Introduction
Fibroblasts are widely distributed throughout the
mesenchyme where they synthesize extracellular matrix (ECM)
proteins that form a structural framework to support tissue
architecture and function in steady-state conditions (1). Skin wound repair is a complex process
that involves inflammation, proliferation and a remolding phase. At
the early proliferation stage, fibroblast migration to the wound
site is important for the formation of provisional ECM, such as
collagen and fibronectin, in which the respective cell migration
and organization takes place in successful repair (2–22).
Various signaling pathways, the transforming growth
factor-β (TGF-β)/SMAD pathway in particular, have been investigated
in the process of wound healing; however, the mechanisms of these
pathways remain to be elucidated. TGF-β, a multifunctional cytokine
secreted by macrophages, and its effect on wounds is controversial
(23,24). TGF-β may stimulate collagen
production in dermal fibroblasts by fibroblast-to-myofibroblast
transition (25), the excess
amount of TGF-β increases collagen deposition, resulting in a
keloid (26). TGF-β that is
produced during inflammatory phase by macrophages is an important
mediator of fibroblast activation and tissue repair (27). Although the effect of TGF-β on
wounds has been previously identified, the complete underlying
effect of TGF-β on fibroblasts at the early proliferation phase of
wound repair remain poorly understood.
Microarray technology has been used to obtain
information on the genetic alteration that occurs during many
diseases (28–30). The current study used
bioinformatics to identify the differentially expressed genes
(DEGs) of fibroblasts treated with TGF-β for 24 h. Then, the gene
ontology (GO) and pathway enrichment were analyzed. By analyzing
their biological function and pathway, we may determine the effect
of TGF-β at the early stage of wound repair.
Materials and methods
Microarray data
Gene expression omnibus (GEO; www.ncbi.nlm.nih.gov/geo) contains original
submitter-supplied records and curated DataSets, which is freely
available to users. Two gene expression profiles (GSE79621 and
GSE27165) were obtained from the GEO database. Data of fibroblast
samples and TGF-β 24 h treated fibroblasts samples from the two
gene expression profiles. The GSE79621 contained 3 fibroblast
samples and 3TGF-β 24 h-treated fibroblast samples. The GSE27165
contained 2 fibroblast samples and 2 TGF-β 24 h-treated fibroblast
samples.
Identification of DEGs
The data was processed using web-based tool Morpheus
(https://software.broadinstitute.org/morpheus/), which
is a matrix visualization and analysis platform designed to support
visual data exploration. The expressions of mRNAs with Signal to
noise >1 or signal to noise <1 was defined as DEGs. The
co-expressed upregulated and downregulated DEGs of the two gene
expression profiles were identified with a Venn Diagram (http://bioinfogp.cnb.csic.es/tools/venny/index.html;
Venny 2.1.0).
GO and Kyoto Encyclopedia of Genes and
Genomes (KEGG) pathway enrichment analysis
The common upregulated and downregulated DEGs were
analyzed using Database for Annotation, Visualization and
Integrated Discovery version 6.7 (DAVID; david.ncifcrf.gov), an online program that provides a
comprehensive set of function annotation tools for researchers to
understand the biological meaning lists of genes. In order to
analyze the DEGs at the function level, GO enrichment and KEGG
pathway analyses were performed using DAVID. P<0.05 was
considered to indicate statistically significant difference.
Protein-protein interaction (PPI)
network analysis
To further understand the functional interactions
between these DEGs a PPI network was used. The DEGs were mapped
with the Search Tool for the Retrieval of Interacting Genes
(STRING; www.string-db.org) and experimentally
validated interactions with a combined score of >0.5 were
