Introduction
Inhibitor of Growth 1 (ING1) was first identified as
a tumor suppressor gene from the comparison with normal mammary
epithelium cells and breast cancer cell lines (1). Following this discovery, ING2, 3, 4,
and 5 were identified (2–5). The family of ING proteins share
similar structural domains including a highly conserved plant
homeodomain (PHD) and nuclear localization sequences (NLS)
(6). ING isoforms are also
generated by alternative RNA splicing (6). These isoforms vary in their function
thereby increasing the diversity of ING biological activities. For
example, the N-terminal of ING1b, but not ING1a, binds to
proliferating cell nuclear antigen (PCNA) and the variants of ING4
acts as a dominant negative (7).
ING2 has the Leucine Zipper motif, suggesting that ING2 might have
the potential ability to bind to DNA. Except for a difference in
the N-terminus, ING2 structure is similar to ING1b. These are both
physically and functionally interacted with HDAC1, mSin3A and sap30
(8–11).
ING2 binds to the histone H3 trimethylated at lysine
4 (H3K4me3) and is involved in chromatin remodeling by forming
complexes with HDAC1 and mSin3A, indicating that ING2 can regulate
gene expression (12–14). H3K4me3 is associated with
transcriptional transactivation, while other histone methylations
are associated with gene suppression (15–17).
We previously reported that overexpression of ING2 induced matrix
metalloproteinase 13 (MMP13) expression (18). MMP13 expression was enhanced under
co-overexpression of ING2 and HDAC1 or mSin3A.
ING2 is also interacted with phosphatidylinositol
5-phosphate (PtdIns5P) (19).
PtdIns5P, which may function in the nuclear signaling pathway, has
been implicated as a critical regulator of nuclear signaling events
during cell-cycle progression (20,21).
Cellular stress, such as UV irradiation or oxidative stress, causes
the accumulation of nuclear PtdIns5P, resulting in the activation
of ING2 (22,23). A recent report has shown that
PtdIns5P binds to the PHD and C-terminal in ING2 (24).
We previously showed that ING2 mRNA expression was
upregulated in colorectal cancer (18). Knockdown of ING2 expression
suppressed cancer cellular growth and inhibited the ability of
cellular invasion (25,26). Therefore, ING2 and the genes of its
downstream may be pivotal target genes of cancer depending on
cancer types.
In the present study, we demonstrated the functional
network of ING2 structure by observing the alteration of ING2
target gene's expression, and further identified the novel
downstream of ING2.
Materials and methods
Cell culture, plasmid, transfection
and reagent
HEK293 was originally obtained from American Type
Culture Collection (Rockville, MD, USA) and was grown at 37°C in
the presence of 5% CO2 in Dulbecco's modified Eagle's
medium (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA,
USA) supplemented with 10% FCS. ING gene's expression vectors were
used as previously described (3–5). In
transfection experiments, cells were plated at 6-cm dish prior to
24 h before transfection. The transfection procedure was performed
by the lipofection method using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) following the
manufacturer's protocol.
RNA extraction and real-time reverse
transcription-PCR (RT-PCR) analysis of mRNA expression
Total RNA from the cells was extracted using TRIzol
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. Five micrograms of total RNA were used for
the synthesis of first-strand cDNA using the SuperScript III First
Strand cDNA Synthesis kit (Invitrogen; Thermo Fisher Scientific,
Inc.) following the manufacturer's instructions. Real-time RT-PCR
analysis was performed using ABI prism 7500 (Applied Biosystems;
Thermo Fisher Scientific, Inc.) with a TaqMan probe provided by the
manufacturer as the following; MMP13 (Id Hs00233992_m1), MMP2 (Id
Hs00234422_m1), PAI-1 (Id Hs00167155_m1), uPA (Id Hs00170182_m1),
HSPA1A (Id Hs00234422_m1), GADD45B (Id Hs00234422_m1), LGALS1 (Id
Hs00234422_m1), and GAPDH (Id Hs99999905_m1). Other genes were
detected by syber green using the following primer sets. C19orf30:
Forward 5′-TGGACATTTTCCCAGAAAGG-3′, and reverse
5′-CTGTCCGGATATTTGGTGCT-3′, CSH1: Forward
5′-TCCGTTATCCAGGCTTTTTG-3′, and reverse 5′-TCATGGTTGTGCGAGTTTGT-3′,
DHRS2: Forward 5′-GCCCTACATGGAGAACAGGA-3′, and reverse
5′-AGCTCCAATGCCAGTGTTCT-3′, MYL1: Forward
5′-GTGCTGACCAGATTGCTGAA-3′, and reverse 5′-TGGCATTCAGCTCTTCATTG-3′,
VGF: Forward 5′-GACCCTCCTCTCCACCTCTC-3′, and reverse
5′-AACCCGTTGATCAGCAGAAG-3′. The relative amount of targeted gene's
transcripts was normalized by the amount of GAPDH mRNA
transcripts.
Nuclear extract preparation, western
blot analysis and immunoprecipitation
The nuclear extraction was performed using NE-PER
Nuclear and Cytoplasmic Extraction Reagents (Pierce; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. For
immunoprecipitation, HEK293 cells transfected with FLAG expression
plasmids were harvested at 48 h post-transfection. The cellular
extracts (800 µg) were immunoprecipitated with FLAG-M2 resin
(Sigma, St. Louis, MO, USA) for 4 h at 4°C. Protein aliquots were
denatured in 2X SDS buffer at 95°C for 5 min separated on
SDS-polyacrylamide gels and transferred onto nitrocellulose
membranes. Membranes were incubated in 5% non-fat dry milk in TBST
(60 mM Tris-base, 120 mM NaCl, 0.05% Tween-20) for 1 h at room
temperature. Blots were probed with a primary antibody for 1 h at
room temperature in TBST, and then incubated with a horseradish
peroxidase-conjugated secondary antibody (Santa Cruz Biotechnology,
Inc., Dallas, TX, USA) in TBST for 30 min at room temperature.
