMed/NCBI
|
|
13
|
Wang LL, Wang L, Wang XY, Shang D, Yin SJ,
Sun LL and Ji HB: MicroRNA-218 inhibits the proliferation,
migration, and invasion and promotes apoptosis of gastric cancer
cells by targeting LASP1. Tumour Biol. 37:15241–15252. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li H, Zhang H, Lu G, Li Q, Gu J, Song Y,
Gao S and Ding Y: Mechanism analysis of colorectal cancer according
to the microRNA expression profile. Oncol Lett. 12:2329–2336.
2016.PubMed/NCBI
|
|
15
|
Peng G, Liao Y and Shen C: miRNA-429
inhibits astrocytoma proliferation and invasion by targeting BMI1.
Pathol Oncol Res. 23:369–376. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wu D, Niu X, Pan H, Zhou Y, Qu P and Zhou
J: MicroRNA-335 is downregulated in bladder cancer and inhibits
cell growth, migration and invasion via targeting ROCK1. Mol Med
Rep. 13:4379–4385. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang Q, Huang Z, Guo W, Ni S, Xiao X, Wang
L, Huang D, Tan C, Xu Q, Zha R, et al: microRNA-202-3p inhibits
cell proliferation by targeting ADP-ribosylation factor-like 5A in
human colorectal carcinoma. Clin Cancer Res. 20:1146–1157. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Josson S, Gururajan M, Hu P, Shao C, Chu
GY, Zhau HE, Liu C, Lao K, Lu CL, Lu YT, et al: miR-409-3p/-5p
promotes tumorigenesis, epithelial-to-mesenchymal transition, and
bone metastasis of human prostate cancer. Clin Cancer Res.
20:4636–4646. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Fan MQ, Huang CB, Gu Y, Xiao Y, Sheng JX
and Zhong L: Decrease expression of microRNA-20a promotes cancer
cell proliferation and predicts poor survival of hepatocellular
carcinoma. J Exp Clin Cancer Res. 32:212013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Du C, Weng X, Hu W, Lv Z, Xiao H, Ding C,
Gyabaah OA, Xie H, Zhou L, Wu J and Zheng S: Hypoxia-inducible
MiR-182 promotes angiogenesis by targeting RASA1 in hepatocellular
carcinoma. J Exp Clin Cancer Res. 34:672015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chuang KH, Whitney-Miller CL, Chu CY, Zhou
Z, Dokus MK, Schmit S and Barry CT: MicroRNA-494 is a master
epigenetic regulator of multiple invasion-suppressor microRNAs by
targeting ten eleven translocation 1 in invasive human
hepatocellular carcinoma tumors. Hepatology. 62:466–480. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liang Z, Ahn J, Guo D, Votaw JR and Shim
H: MicroRNA-302 replacement therapy sensitizes breast cancer cells
to ionizing radiation. Pharm Res. 30:1008–1016. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liang Z, Bian X and Shim H: Inhibition of
breast cancer metastasis with microRNA-302a by downregulation of
CXCR4 expression. Breast Cancer Res Treat. 146:535–542. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liu L, Lian J, Zhang H, Tian H, Liang M,
Yin M and Sun F: MicroRNA-302a sensitizes testicular embryonal
carcinoma cells to cisplatin-induced cell death. J Cell Physiol.
228:2294–2304. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yamaguchi R, Yano H, Iemura A, Ogasawara
S, Haramaki M and Kojiro M: Expression of vascular endothelial
growth factor in human hepatocellular carcinoma. Hepatology.
28:68–77. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Miura H, Miyazaki T, Kuroda M, Oka T,
Machinami R, Kodama T, Shibuya M, Makuuchi M, Yazaki Y and Ohnishi
S: Increased expression of vascular endothelial growth factor in
human hepatocellular carcinoma. J Hepatol. 27:854–861. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mao B and Wang G: MicroRNAs involved with
hepatocellular carcinoma (Review). Oncol Rep. 34:2811–2820. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Giordano S and Columbano A: MicroRNAs: New
tools for diagnosis, prognosis, and therapy in hepatocellular
carcinoma? Hepatology. 57:840–847. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jin H, Yu M, Lin Y, Hou B, Wu Z, Li Z and
Sun J: MiR-502-3P suppresses cell proliferation, migration, and
invasion in hepatocellular carcinoma by targeting SET. Onco Targets
Ther. 9:3281–3289. 2016.PubMed/NCBI
|
|
31
|
Guo T, Yu W, Lv S, Zhang C and Tian Y:
MiR-302a inhibits the tumorigenicity of ovarian cancer cells by
suppression of SDC1. Int J Clin Exp Pathol. 8:4869–4880.
2015.PubMed/NCBI
|
|
32
|
Zhang GM, Bao CY, Wan FN, Cao DL, Qin XJ,
Zhang HL, Zhu Y, Dai B, Shi GH and Ye DW: MicroRNA-302a suppresses
tumor cell proliferation by inhibiting AKT in prostate cancer. PLoS
One. 10:e01244102015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wei ZJ, Tao ML, Zhang W, Han GD, Zhu ZC,
Miao ZG, Li JY and Qiao ZB: Up-regulation of microRNA-302a
inhibited the proliferation and invasion of colorectal cancer cells
by regulation of the MAPK and PI3K/Akt signaling pathways. Int J
Clin Exp Pathol. 8:4481–4491. 2015.PubMed/NCBI
|
|
34
|
Fan L, Wu Q, Xing X, Wei Y and Shao Z:
MicroRNA-145 targets vascular endothelial growth factor and
inhibits invasion and metastasis of osteosarcoma cells. Acta
Biochim Biophys Sin (Shanghai). 44:407–414. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ji YN, Wang Q, Li Y and Wang Z: Prognostic
value of vascular endothelial growth factor A expression in gastric
cancer: A meta-analysis. Tumour Biol. 35:2787–2793. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Huang YJ, Qi WX, He AN, Sun YJ, Shen Z and
Yao Y: Prognostic value of tissue vascular endothelial growth
factor expression in bladder cancer: A meta-analysis. Asian Pac J
Cancer Prev. 14:645–649. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Youssef NS and Said AM:
Immunohistochemical expression of CD117 and vascular endothelial
growth factor in retinoblastoma: Possible targets of new therapies.
Int J Clin Exp Pathol. 7:5725–5737. 2014.PubMed/NCBI
|
|
38
|
Wen L, Wang R, Lu X and You C: Expression
and clinical significance of vascular endothelial growth factor and
fms-related tyrosine kinase 1 in colorectal cancer. Oncol Lett.
9:2414–2418. 2015.PubMed/NCBI
|
|
39
|
Zhuang Y and Wei M: Impact of vascular
endothelial growth factor expression on overall survival in
patients with osteosarcoma: A meta-analysis. Tumour Biol.
35:1745–1749. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Liu Y, Zheng Q, Wu H, Guo X, Li J and Hao
S: Rapamycin increases pCREB, Bcl-2, and VEGF-A through ERK under
normoxia. Acta Biochim Biophys Sin (Shanghai). 45:259–267. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wiszniak S, Mackenzie FE, Anderson P,
Kabbara S, Ruhrberg C and Schwarz Q: Neural crest cell-derived VEGF
promotes embryonic jaw extension. Proc Natl Acad Sci USA.
112:6086–6091. 2015; View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wu YY, Chen L, Wang GL, Zhang YX, Zhou JM,
He S, Qin J and Zhu YY: Inhibition of hepatocellular carcinoma
growth and angiogenesis by dual silencing of NET-1 and VEGF. J Mol
Histol. 44:433–445. 2013. View Article : Google Scholar : PubMed/NCBI
|