selected as significant. Then, the PPI network was constructed and
visualized using Cytoscape software (version 3.4.0). The top 10
essential nodes ranked by degree were selected. The plug-in
Molecular Complex Detection (MCODE) was used to identify the
modules of the PPI network in Cytoscape. The criteria were set as
follows: MCODE score ≥4 and number of nodes >4. The function
enrichment analysis of DEGs in the top module was performed using
DAVID.
Results
Identification of DEGs
A total number of samples analyzed were 5 fibroblast
samples and 5 TGF-β 24 h-treated fibroblast samples. The gene
expression profiles were analyzed separately using Morpheus
software. Then, a total of 4,211, 2,433 upregulated and 4,340,
2,032 downregulated DEGs were identified from the GSE79621 and
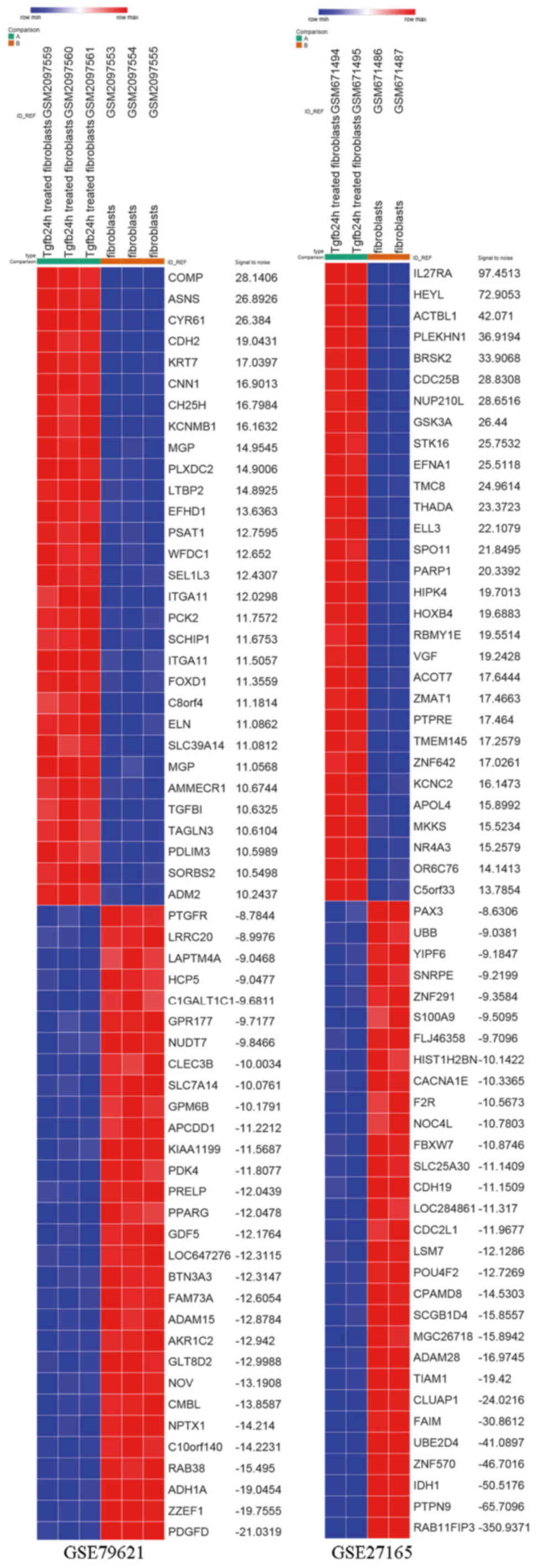
GSE27165 datasets, respectively. DEG expression heat map of the top
30 upregulated and downregulated genes of the two gene expression
profiles are shown in Fig. 1. The
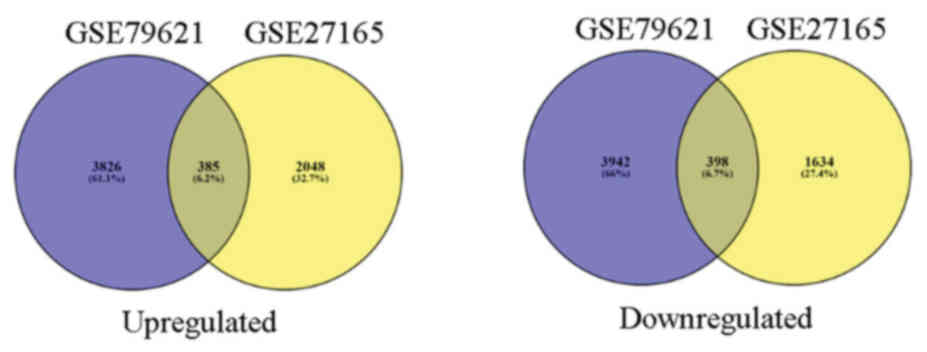
385 upregulated and 398 downregulated co-expressed genes were
identified in the two gene expression profiles (Fig. 2).
GO term enrichment
The upregulated and downregulated DEGs were loaded
to the DAVID software to identify GO categories and KEGG pathways.
GO analysis results showed that in biological process, upregulated
DEGs were significantly enriched in Arp2/3 complex-mediated actin
nucleation, positive regulation of hyaluronan cable assembly,
purine nucleobase biosynthetic process, de novo IMP
biosynthetic process, and positive regulation of epithelial cell
proliferation, whereas the downregulated DEGs were enriched in
regulation of blood pressure, negative regulation of cell
proliferation, ossification, negative regulation of gene expression
and the type I interferon signaling pathway (Table I).
 | Table I.GO analysis of upregulated and
downregulated differentially expressed genes in biological
processes. |
Table I.
GO analysis of upregulated and
downregulated differentially expressed genes in biological
processes.
| A, Upregulated |
|---|
|
|---|
| Term | Function | Count | P-value |
|---|
| GO:0034314 | Arp2/3
complex-mediated actin nucleation | 5 |
6.98×10−4 |
| GO:1900106 | Positive regulation
of hyaluranon cable assembly | 3 | 0.001053124 |
| GO:0009113 | Purine nucleobase
biosynthetic process | 3 | 0.003423324 |
| GO:0006189 | De novo IMP
biosynthetic process | 3 | 0.005071039 |
| GO:0050679 | Positive regulation
of epithelial cell proliferation | 6 | 0.006688708 |
|
| B,
Downregulated |
|
| Term | Function | Count | P-value |
|
| GO:0008217 | Regulation of blood
pressure | 9 | 5.66
×10−5 |
| GO:0008285 | Negative regulation
of cell proliferation | 18 | 0.00333738 |
| GO:0001503 | Ossification | 7 | 0.005354786 |
| GO:0010629 | Negative regulation
of gene expression | 9 | 0.006461428 |
| GO:0060337 | Type I interferon
signaling pathway | 6 | 0.00863459 |
KEGG pathway analysis
The KEGG pathway analysis of upregulated and
downregulated DEGs was performed using DAVID is presented in
Table II. The top 5 KEGG pathways
of upregulated DEGs were shigellosis, pathogenic Escherichia
coli infection, mitogen-activated protein kinase (MAPK)
signaling pathway, Ras signaling pathway and bacterial invasion of
epithelial cells. The top 5 KEGG pathways of downregulated DEGs
were systemic lupus erythematosus, lysosome, arachidonic acid
metabolism, thyroid cancer and allograft rejection (Table II).
 | Table II.KEGG pathway analysis of upregulated
and downregulated differentially expressed genes. Top 5 terms were
selected according to P-value when more than five terms enriched
terms were identified in each category. |
Table II.
KEGG pathway analysis of upregulated
and downregulated differentially expressed genes. Top 5 terms were
selected according to P-value when more than five terms enriched
terms were identified in each category.