Bound antibodies were detected by the enhanced chemiluminescence
(ECL) detection reagents (Amersham Pharmacia Biotech, Inc.,
Piscataway, NJ, USA) and visualized by autoradiography. The primary
antibodies used for western blot analysis were: Rabbit polyclonal
anti-mSin3A, rabbit polyclonal anti-HDAC1, goat anti-lamin A/C
(Santa Cruz Biotechnology, Inc.), rabbit polyclonal anti-sap30 (EMD
Millipore, Billerica, MA, USA), mouse monoclonal anti-FLAG-M2
(Sigma), and rabbit polyclonal anti-ING2, which was previously
described (3).
ING2 mutant constructs
The construction of mutated and deletion ING2 was
generated by PCR method and was inserted into pcDNA3.1 (Invitrogen;
Thermo Fisher Scientific, Inc.) and pFLAG-CMV-6 (Sigma) vectors.
Site-directed mutagenesis was carried out using QuickChange kit
from Strategene according to manufactory's instructions.
Microarray analysis
Microarray experiments were performed as previously
described (27). Samples for the
microarray experiments were prepared as the following; the ING2
and/or HDAC1 expression vectors were transfected into HEK293 cells
by Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). Cells were harvested at 48 h after the
transfection. The basic raw data and derived ratio measurements
were then uploaded to the National Cancer Institute MicroArray
Database system managed by Center for Information Technology (CIT)
(https://madb.nci.nih.gov/index.shtml), which provides
the bioinformatics and analysis tools necessary for the
interpretation of gene expression data.
Results
MMP13 and PAI-1 mRNA transcripts were
increased by the overexpression of ING1b and ING2
We first investigated whether each ING gene has the
potential function to regulate MMP13 expression. MMP2, which
belongs to the collagenase family as well as MMP13, was examined as
a negative control. MMP13 mRNA expression was remarkably increased
by either ING1b or ING2 overexpression, while MMP2 mRNA expression
was not changed by all of the ING genes (Fig. 1A). Several reports (28–30)
indicated the association with MMP13 and plasminogen activator
inhibitor 1 (PAI-1) or urokinase-type plasminogen activator (uPA).
Thus, we next investigated the PAI-1 and uPA expression by the
overexpression of ING genes. The PAI-1 mRNA level was significantly
increased by either ING1b or ING2 overexpression, whereas uPA
expression was not induced by all of ING genes (Fig. 1B).
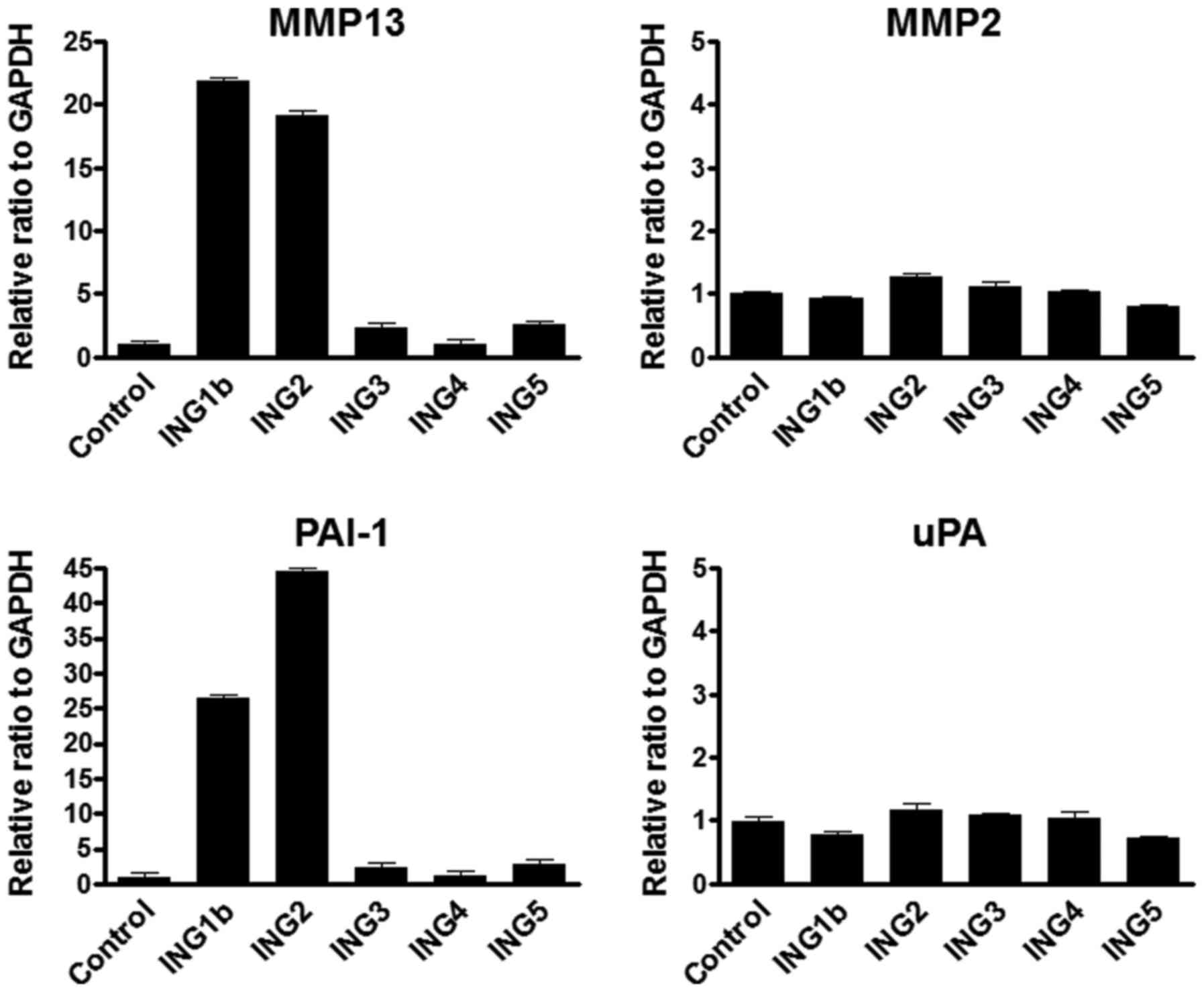 | Figure 1.MMP13 and PAI-1 mRNA was upregulated
under overexpressing ING1b and 2. (A) Results of real-time RT-PCR
analyses of mRNA levels of MMP13 and MMP2 mRNA in ING1b, 2, 3, 4 or
5 overexpressed HEK293 cells. Each ING gene (pcDNA3.1 vector 8 µg)
was transfected into HEK293 cells using the
Lipofectamine® 2000 following manufacturer's protocol.