| A, Upregulated |
|---|
|
|---|
| Pathway ID | Name | Count | P-value | Genes |
|---|
| hsa05131 | Shigellosis | 8 |
5.30×10−4 | ACTB, ARPC2,
NFKBIB, ARPC5L, ARPC4, MAPK11, MAPK10, FBXW11 |
| hsa05130 | Pathogenic
Escherichia coli infection | 6 | 0.005446999 | ACTB, TUBB, ARPC2,
ARPC5L, TUBA4A, ARPC4 |
| hsa04010 | MAPK signaling
pathway | 13 | 0.011844961 | NTF3, MRAS, MAPK11,
MAPK10, ARRB2, RASGRP1, RASGRP2, PRKACB, FGF1, MYC, GADD45A, RASA1,
NFATC1 |
| hsa04014 | Ras signaling
pathway | 12 | 0.012462871 | LAT, GRIN2B, MRAS,
HTR7, RASGRP1, VEGFA, RASGRP2, GNB4, PRKACB, MAPK10, FGF1,
RASA1 |
| hsa05100 | Bacterial invasion
of epithelial cells | 6 | 0.030311184 | ACTB, ARPC2,
ARPC5L, ARPC4, CD2AP, FN1 |
|
| B,
Downregulated |
| Pathway ID | Name | Count | P-value | Genes |
|
| hsa05322 | Systemic lupus
erythematosus | 10 | 0.006763183 | HIST2H2AA3,
HIST1H2AC, HIST1H2BD, HIST1H2BK, HIST2H2BE, HIST2H2AC, HLA-DPA1,
C1R, C1S, CD40 |
| hsa04142 | Lysosome | 9 | 0.011440506 | CTSK, LAMP2, TPP1,
GUSB, SMPD1, PPT2, NEU1, ATP6V0A4, IDUA |
| hsa00590 | Arachidonic acid
metabolism | 6 | 0.019902025 | PLA2G4A, TBXAS1,
PTGS1, EPHX2, LTA4H, PLA2G4C |
| hsa05216 | Thyroid cancer | 4 | 0.036016354 | TCF7, RXRA, TFG,
TCF7L1 |
| hsa05330 | Allograft
rejection | 4 | 0.066232041 | HLA-C, HLA-DPA1,
HLA-B, CD40 |
PPI network construction and module
analysis
Based on the information in the STRING and Cytoscape
databases, the top 10 hub nodes with high degrees were identified.
These hub genes were GAPDH, MYC, CDK1, ACTB, APP, PRKACB, MAKP11,
ACACB, HSPA4, CAT (Table III).
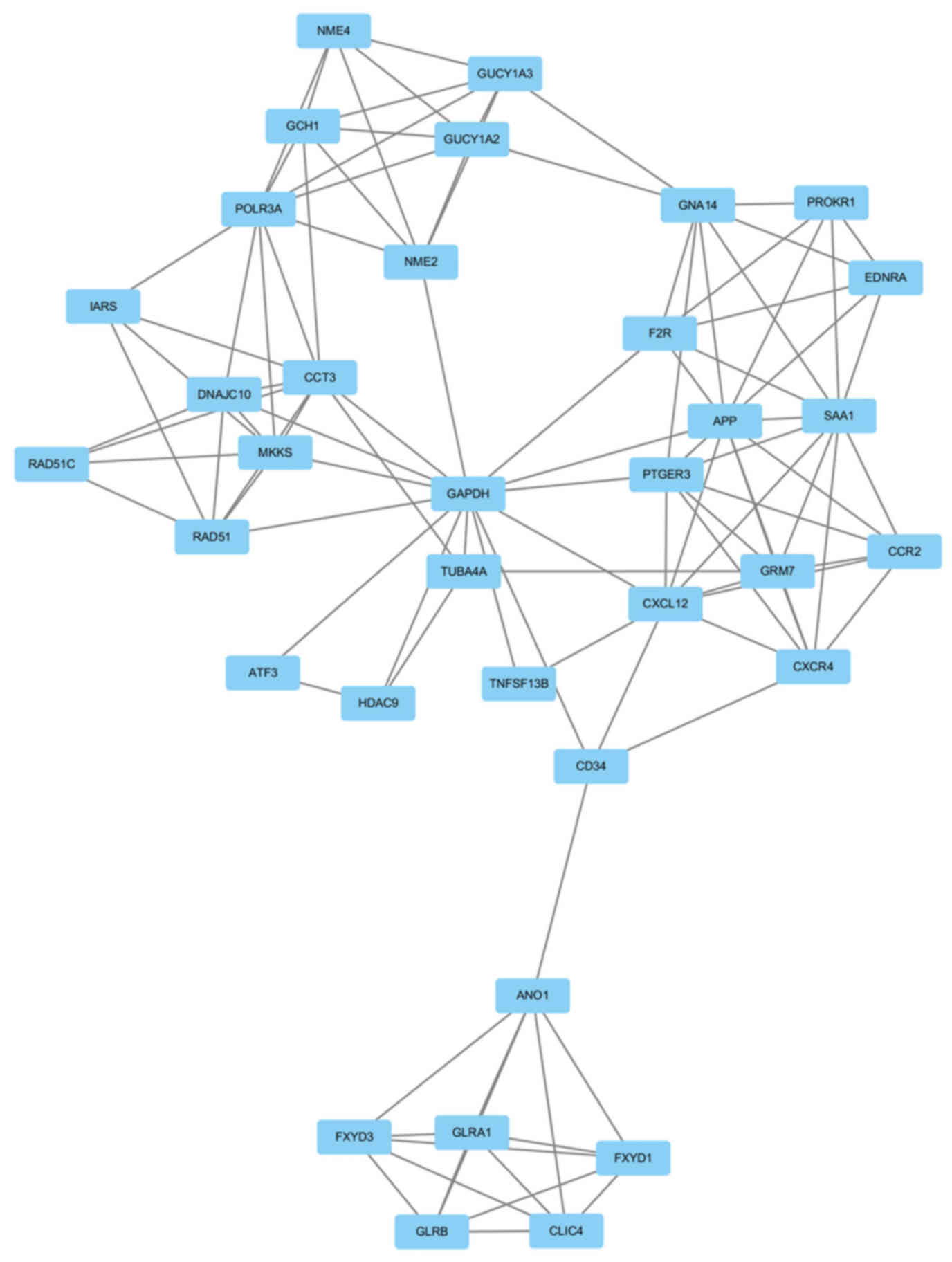
GAPDH had the highest node degree, which was 83. A total of 6
modules from the PPI network satisfied the criteria of MCODE scores
≥4 and number of nodes >4 (Table
IV). The GO and KEGG pathway enrichment of the genes were
included in the top module (Fig.
3) revealed that these genes were primarily associated with
chloride transmembrane transport, chloride transport, positive
regulation of cytosolic calcium ion concentration, chloride channel
complex, chloride channel activity, GTP binding,
extracellular-glycine-gated chloride channel activity, neuroactive
ligand-receptor interaction, purine metabolism and intestinal
immune network for IgA production (Table V).
 | Table III.Degree of top 10 genes. |
Table III.
Degree of top 10 genes.
| Gene ID | Gene name | Degree | Expression |
|---|
| GAPDH |
Glyceraldehyde-3-phosphate
dehydrogenase | 83 | Upregulation |
| MYC | Myc proto-oncogene
protein | 55 | Upregulation |
| CDK1 | Cyclin-dependent
kinase 1 | 51 | Downregulation |
| ACTB | Actin, cytoplasmic
1 | 50 | Upregulation |
| APP | Amyloid beta A4
protein | 40 | Downregulation |
| PRKACB | cAMP-dependent
protein kinase catalytic subunit beta | 36 | Upregulation |
| MAKP11 | Mitogen-activated
protein kinase 11 | 35 | Upregulation |
| ACACB | Acetyl-CoA
carboxylase 2 | 34 | Downregulation |
| HSPA4 | Heat shock 70 kDa
protein 4 | 31 | Upregulation |
| CAT | Catalase | 31 | Downregulation |
 | Table IV.Six modules from the protein-protein
interaction network satisfied the criteria of MCODE scores ≥4 and
number of nodes >4. |
Table IV.
Six modules from the protein-protein
interaction network satisfied the criteria of MCODE scores ≥4 and
number of nodes >4.
| Cluster | Score | Nodes | Edges | Node IDs |
|---|
| 1 |
6.353 | 35 | 108 | FXYD1, ANO1, FXYD3,
CD34, POLR3A, GCH1, GNA14, CCR2, NME4, EDNRA, NME2, GUCY1A3,
GUCY1A2, PTGER3, PROKR1, HDAC9, TNFSF13B, TUBA4A, DNAJC10, CCT3,
GLRB, GLRA1, IARS, CLIC4, CXCL12, CXCR4, RAD51C, GRM7, F2R, RAD51,
GAPDH, MKKS, APP, SAA1, ATF3 |
| 2 | 4.8 | 6 | 12 | MRAS, LRRC2, FMOD,
RAP2C, MST4, PDE7B |
| 3 | 4.5 | 9 | 18 | HLA-B, IRF4, GLDC,
ACACB, OAS3, GART, PMPCB, IFIT3, HLA-C |
| 4 | 4.5 | 5 | 9 | WDR4, CECR1, WDR12,
PUS1, PUS7 |
| 5 | 4.3 | 21 | 43 | MYC, DHX9, MDM2,
MCL1, VAMP1, YWHAG, RABL6, UPF2, STX16, TUBB, NUDT21, RAB6A, NSF,
RPL37, RPL22, TUBB4Q, PHF5A, ACTB, VAMP4, STX6, AR |
| 6 | 4 | 4 | 6 | COL6A2, COL13A1,
COL4A4, COL11A1 |
 | Table V.Functional and pathway enrichment
analysis of the genes in module. Top 3 terms were selected
according to P-value when more than 3 terms enriched terms were
identified in each category. |
Table V.
Functional and pathway enrichment
analysis of the genes in module. Top 3 terms were selected
according to P-value when more than 3 terms enriched terms were
identified in each category.
| A, Biological
processes |
|---|
|
|---|
| Term | Name | Count | P-value | Genes |
|---|
| GO:1902476 | Chloride
transmembrane transport | 6 |
1.15×10−6 | FXYD1, GLRB, FXYD3,
GLRA1, CLIC4, ANO1 |
| GO:0006821 | Chloride
transport | 5 |
1.22×10−6 | FXYD1, FXYD3,
GLRA1, CLIC4, ANO1 |
| GO:0007204 | Positive regulation
of cytosolic calcium ion concentration | 6 |
8.60×10−6 | EDNRA, PTGER3,
CXCR4, SAA1, CCR2, F2R |
|
| B, Molecular
functions |
|
| Term | Name | Count | P-value | Genes |
|
| GO:0005254 | Chloride channel
activity | 4 |
2.02×10−4 | FXYD1, FXYD3,
CLIC4, ANO1 |
| GO:0005525 | GTP binding | 5 | 0.007953298 | GNA14, GUCY1A2,
GUCY1A3, TUBA4A, GCH1 |
| GO:0016934 |
Extracellular-glycine-gated chloride
channel activity | 2 | 0.010379054 | GLRB, GLRA1 |
|
| C, KEGG
pathways |
|
| Term | Name | Count | P-value | Genes |
|
| hsa04080 | Neuroactive
ligand-receptor interaction | 6 | 0.003899897 | EDNRA, GLRB,
PTGER3, GLRA1, GRM7, F2R |
| hsa00230 | Purine
metabolism | 5 | 0.00451383 | NME4, NME2,
GUCY1A2, GUCY1A3, POLR3A |
| hsa04672 | Intestinal immune
network for IgA production | 3 | 0.014264846 | TNFSF13B, CXCR4,
CXCL12 |
Discussion
Despite advancements in the understanding of the
mechanism of wound repair, an effective method for accelerating the
process remains to be identified. Investigation of the complicated
molecular mechanism of wound repair is of important for treatment,
particularly of chronic traumatic wounds, diabetic foot and
bedsores. Previous studies (31,32)
have focused on the effect of vascular endothelial cells,
epithelial cells and bone marrow mesenchymal stem cells in wound
and the effect of fibroblasts remains to be determined. The
proliferation phase ensues with fibroblast migration, which then
synthesize ECM components, which have a significant role in each
stage of the healing process. TGF-β contributes to the function of
fibroblasts, including cell migration, collagen synthesis and cell
proliferation. Therefore, understanding the effect of TGF-β on
fibroblasts at the early proliferation stage is essential to
identify an effective way to improve wound healing. The present
study used bioinformatics analysis to investigate the effect of
TGF-β on fibroblasts at the early proliferation stage of wound
healing. Data from GSE79621 and GSE27165 was obtained and
identified 385 upregulated and 398 downregulated overlapped DEGs
between normal fibroblasts and TGF-β 24 h treated fibroblasts. In
order to understand the interactions of DEGs, GO and KEGG pathway
analyses were performed.