Empty vector (8 µg) was used for a control. Cells were harvested at
48 h post-transfection. GAPDH mRNA level was used as the internal
control. Columns, average of three independent experiments; Bars,
SD. (B) Real-time RT-PCR was performed for detecting PAI-1 and uPA
mRNA transcripts using the same samples as above. PAI-1 and uPA
mRNA levels were normalized by GAPHD mRNA transcripts. Columns,
average of three independent experiments; Bars, SD. MMP, matrix
metalloproteinase; PAI-1, plasminogen activator inhibitor-1; ING,
inhibitor of growth. |
The alteration of MMP13 and PAI-1
expression by ING2 point mutation
Based on the information of the ING1 mutation
(11,31), the point mutated constructs of ING2
were generated as the following; A194D: 194 (Ala→Asp), N218S: 218
(Asn→Ser), E224K: 224 (Glu→Lys), N262: 262 (Asn→Ser), and K272N:
272 (Lys→Asn) (Fig. 2A). The ING2
protein expressions of these constructs were confirmed by western
blotting using the nuclear extract from HEK293 cells transfected
with mutational ING2 expression vectors (Fig. 2B). The detected band in
E224K-overexpressing cells was slightly smaller than in other
mutated ING2. Both MMP13 and PAI-1 mRNA levels in A194D mutation
overexpressing cells were same as in normal ING2-overexpressed
cells (Fig. 2C). The mutation of
PHD domain at 218 significantly attenuated the MMP13 and PAI-1
expression compared with normal ING2, while the mutation at 224 led
to increase it (Fig. 2C).
Furthermore, MMP13 and PAI-1 expressions were slightly reduced by
the mutation of the C-terminal at either 262 or 272.
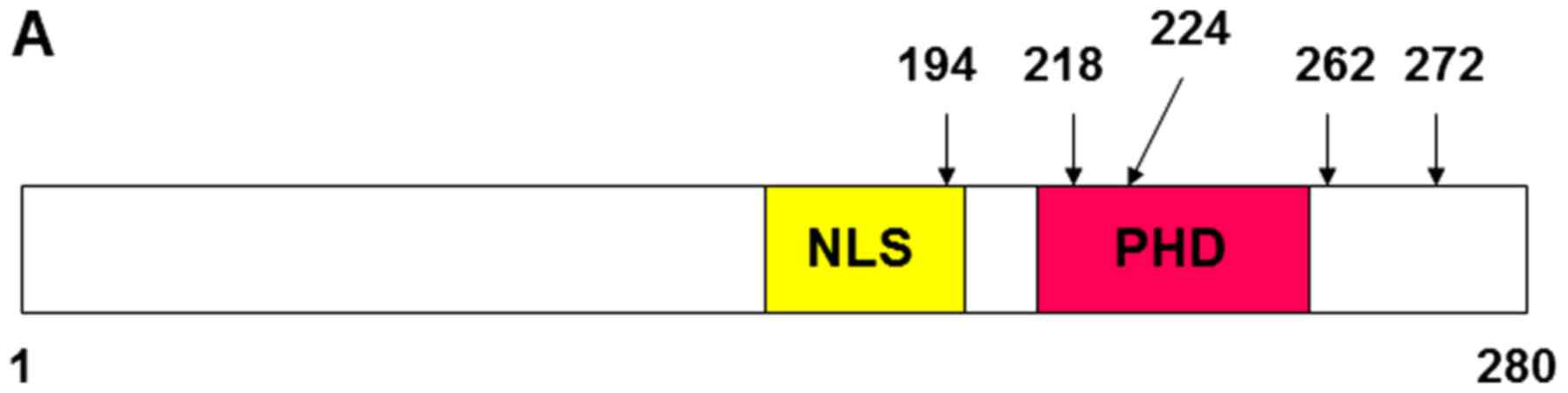 | Figure 2.MMP13 and PAI-1 mRNA expression was
altered by the ING2 mutation. (A) Structural feature of ING2
protein and the location of the mutation site. Vertical bars
indicate mutation sites. (B) Nuclear ING2 expression was determined
by western blotting. The lysates were prepared from HEK293 cells,
in which each 8 µg pcDNA3.1 vectors inserted ING2, A149D, N218S,
E224K, N262S, or K272N were transfected and incubated for 48 h.