The GO term analysis showed that the upregulated
DEGs were mainly involved in Arp2/3 complex-mediated actin
nucleation, positive regulation of hyaluronan cable assembly,
purine nucleobase biosynthetic process, de novo IMP
biosynthetic process and positive regulation of epithelial cell
proliferation. Previous studies have found that actin has an
important role in cell migration (33–35).
The GO term analysis in biological processes suggested that at the
proliferation phase, TGF-β may regulate actin-based fibroblast
migration to the wound site by Arp2/3 complex. Previous studies
have demonstrated that fetal wounds consist of ECM with an
abundance of hyaluronan (36,37).
Hyaluronan is produced by fibroblasts and promotes fibroblasts
migration and proliferation early in the repair process (37). Therefore, TGF-β-mediated hyaluronan
synthesis may be essential for the migration and proliferation of
fetal fibroblasts. In addition, the findings of the present study
also suggest that TGF-β may regulate the biological activity of
fibroblasts, purine nucleobase biosynthetic process and de
novo IMP biosynthetic process. Furthermore, TGF-β is the only
growth factor, which accelerates ‘maturation’ of epithelial cell
layers (38), which is accordance
with the current findings that TGF-β is related to the positive
regulation of epithelial cell proliferation. The GO term analysis
revealed that the downregulated DEGs were primarily involved in the
regulation of blood pressure, negative regulation of cell
proliferation, ossification, gene expression and the type I
interferon signaling pathway. The regulation of blood pressure is a
process controlled by a balance of processes that increase pressure
and reduce pressure. A previous study determined that blood
pressure is negatively regulated by TGF-β (39), which was consistent the current
findings. Ossification is the conversion of fibrous tissue into
bone or a bony substance. Inhibition of ossification by
downregulation of genes (Bone morphogenic protein 1, Ras
association domain-containing protein 2, neuronal membrane
glycoprotein M6-b, atrial natriuretic peptide receptor 2,
exostosin-1, V-type proton ATPase 116 kDa subunit a isoform 4 and
extracellular matrix protein1) may imply that the early stage of
wound healing is mainly associated with fibroblast proliferation
and collagen synthesis. Increased levels of genes in the type I
interferon pathway have been observed in dermal fibrosis (40). The regulation mechanism of type I
interferon of dermal fibroblasts and its participation in the
development of dermal fibrosis remains to be elucidated. The
current findings suggest that TGF-β may inhibit the interferon
signaling pathway of fibroblasts by downregulation of gene
expression levels, thus preventing wound fibrosis at early stage.
Previous studies have found that TGF-β could mediate cell
proliferation (41,42) and collagen production (43,44).
The GO term analysis demonstrated that downregulated genes were
mainly enriched in negative regulation of cell proliferation and
gene expression, suggesting that TGF-β could promote fibroblast
proliferation and collagen synthesis at the early stage of wound
healing by downregulating related genes. In addition, the enriched
KEGG pathways of upregulated DEGs included shigellosis, pathogenic
Escherichia coli infection, MAPK signaling pathway, Ras
signaling pathway and bacterial invasion of epithelial cells. The
pathway of shigellosis, pathogenic Escherichia coli
infection and bacterial invasion of epithelial cells was associated
with infection, suggesting that TGF-β has an antibacterial effect
on the healing of infected wounds. Previous studies have
demonstrated that activation the MAPK and Ras signaling pathways
could promote cell proliferation, differentiation and migration
(45,46). In addition, the top 5 KEGG pathways
of the downregulated DEGs were systemic lupus erythematosus,
lysosome, arachidonic acid metabolism, thyroid cancer and allograft
rejection. The systemic lupus erythematosus pathway and allograft
rejection pathways are associated with autoantibodies and could
cause tissue injury. Lysosomes serve as the cell's main digestive
compartment and macromolecules are delivered for degradation.
However, previous studies have found (47) that ingested microparticles may
induce cellular damage in phagocytes through the release of
lysosomal enzymes following lysosome rupture. Arachidonic acid is
oxygenated and further transformed into a variety of products,
which mediate inflammatory reactions (48).
A PPI network was constructed from the DEGs by
Cytoscape and the 10 genes exhibiting the highest degree of
connectivity. From these 10 genes, 6 genes were upregulated (GAPDH,
MYC, ACTB, PRKACB, MAKP11, HSPA4) and 4 genes (CDK1, APP, ACACB,
CAT) were downregulated. The top hub gene GAPDH, encodes a member
of the glyceraldehyde-3-phosphate dehydrogenase protein family,
which catalyzes an important energy-yielding step in carbohydrate
metabolism. A previous study has determined that GAPDH could
maintain the integrity of a protein (49). Lin et al (50) reported that MYC plays a positive
role in regulation of fibroblasts proliferation. ACTB encodes one
of actin proteins, which are highly conserved proteins involved in
various types of cell motility. A previous study determined that
β-actin has a key role in cell growth and migration (51). The protein encoded by PRKACB is a
member of the serine/threonine protein kinase family. The encoded
protein is a catalytic subunit of cAMP-dependent protein kinase,
which mediates signaling though cAMP. cAMP signaling is important
for various processes, including cell proliferation and
differentiation (52). MAKP11
encodes mitogen-activated protein kinase 11, which is a member of
the protein kinases family. MAKP11 is involved in the integration
of biochemical signals for a wide variety of cellular processes,
including cell proliferation, differentiation, transcriptional
regulation and development (53,54).
The activity of the heat shock protein A4 (HSPA4), a member of the
HSP110 family, is inducible under various conditions (55). However, a previous study revealed
that overexpression of HSPA4 may inhibit the migration of
fibroblasts cells (56). The 4
downregulated hub genes were CDK1, APP, ACACB and CAT. CDK1, also
termed CDC2, has a key role in the control of cell cycles and
promotes cell migration and proliferation (49,57,58).