Lamin A/C was probed as an internal marker for indicating the
nuclear fraction. (C) The MMP13 and PAI-1 mRNA transcripts were
analyzed by realtime RT-PCR using the same samples as above.
Columns, average of three independent experiments; Bars, SD. MMP,
matrix metalloproteinase; PAI-1, plasminogen activator inhibitor-1;
ING, inhibitor of growth. |
PHD domain is essential for gene
regulation
We examined whether the interaction with HDAC1 and
mSin3A could enhance the MMP13 and PAI-1 expression when ING2
mutated at 218 was overexpressed. The MMP13 and PAI-1 mRNA levels
were enhanced by the each combination with normal ING2-HDAC1 or
normal ING2-mSin3A compared with normal ING2 only (Fig. 3A). However, we couldn't observe
significant induction of MMP13 and PAI-1 expression in the
combination of ING2 mutated at 218 and HDAC1 compared with the
combination of normal ING2 and HDAC1, while the pair of ING2
mutated at 218 and mSin3A could (Fig.
3A). Next, the immunoprecipitation was performed to know the
interaction with ING2, HDAC1 and mSin3A under this condition. The
result of the immunoprecipitation with ING2 showed that the binding
of ING2 with HDAC1 was attenuated by the 218 mutation (Fig. 3B).
The interaction region of ING2
structure with HDAC1, mSin3A and sap30
ING gene family has shared the similar structure
including PHD domain and NLS. However, only ING1b and 2 have the
potential function to regulate MMP13 and PAI-1 genes (Fig. 1A and B). To elucidate the source of
making the distinction between ING1b and 2 and other ING genes, the
five difference deletion constructs of ING2 were generated as shown
in Fig. 4A and inserted into the
FLAG vector. ING2 protein expression in the nucleus was probed by
anti-FLAG antibody and confirmed using the nuclear lysates from the
cells, into which each deletion expression vector was transfected
(Fig. 4B). MMP13 and PAI-1 mRNA
expression was analyzed under overexpression of these deletion
constructs. No induction of MMP13 and PAI-1 expression was observed
in the PHD and C-terminal deletion constructs including ING2 1–196,
1–214, and 1–258 (Fig. 4C). ING2
82–280 and 143–280, which were lacking of the N-terminal, could
induce the MMP13 and PAI-1 expression, thought the levels were not
full compared with ING2 full length (Fig. 4C). The ING2 interaction with
HDAC complexes were further investigated using these ING2 deletion
constructs by the immunoprecipitation method. The N-terminal of
ING2 interacted with mSin3A, sap30, and HDAC1 (Fig. 4D).
 | Figure 4.The C-terminal in ING2 structure was
essential to induce MMP13 and PAI-1 expression. (A) Structural
feature of ING2 protein by the deletion ING2 construct. The numbers
indicate amino acid sites. These constructs were inserted into
pFLAG vectors. (B) Nuclear ING2 expression was determined by
western blotting. Nuclear extracts were prepared from HEK293 cells,
in which each 8 µg pFLAG expression vectors inserted indicated
amino acids fraction of ING2 protein were transfected and incubated
for 48 h. Lamin A/C was probed as an internal marker for indicating
the nuclear fraction. (C) The MMP13 and PAI-1 mRNA transcripts were
analyzed by realtime RT-PCR using the same samples as above.
Columns, average of three independent experiments; Bars, SD. (D)
Immunoprecipitation (IP) was performed for detecting the ING2
interacting proteins, HDAC1, sap30 and mSin3A. Cell lysate was
prepared from HEK293 cells, to which indicated pFLAG-ING2 vector
was transfected and incubated for 48 h. MMP, matrix
metalloproteinase; PAI-1, plasminogen activator inhibitor-1; ING,
inhibitor of growth; HDAC1, histone deacetylase 1 |
Novel identification of ING2-related
genes by microarray experiments
Although HDAC1 is an intrinsically corepressor
(31), HDAC1 could induce the
MMP13 and PAI-1 expression with ING2. In addition to these genes,
to find the ING2/HDAC1-related genes, microarray experiments were
performed using 293 cells overexpressed ING2 and/or HDAC1 (Fig. 5A). The candidate genes were
extracted from the data that the levels in overexpressing ING2 were
two times and higher than in control. Eight genes, which were
HSPA1A, C19orf30, CSH1, GADD45B, DHRS2, LGALS1, MYL1 and VGF, were
upregulated in overexpressing ING2 (Table I). These genes were also
upregulated by the combination of ING2 and HDAC1. To verify the
microarray data, realtime RT-PCR was performed using same samples,
which were used for microarray experiments. The mRNA levels of
eight genes were consistently upregulated in ING2-overexpressed
cells (Fig. 5B). Three genes,
HSPA1A, CSH1 and GADD45B, were also upregulated in
HDAC1-overexpressed cells and enhanced by the combination of ING2
and HDAC1. Other five genes were not significant change in
HDAC1-overexpressed cells. Moreover, these genes were suppressed by
the combination of ING2 and HDAC1.
 | Table I.Upregulated genes in ING2
overexpressed 293 cells. |
Table I.
Upregulated genes in ING2
overexpressed 293 cells.
| Gene symbol | Gene Bank Accession
number | ING2 exp.1 | ING2 exp.2 | ING2+HDAC1
exp.1 | ING2+HDAC1
exp.2 |
|---|
| ING2 | NM_001564 | 31.7297 | 41.2179 | 60.7743 | 40.8336 |
| HDAC1 | NM_004964 | 1.5156 | 1.2142 | 48.1262 | 37.364 |
| HSPA1A | NM_005345 | 3.6251 | 3.147 | 31.2284 | 27.4726 |
| C19orf30 | NM_174947 | 3.4867 | 4.1503 | 11.2609 | 5.9134 |
| CSH1 | NM_022641 | 2.7464 | 3.137 | 6.3723 | 7.5867 |
| GADD45B | NM_015675 | 3.6172 | 3.9141 | 4.7017 | 3.9916 |
| DHRS2 | NM_005794 | 2.0464 | 2.1857 | 4.4004 | 2.3702 |
| LGALS1 | NM_002305 | 3.1078 | 4.3551 | 2.7024 | 2.2653 |
| MYL1 | NM_079422 | 2.2785 | 3.7074 | 2.3336 | 1.9714 |
| VGF | NM_003378 | 2.3762 | 3.3473 | 3.1858 | 2.8928 |
Discussion
ING family genes, which have the similar structure,
possess the HDAC and HAT activity so that they may have the
potential ability of gene regulation cooperating with histone
chromatin related molecules. ING2 has the strong affinity to bind
to the H3K4me3, suggesting that ING2 is implicated with chromatin
remodeling (10–12). We have reported previously that
ING2 was upregulated in colorectal cancer and overexpressed ING2
induced MMP13 expression (18).
Knockdown of ING2 expression suppressed cancer cellular growth and
inhibited the ability of cellular invasion (25,26).
Therefore, ING2 may be pivotal a target gene of cancer depending on
cancer types. In the present study, we focused on novel target
genes of ING2 as a cancer-related gene and the association between
ING2 structure and target gene's regulation.
The induction of MMP13 expression was specific to
the ING1b and 2 genes. Although MMP2 and 13 are classified as the
collagenase group, ING1b and 2 could activate MMP13, not MMP2.
Since several reports (28–30)
indicated the association with MMP13 and PAI-1 or uPA, we
investigated the PAI-1 and uPA expression under the overexpression
of ING genes. We newly found that PAI-1 was a target gene of ING1b
and 2. Basically, ING1b and ING2 proteins have the highly homology
among ING family genes. Therefore, there is the possibility that
both ING1b and ING2 have the similar function.
ING1b gene expression was altered in several cancer
types (9). According to ING1b
mutation analysis in cancers, ING1b mutations were almost detected
in the PHD and C-terminal of ING1b gene (31). Thus, we generated point mutant
constructs based on the ING1b mutations, which were missense, to
know the influence on MMP13 and PAI-1 gene regulation of ING2.
Amino acid 218 and 224 in ING2 are close location within the PHD
domain, but these mutations showed opposite response to the
regulation of MMP13 and PAI-1 expression. A previous report has
shown that the ING2 PHD domain is essential to bind to the H3K4me3,
and the mutation of this region inhibits the binding to the H3K4me3
(11). Our results indicated that
the mutation of ING2 at 218 attenuated the transcriptional ability
due to the binding inhibition to the H3K4me3. On the other hand,
the 224 site mutation, which changed the amino acid from Glu to
Lys, enhanced the MMP13 and PAI-1 expression, speculating that this
missense mutation could acquire the strong binding affinity to the
H3K4me3. We further investigated the association with ING2 218
mutation and the interaction with HDAC1 and mSin3A. Although the
combination of normal ING2 and HDAC1 remarkably induced the MMP13
and PAI-1 mRNA levels compared with only ING2 overexpression,
co-transfection of ING2 218 mutation and HDAC1 couldn't enhance
both MMP13 and PAI-1 expression. Under this mutational condition,
we confirmed the binding ability of HDAC1 to ING2 appeared to be
decreasing by immunoprecipitation experiments. Taken together, the
HDAC1 binding around the PHD domain may contribute to the
interaction with ING2 and H3K4me3.
We understand that the ING2 PHD domain is essential
to regulate the ING2 target genes. Our data demonstrated that the
overexpression of ING1b and 2 increased MMP13 and PAI-1 expression,
not ING3, 4, and 5, though ING family gene has common PHD domain on
the structure. To elucidate the specificity of MMP13 and PAI-1
regulation by the ING1b and 2, we generated several types of ING2
deletion constructs. As shown in Fig.
4C, in addition to the lack of the PHD domain, the lack of the
C-terminal in ING2 also lost the essential function to induce MMP13
and PAI-1 expression. Indeed, the C-terminal structure in ING1b and
2, which has shared the similarity, differed from in ING3, 4, and
5. These results led us to conclude that the C-terminal in ING1b
and 2 is essential to possess the specific function.