APP encodes amyloid β A4 protein, an integral membrane protein
largely known for its role as the precursor of A β peptides and for
its involvement in the pathogenesis of Alzheimer's disease
(59). Previous studies have
determined that sAPP, the secretory domain of the APP has been
observed to reestablish cell growth in APP-deficient fibroblasts
(60). Acetyl-CoA carboxylase β is
encoded by ACACB regulates cellular metabolic processes and may be
involved in the regulation of fatty acid oxidation. CAT encodes
catalase, a key antioxidant enzyme in the defense against oxidative
stress. Downregulation of ACACB and CAT may promote wound repair
through prevention of oxidative stress (61). It is of note that at the early
proliferation stage of wound repair, fibroblasts migrate to the
wound site from surrounding tissue, and contribute to the
proliferation and synthesis ECM components. However, the current
study revealed that TGF-β upregulated the expression level of HSPA4
and downregulated the level of CDK1 at the early wound healing
stage. It is of note that previous studies determined that
overexpression of HSPA4 and reduced expression of CDK1 may inhibit
cell migration and proliferation (56,58).
Therefore, TGF-β may inhibit excessive fibroblast migration and
proliferation by meditating the expression level of different
genes. Therefore, further investigation is required in order to
clarify the underlying biological mechanisms of HSPA4 and CDK1 on
fibroblasts at the early wound healing stage.
A previous study revealed that ion channels and
transporters have a key role in cellular functions (62). Their physiological roles in cell
proliferation have been considered, as cell volume changes, which
involves the movement of ions across the cell membrane, which is
essential for cell-cycle progression (63,64).
The biological process and molecular function analysis identified
one module that contained genes mainly enriched in chloride and
calcium channels, which also suggested that ions are important in
fibroblast migration and proliferation. However, the mechanism
behind this the regulation of this process by ion channels is still
unclear. Therefore, future studies should investigate the genes
associated with ion channels to elucidate how TGF-β mediated ion
channels and in turn cellular functions. Furthermore, the pathway
analysis revealed that the effect of TGF-β on fibroblasts was
associated with neuroactive ligand-receptor interaction, purine
metabolism and intestinal immune network for IgA production. A
previous study determined that fibroblasts could interact with
other cells during the process of wound repair (65). It is of note that, tenascin-C
derived from fibroblasts could promote Schwann cell migration to
the wound site and peripheral nerve regeneration (66). It has been established that purine
metabolites provide a cell with the necessary energy to promote
cell progression (56,67). One feature of intestinal immunity
is its ability to generate non-inflammatory immunoglobulin A
antibodies that act as the first line of defense against
microorganisms. The aforementioned GO and KEGG pathway analyses of
the top module suggest that TGF-β could promote fibroblast
proliferation, migration and have an antimicrobial effect.
The current study has several limitations. Firstly,
the sample size was small and samples were not directly obtained
from the wound. Wound repair is a complex interaction process and
except TGF-β, other cytokines also have effect on fibroblasts.
Secondly, the proliferation stage may continue for 3–10 days and
TGF-β may regulate other genes included in fibroblasts. Thirdly,
these finding would benefit from a validation by western blotting
and polymerase chain reaction. However, the present findings may
suggest potential methods to investigate the effect of TGF-β on
fibroblasts. In addition, future studies will be designed to
clarify the role of TGF-β on fibroblasts during the early stage of
wound repair.
In conclusion, the current study provided a
comprehensive bioinformatics analysis of DEGs, which was associated
with the effect of TGF-β on fibroblasts at the early proliferation
phase of wound repair. The current study provided a set of useful
target genes and pathways for further investigation into the
molecular mechanisms of wound repair. In addition, the current
study may provide a novel research method, which is rarely used in
wound repair. Additional clinical samples are required to confirm
the DEGs of fibroblasts affected by TGF-β during wound repair.
Acknowledgements
The authors would like to thank Mr Liming Xiong for
the support and the design idea for the current study.
References
|
1
|
Ueha S, Shand FH and Matsushima K:
Cellular and molecular mechanisms of chronic
inflammation-associated organ fibrosis. Front Immunol. 3:712012.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jacinto A, Martinez-Arias A and Martin P:
Mechanisms of epithelial fusion and repair. Nat Cell Biol.
3:E117–E123. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Baer ML and Colello RJ: Endogenous
bioelectric fields: A putative regulator of wound repair and
regeneration in the central nervous system. Neural Regen Res.
11:861–864. 2016.PubMed/NCBI
|
|
4
|
Xie SY, Peng LH, Shan YH, Niu J, Xiong J
and Gao JQ: Adult stem cells seeded on electrospinning silk fibroin
nanofiberous scaffold enhance wound repair and regeneration. J
Nanosci Nanotechnol. 16:5498–5505. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Smith-Bolton R: Drosophila imaginal discs
as a model of epithelial wound repair and regeneration. Adv Wound
Care (New Rochelle). 5:251–261. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yang S, Ma K, Geng Z, Sun X and Fu X:
Oriented cell division: New roles in guiding skin wound repair and
regeneration. Biosci Rep. 35:pii: e002802015. View Article : Google Scholar
|
|
7
|
Peng LH, Wei W, Shan YH, Zhang TY, Zhang
CZ, Wu JH, Yu L, Lin J, Liang WQ, Khang G and Gao JQ:
β-Cyclodextrin-linked polyethylenimine nanoparticles facilitate
gene transfer and enhance the angiogenic capacity of mesenchymal
stem cells for wound repair and regeneration. J Biomed Nanotechnol.
11:680–690. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kotwal GJ, Sarojini H and Chien S: Pivotal
role of ATP in macrophages fast tracking wound repair and
regeneration. Wound Repair Regen. 23:724–727. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shingyochi Y, Orbay H and Mizuno H:
Adipose-derived stem cells for wound repair and regeneration.
Expert Opin Biol Ther. 15:1285–1292. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Eming SA, Martin P and Tomic-Canic M:
Wound repair and regeneration: Mechanisms, signaling, and
translation. Sci Transl Med. 6:265sr62014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xu S, Sang L, Zhang Y, Wang X and Li X:
Biological evaluation of human hair keratin scaffolds for skin
wound repair and regeneration. Mater Sci Eng C Mater Biol Appl.
33:648–655. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sen CK and Roy S: OxymiRs in cutaneous
development, wound repair and regeneration. Semin Cell Dev Biol.
23:971–980. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Reinke JM and Sorg H: Wound repair and
regeneration. Eur Surg Res. 49:35–43. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li H and Fu X: Mechanisms of action of
mesenchymal stem cells in cutaneous wound repair and regeneration.
Cell Tissue Res. 348:371–377. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Eldardiri M, Martin Y, Roxburgh J,
Lawrence-Watt DJ and Sharpe JR: Wound contraction is significantly
reduced by the use of microcarriers to deliver keratinocytes and
fibroblasts in an in vivo pig model of wound repair and
regeneration. Tissue Eng Part A. 18:587–597. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Peng LH, Tsang SY, Tabata Y and Gao JQ:
Genetically-manipulated adult stem cells as therapeutic agents and
gene delivery vehicle for wound repair and regeneration. J Control
Release. 157:321–330. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Fu X and Li H: Mesenchymal stem cells and
skin wound repair and regeneration: Possibilities and questions.
Cell Tissue Res. 335:317–321. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gurtner GC, Werner S, Barrandon Y and
Longaker MT: Wound repair and regeneration. Nature. 453:314–321.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yao F and Eriksson E: Gene therapy in
wound repair and regeneration. Wound Repair Regen. 8:443–451. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Puchelle E: Airway epithelium wound repair
and regeneration after injury. Acta Otorhinolaryngol Belg.
54:263–270. 2000.PubMed/NCBI
|
|
21
|
Clark LD, Clark RK and Heber-Katz E: A new
murine model for mammalian wound repair and regeneration. Clin
Immunol Immunopathol. 88:35–45. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sicard RE and Nguyen LM: An in vivo model
for evaluating wound repair and regeneration microenvironments. In
Vivo. 10:477–481. 1996.PubMed/NCBI
|
|
23
|
Suga H, Sugaya M, Fujita H, Asano Y, Tada
Y, Kadono T and Sato S: TLR4, rather than TLR2, regulates wound
healing through TGF-β and CCL5 expression. J Dermatol Sci.
73:117–124. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lee MJ, Shin JO and Jung HS: Thy-1
knockdown retards wound repair in mouse skin. J Dermatol Sci.
69:95–104. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu J, Wang Y, Pan Q, Su Y, Zhang Z, Han
J, Zhu X, Tang C and Hu D: Wnt/β-catenin pathway forms a negative
feedback loop during TGF-β1 induced human normal skin
fibroblast-to-myofibroblast transition. J Dermatol Sci. 65:38–49.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen J, Zeng B, Yao H and Xu J: The effect
of TLR4/7 on the TGF-β-induced Smad signal transduction pathway in
human keloid. Burns. 39:465–472. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Diegelmann RF and Evans MC: Wound healing:
An overview of acute, fibrotic and delayed healing. Front Biosci.
9:283–289. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Elgaaen BV, Olstad OK, Sandvik L, Odegaard
E, Sauer T, Staff AC and Gautvik KM: ZNF385B and VEGFA are strongly
differentially expressed in serous ovarian carcinomas and correlate
with survival. PLoS One. 7:e463172012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li C, Zhen G, Chai Y, Xie L, Crane JL,
Farber E, Farber CR, Luo X, Gao P, Cao X and Wan M: RhoA determines
lineage fate of mesenchymal stem cells by modulating CTGF-VEGF
complex in extracellular matrix. Nat Commun. 7:114552016.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Al-Rekabi Z, Wheeler MM, Leonard A, Fura
AM, Juhlin I, Frazar C, Smith JD, Park SS, Gustafson JA, Clarke CM,
et al: Activation of the IGF1 pathway mediates changes in cellular
contractility and motility in single-suture craniosynostosis. J
Cell Sci. 129:483–491. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chigurupati S, Mughal MR, Okun E, Das S,
Kumar A, McCaffery M, Seal S and Mattson MP: Effects of cerium
oxide nanoparticles on the growth of keratinocytes, fibroblasts and
vascular endothelias cells in cutaneous wound healing.
Biomaterials. 34:2194–2201. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhao H, Han T, Hong X and Sun D: Adipose
differentiation-related protein knockdown inhibits vascular smooth
muscle cell proliferation and migration and attenuates neointima
formation. Mol Med Rep. 16:3079–3086. 2017.PubMed/NCBI
|
|
33
|
Ridley AJ, Schwartz MA, Burridge K, Firtel
RA, Ginsberg MH, Borisy G, Parsons JT and Horwitz AR: Cell
migration: Integrating signals from front to back. Science.
302:1704–1709. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Le Clainche C and Carlier MF: Regulation
of actin assembly associated with protrusion and adhesion in cell
migration. Physiol Rev. 88:489–513. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Fotedar R and Margolis RL: WISp39 and
Hsp90: Actin' together in cell migration. Oncotarget.
6:17871–17872. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Mast BA, Diegelmann RF, Krummel TM and
Cohen IK: Hyaluronic acid modulates proliferation, collagen and
protein synthesis of cultured fetal fibroblasts. Matrix.
13:441–446. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Balaji S, King A, Marsh E, LeSaint M,
Bhattacharya SS, Han N, Dhamija Y, Ranjan R, Le LD, Bollyky PL, et
al: The role of interleukin-10 and hyaluronan in murine fetal
fibroblast function in vitro: Implications for recapitulating fetal
regenerative wound healing. PLoS One. 10:e1243022015. View Article : Google Scholar
|
|
38
|
Wu YS and Chen SN: Apoptotic cell: Linkage
of inflammation and wound healing. Front Pharmacol. 5:12014.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Matsuki K, Hathaway CK, Lawrence MG,
Smithies O and Kakoki M: The role of transforming growth factor β1
in the regulation of blood pressure. Curr Hypertens Rev.
10:223–238. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Agarwal SK, Wu M, Livingston CK, Parks DH,
Mayes MD, Arnett FC and Tan FK: Toll-like receptor 3 upregulation
by type I interferon in healthy and scleroderma dermal fibroblasts.
Arthritis Res Ther. 13:R32011. View
Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhang Y, Alexander PB and Wang XF: TGF-β
family signaling in the control of cell proliferation and survival.