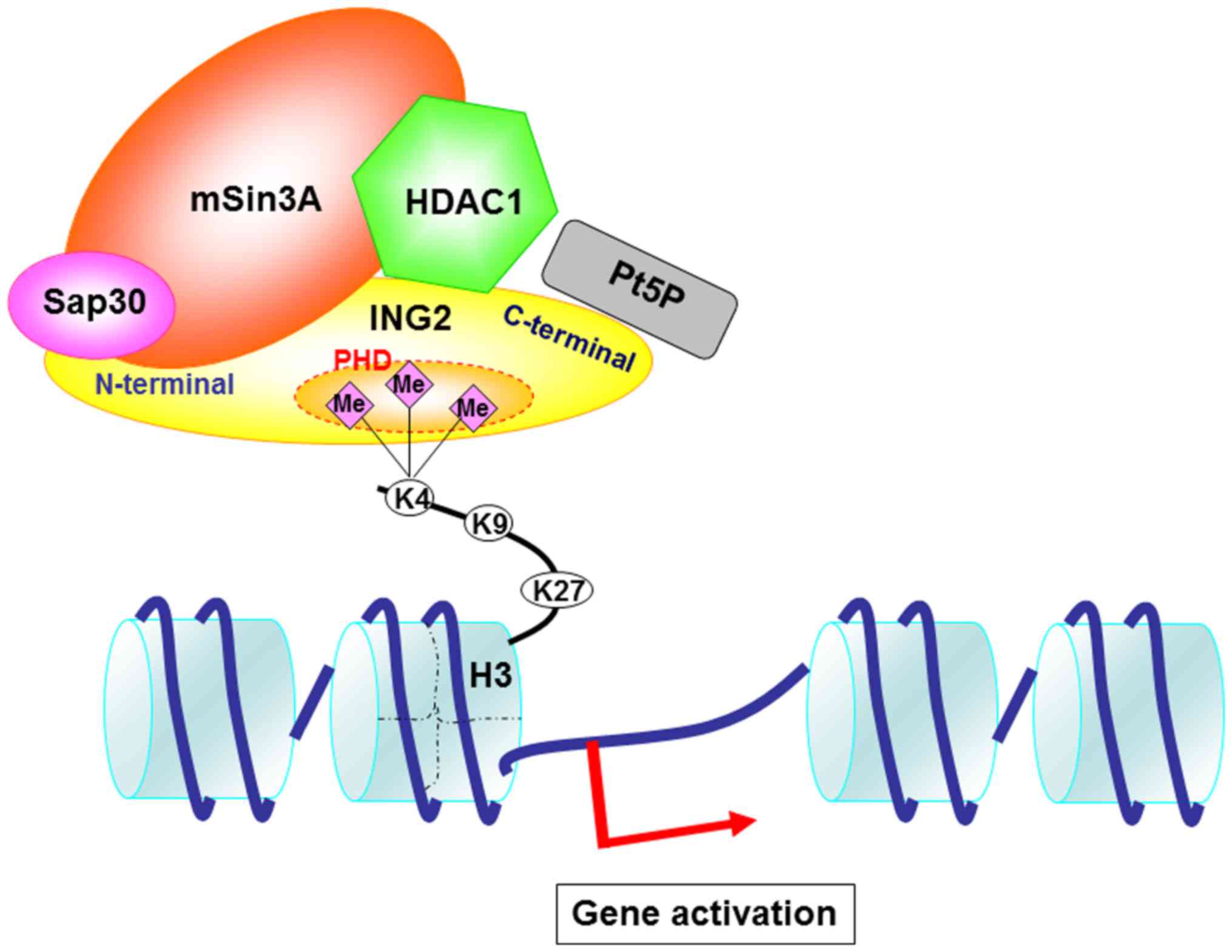
Based on these findings and previous findings by us
and others, we speculated on the mechanism of gene regulation
through the ING2 and HDAC complexes (Fig. 6). HDAC complexes including HDAC1,
sap30 and mSin3A bind to the N-terminal of ING2. HDAC1 structure
may also interact with the PHD domain. The PHD domain is essential
to bind to the H3K4me3. PtdIns5P binds to the partial PHD domain
and C-terminal in ING2. Basically, H3K4me3 is implicated with the
gene activation. Therefore, we focused on the upregulated genes and
have shown that MMP13 and PAI-1 mRNA expression was increased by
the overexpressing ING2.
Although HDAC complexes are generally associated
with gene repression, our results indicate that HDAC1 has the
potential to activate gene expression. According to a recent
report, HDAC1 serves as a coactivator for the glucocorticoid
receptor (GR) and is required for the induction of some genes by
the GR (32). There are many
evidences that HDAC complexes appear to be required for gene
activation (33,34). In the present study, in addition to
MMP13 and PAI-1, we found eight ING2-related genes including
HSPA1A, C19orf30, CSH1, GADD45B, DHRS2, LGALS1, MYL1 and VGF. A
previous report showed that HSPA1A expression was increased by the
ING1b or ING2 overexpression (35). HSAP1A expression was remarkably
induced by HDAC1 overexpression and further enhanced by the
combination with the co-overexpression of ING2. The expression of
DHRS2, LGALS1, MYL1 and VGF genes was suppressed by the
co-overexpression of HDAC1, though these genes expression was
upregulated by the overexpressing ING2. This result indicates that
the ING2 regulation mechanism for these genes may be different with
that for MMP13, PAI-1 and HSPA1A.
Previous reports demonstrated that these genes,
including PAI-1 (36), GADD45B
(37), and LGALS1 (38), were upregulated in colorectal
cancer. We previously found that upregulated ING2 in colorectal
cancer enhanced MMP13 (18). It
has been reported that ING2 was upregulated under the cellular
stress, such as UV irradiation or oxidative stress through the
accumulation of nuclear PtdIns5P (20,21).
PAI-1 and MMP13 expressions were remarkably induced under the
hypoxic stress (34). Of the
target genes of ING2 we found in the present study, GADD45B
expression is associated with oxidative stress (39) and LGALS1 expression is regulated by
hypoxia-inducible factor-1alpha (HIF-1α) (40). Therefore, ING2 might be associated
with the gene regulation upon the hypoxic stress in cancer
cells.