Cold Spring Harb Perspect Biol. 9:pii: a0221452017. View Article : Google Scholar
|
|
42
|
DiRenzo DM, Chaudhary MA, Shi X, Franco
SR, Zent J, Wang K, Guo LW and Kent KC: A crosstalk between
TGF-β/Smad3 and Wnt/beta-catenin pathways promotes vascular smooth
muscle cell proliferation. Cell Signal. 28:498–505. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ghosh AK, Mori Y, Dowling E and Varga J:
Trichostatin A blocks TGF-beta-induced collagen gene expression in
skin fibroblasts: Involvement of Sp1. Biochem Biophys Res Commun.
354:420–426. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Lu J, Shi J, Li M, Gui B, Fu R, Yao G,
Duan Z, Lv Z, Yang Y, Chen Z, et al: Activation of AMPK by
metformin inhibits TGF-β-induced collagen production in mouse renal
fibroblasts. Life Sci. 127:59–65. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhang W and Liu HT: MAPK signal pathways
in the regulation of cell proliferation in mammalian cells. Cell
Res. 12:9–18. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Takino K, Ohsawa S and Igaki T: Loss of
Rab5 drives non-autonomous cell proliferation through TNF and Ras
signaling in Drosophila. Dev Biol. 395:19–28. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Gause WC, Wynn TA and Allen JE: Type 2
immunity and wound healing: Evolutionary refinement of adaptive
immunity by helminths. Nat Rev Immunol. 13:607–614. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Samuelsson B: Arachidonic acid metabolism:
Role in inflammation. Z Rheumatol. 50 Suppl 1:S3–S6. 1991.
|
|
49
|
Chen X, Stauffer S, Chen Y and Dong J:
Ajuba Phosphorylation by CDK1 Promotes Cell Proliferation and
Tumorigenesis. J Biol Chem. 291:14761–14772. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Lin L, Zhang JH, Panicker LM and Simonds
WF: The parafibromin tumor suppressor protein inhibits cell
proliferation by repression of the c-myc proto-oncogene. Proc Natl
Acad Sci USA. 105:pp. 17420–17425. 2008; View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Bunnell TM, Burbach BJ, Shimizu Y and
Ervasti JM: β-Actin specifically controls cell growth, migration,
and the G-actin pool. Mol Biol Cell. 22:4047–4058. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Liu D, Huang Y, Bu D, Liu AD, Holmberg L,
Jia Y, Tang C, Du J and Jin H: Sulfur dioxide inhibits vascular
smooth muscle cell proliferation via suppressing the Erk/MAP kinase
pathway mediated by cAMP/PKA signaling. Cell Death Dis.
5:e12512014. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Jiang Y, Chen C, Li Z, Guo W, Gegner JA,
Lin S and Han J: Characterization of the structure and function of
a new mitogen-activated protein kinase (p38beta). J Biol Chem.
271:17920–17926. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Hsu YL, Wang MY, Ho LJ, Huang CY and Lai
JH: Up-regulation of galectin-9 induces cell migration in human
dendritic cells infected with dengue virus. J Cell Mol Med.
19:1065–1076. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Adachi T, Sakurai T, Kashida H, Mine H,
Hagiwara S, Matsui S, Yoshida K, Nishida N, Watanabe T, Itoh K, et
al: Involvement of heat shock protein a4/apg-2 in refractory
inflammatory bowel disease. Inflamm Bowel Dis. 21:31–39. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Sakurai T, Kashida H, Hagiwara S, Nishida
N, Watanabe T, Fujita J and Kudo M: Heat shock protein A4 controls
cell migration and gastric ulcer healing. Dig Dis Sci. 60:850–857.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Wang Z, Fan M, Candas D, Zhang TQ, Qin L,
Eldridge A, Wachsmann-Hogiu S, Ahmed KM, Chromy BA, Nantajit D, et
al: Cyclin B1/Cdk1 coordinates mitochondrial respiration for
cell-cycle G2/M progression. Dev Cell. 29:217–232. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Han IS, Seo TB, Kim KH, Yoon JH, Yoon SJ
and Namgung U: Cdc2-mediated Schwann cell migration during
peripheral nerve regeneration. J Cell Sci. 120:246–255. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Dawkins E and Small DH: Insights into the
physiological function of the β-amyloid precursor protein: Beyond
Alzheimer's disease. J Neurochem. 129:756–769. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Saitoh T, Sundsmo M, Roch JM, Kimura N,
Cole G, Schubert D, Oltersdorf T and Schenk DB: Secreted form of
amyloid beta protein precursor is involved in the growth regulation
of fibroblasts. Cell. 58:615–622. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Ponugoti B, Xu F, Zhang C, Tian C, Pacios
S and Graves DT: FOXO1 promotes wound healing through the
up-regulation of TGF-β1 and prevention of oxidative stress. J Cell
Biol. 203:327–343. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Wang YL, Sun GY, Zhang Y, He JJ, Zheng S
and Lin JN: Tormentic acid inhibits H2O2-induced oxidative stress
and inflammation in rat vascular smooth muscle cells via inhibition
of NF-kB signaling pathway. Mol Med Rep. 14:3559–3564. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Ohsawa R, Miyazaki H, Niisato N, Shiozaki
A, Iwasaki Y, Otsuji E and Marunaka Y: Intracellular chloride
regulates cell proliferation through the activation of
stress-activated protein kinases in MKN28 human gastric cancer
cells. J Cell Physiol. 223:764–770. 2010.PubMed/NCBI
|
|
64
|
Bear CE: Phosphorylation-activated
chloride channels in human skin fibroblasts. Febs Lett.
237:145–149. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Sorrell JM and Caplan AI: Fibroblasts-a
diverse population at the center of it all. Int Rev Cell Mol Biol.
276:161–214. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Zhang Z, Yu B, Gu Y, Zhou S, Qian T, Wang
Y, Ding G, Ding F and Gu X: Fibroblast-derived tenascin-C promotes
Schwann cell migration through β1-integrin dependent pathway during
peripheral nerve regeneration. Glia. 64:374–385. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Pedley AM and Benkovic SJ: A new view into
the regulation of purine metabolism: The purinosome. Trends Biochem
Sci. 42:141–154. 2017. View Article : Google Scholar : PubMed/NCBI
|