In summary, we demonstrated that overexpression of
ING2 upregulated the expression of PAI-1, HSPA1A, C19orf30, CSH1,
GADD45B, DHRS2, LGALS1, MYL1 and VGF, as well as MMP13 expression.
Among those genes, the expression of PAI-1, HSPA1A, CSH1, and
GADD45B was significantly enhanced under co-overexpression of ING2
and HDAC1. We further confirmed that the PHD domain and the
C-terminal of ING2, which were binding sites of HDAC1 and mSin3A,
were essential regions for the regulation of the gene
expression.
References
|
1
|
Garkavtsev I, Kazarov A, Gudkov A and
Riabowol K: Suppression of the novel growth inhibitor p33ING1
promotes neoplastic transformation. Nat Genet. 14:415–420. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Shimada Y, Saito A, Suzuki M, Takahashi E
and Horie M: Cloning of a novel gene (ING1L) homologous to ING1, a
candidate tumor suppressor. Cytogenet Cell Genet. 83:232–235. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Nagashima M, Shiseki M, Miura K, Hagiwara
K, Linke SP, Pedeux R, Wang XW, Yokota J, Riabowol K and Harris CC:
DNA damage-inducible gene p33ING2 negatively regulates cell
proliferation through acetylation of p53. Proc Natl Acad Sci USA.
98:9671–9676. 2001; View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nagashima M, Shiseki M, Pedeux RM, Okamura
S, Kitahama-Shiseki M, Miura K, Yokota J and Harris CC: A novel
PHD-finger motif protein, p47ING3, modulates p53-mediated
transcription, cell cycle control, and apoptosis. Oncogene.
22:343–350. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shiseki M, Nagashima M, Pedeux RM,
Kitahama-Shiseki M, Miura K, Okamura S, Onogi H, Higashimoto Y,
Appella E, Yokota J and Harris CC: p29ING4 and p28ING5 bind to p53
and p300, and enhance p53 activity. Cancer Res. 63:2373–2378.
2003.PubMed/NCBI
|
|
6
|
He GH, Helbing CC, Wagner MJ, Sensen CW
and Riabowol K: Phylogenetic analysis of the ING family of PHD
finger proteins. Mol Biol Evol. 22:104–116. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Unoki M, Shen JC, Zheng ZM and Harris CC:
Novel splice variants of ING4 and their possible roles in the
regulation of cell growth and motility. J Biol Chem.
281:34677–34686. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Scott M, Bonnefin P, Vieyra D, Boisvert
FM, Young D, Bazett-Jones DP and Riabowol K: UV-induced binding of
ING1 to PCNA regulates the induction of apoptosis. J Cell Sci.
114:3455–3462. 2001.PubMed/NCBI
|
|
9
|
Skowyra D, Zeremski M, Neznanov N, Li M,
Choi Y, Uesugi M, Hauser CA, Gu W, Gudkov AV and Qin J:
Differential association of products of alternative transcripts of
the candidate tumor suppressor ING1 with the mSin3/HDAC1
transcriptional corepressor complex. J Biol Chem. 276:8734–8739.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kuzmichev A, Zhang Y, Erdjument-Bromage H,
Tempst P and Reinberg D: Role of the Sin3-histone deacetylase
complex in growth regulation by the candidate tumor suppressor
p33(ING1). Mol Cell Biol. 22:835–848. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Feng X, Hara Y and Riabowol K: Different
HATS of the ING1 gene family. Trends Cell Biol. 12:532–538. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Doyon Y, Cayrou C, Ullah M, Landry AJ,
Côté V, Selleck W, Lane WS, Tan S, Yang XJ and Côté J: ING tumor
suppressor proteins are critical regulators of chromatin
acetylation required for genome expression and perpetuation. Mol
Cell. 21:51–64. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Peña PV, Davrazou F, Shi X, Walter KL,
Verkhusha VV, Gozani O, Zhao R and Kutateladze TG: Molecular
mechanism of histone H3K4me3 recognition by plant homeodomain of
ING2. Nature. 442:100–103. 2006.PubMed/NCBI
|
|
14
|
Shi X, Hong T, Walter KL, Ewalt M,
Michishita E, Hung T, Carney D, Peña P, Lan F, Kaadige MR, et al:
ING2 PHD domain links histone H3 lysine 4 methylation to active
gene repression. Nature. 442:96–99. 2006.PubMed/NCBI
|
|
15
|
Santos-Rosa H, Schneider R, Bannister AJ,
Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J and
Kouzarides T: Active genes are tri-methylated at K4 of histone H3.
Nature. 419:407–411. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bernstein BE, Kamal M, Lindblad-Toh K,
Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK,
Kulbokas EJ III, Gingeras TR, et al: Genomic maps and comparative
analysis of histone modifications in human and mouse. Cell.
120:169–181. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Berger SL: The complex language of
chromatin regulation during transcription. Nature. 447:407–412.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kumamoto K, Fujita K, Kurotani R, Saito M,
Unoki M, Hagiwara N, Shiga H, Bowman ED, Yanaihara N, Okamura S, et
al: ING2 is upregulated in colon cancer and increases invasion by
enhanced MMP13 expression. Int J Cancer. 125:1306–1315. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gozani O, Karuman P, Jones DR, Ivanov D,
Cha J, Lugovskoy AA, Baird CL, Zhu H, Field SJ, Lessnick SL, et al:
The PHD finger of the chromatin-associated protein ING2 functions
as a nuclear phosphoinositide receptor. Cell. 114:99–111. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Clarke JH, Letcher AJ, D'santos CS,
Halstead JR, Irvine RF and Divecha N: Inositol lipids are regulated
during cell cycle progression in the nuclei of murine
erythroleukaemia cells. Biochem J. 357:905–910. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Roberts HF, Clarke JH, Letcher AJ, Irvine
RF and Hinchliffe KA: Effects of lipid kinase expression and
cellular stimuli on phosphatidylinositol 5-phosphate levels in
mammalian cell lines. FEBS Lett. 579:2868–2872. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bunce MW, Gonzales ML and Anderson RA:
Stress-ING out: Phosphoinositides mediate the cellular stress
response. Sci STKE. 2006:pe462006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jones DR, Bultsma Y, Keune WJ, Halstead
JR, Elouarrat D, Mohammed S, Heck AJ, D'Santos CS and Divecha N:
Nuclear PtdIns5P as a transducer of stress signaling: An in vivo
role for PIP4Kbeta. Mol Cell. 23:685–695. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Huang W, Zhang H, Davrazou F, Kutateladze
TG, Shi X, Gozani O and Prestwich GD: Stabilized
phosphatidylinositol-5-phosphate analogues as ligands for the
nuclear protein ING2: Chemistry, biology, and molecular modeling. J
Am Chem Soc. 129:6498–6506. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Unoki M, Kumamoto K and Harris CC: ING
proteins as potential anticancer drug targets. Curr Drug Targets.
10:442–454. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhong J, Yang L, Liu N, Zheng J and Lin
CY: Knockdown of inhibitor of growth protein 2 inhibits cell
invasion and enhances chemosensitivity to 5-FU in human gastric
cancer cells. Dig Dis Sci. 58:3189–3197. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Staib F, Robles AI, Varticovski L, Wang
XW, Zeeberg BR, Sirotin M, Zhurkin VB, Hofseth LJ, Hussain SP,
Weinstein JN, et al: The p53 tumor suppressor network is a key
responder to microenvironmental components of chronic inflammatory
stress. Cancer Res. 65:10255–10264. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Koong AC, Denko NC, Hudson KM, Schindler
C, Swiersz L, Koch C, Evans S, Ibrahim H, Le QT, Terris DJ and
Giaccia AJ: Candidate genes for the hypoxic tumor phenotype. Cancer
Res. 60:883–887. 2000.PubMed/NCBI
|
|
29
|
Higuchi C, Tanihata Y, Nishimura H, Naito
T and Sanaka T: Effects of glucose and plasminogen activator
inhibitor-1 on collagen metabolism in the peritoneum. Ther Apher
Dial. 9:173–181. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Diehl P, Hantke B, Hennig M, Tschesche H,
Mittelmeier W, Schmitt M and Muehlenweg B: Protein expression of
MMP-13, uPA, and PAI-1 in pseudocapsular and interface tissue
around implants of loose artificial hip joints and in
osteoarthritis. Int J Mol Med. 13:711–715. 2004.PubMed/NCBI
|
|
31
|
Nouman GS, Anderson JJ, Lunec J and Angus
B: The role of the tumour suppressor p33 ING1b in human neoplasia.
J Clin Pathol. 56:491–496. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Qiu Y, Zhao Y, Becker M, John S, Parekh
BS, Huang S, Hendarwanto A, Martinez ED, Chen Y, Lu H, et al: HDAC1
acetylation is linked to progressive modulation of steroid
receptor-induced gene transcription. Mol Cell. 22:669–679. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang XQ, Alfaro ML, Evans GF and Zuckerman
SH: Histone deacetylase inhibition results in decreased macrophage
CD9 expression. Biochem Biophys Res Commun. 294:660–666. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ferguson M, Henry PA and Currie RA:
Histone deacetylase inhibition is associated with transcriptional
repression of the Hmga2 gene. Nucleic Acids Res. 31:3123–3133.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Feng X, Bonni S and Riabowol K: HSP70
induction by ING proteins sensitizes cells to tumor necrosis factor
alpha receptor-mediated apoptosis. Mol Cell Biol. 26:9244–9255.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Fujii T, Obara T, Tanno S, Ura H and Kohgo
Y: Urokinase-type plasminogen activator and plasminogen activator
inhibitor-1 as a prognostic factor in human colorectal carcinomas.
Hepatogastroenterology. 46:2299–2308. 1999.PubMed/NCBI
|
|
37
|
Wang L, Xiao X, Li D, Chi Y, Wei P, Wang
Y, Ni S, Tan C, Zhou X and Du X: Abnormal expression of GADD45B in
human colorectal carcinoma. J Transl Med. 10:2152012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Hittelet A, Legendre H, Nagy N, Bronckart
Y, Pector JC, Salmon I, Yeaton P, Gabius HJ, Kiss R and Camby I:
Upregulation of galectins-1 and −3 in human colon cancer and their
role in regulating cell migration. Int J Cancer. 103:370–379. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kim JH, Qu A, Reddy JK, Gao B and Gonzalez
FJ: Hepatic oxidative stress activates the Gadd45b gene by way of
degradation of the transcriptional repressor STAT3. Hepatology.
59:695–704. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhao XY, Chen TT, Xia L, Guo M, Xu Y, Yue
F, Jiang Y, Chen GQ and Zhao KW: Hypoxia inducible factor-1
mediates expression of galectin-1: The potential role in
migration/invasion of colorectal cancer cells. Carcinogenesis.
31:1367–1375. 2010. View Article : Google Scholar : PubMed/NCBI
|