Introduction
Rheumatoid arthritis (RA) is a systemic autoimmune
disease characterized by chronic synovial inflammation and finally
leads to variable degrees of bone and cartilage erosion (1). Presently, the mechanisms and etiology
underlying this disease is still not fully understood, besides,
there are no satisfactory therapeutic strategies to cure RA
(2,3). Patients untreated may suffer short-
and long-term joint destruction and functional disability for
progressive course including inflammation and antibody attack. The
symptoms that allow recognized and diagnosed mainly according to
new classification criteria for RA in 2010 developed by The
American College of Rheumatology (ACR) and European League Against
Rheumatism (EULAR) (4). If the
anti-citrullinated antibody (ACPA) and rheumatoid factor (RF) are
negative or affected joints are indiscoverable, patients with RA in
an earlier phase maybe misdiagnose. And untimely treatment of early
RA would lead to a worse clinical outcome (5). Therefore, new and sensitive
biomarkers to recognize RA and to give specific treatment as early
as possible are necessary for positive therapeutic outcome.
miRNA is one class of the ncRNA and is extensively
studied in eukaryotic gene regulation, which plays a significant
role in the development of RA (6).
Recently, circRNAs can inhibit the function of miRNAs by binding to
miRNAs as sponges was reported (7). circRNAs, form as integral and
conserved closed continuous loops structures wherein the 3′ and 5′
ends are interconnected, are a large novel class of non-coding RNAs
that is highly exist in the eukaryotic transcriptome (8,9). It
could resistance to RNase R conferred by its structure of
covalently closed loops (10). In
the past, its existence was ignored and was regarded as wrong coded
(11). However, it was abundant
and stable and involves transcriptional and post-transcriptional
gene expression regulation (12).
The representative example is mammalian circRNA
called CDR1 antisense (CDR1as). CDR1as is a circRNA sponge of miR-7
that provides more than 70 miR-7 seed binding sites, which shows
high-frequency interactions between circRNAs and miRNAs (13). Recently, there is growing evidence
shows that circRNAs can regulate the expression of miRNAs by sponge
adsorption, which affects the development of the diseases. circRNAs
were found to be important for gastric cancer, colorectal cancer,
osteoarthritis and neurodegenerative pathologies (14), which provide a new directions to
explore the new diagnosis of these diseases. With the advent of
novel biochemical and computational approaches, circRNAs have been
becoming a research hotspot in the RNA field (15).
CircRNA has been measured in tissue, serum, exosomes
and other body fluids in various diseases (16,17),
but the relation between circRNAs and RA remain unclear. Therefore
we assessed the differentially expression ofcircRNA in PBMC of RA
patients and analyzed the symbol genes that potentially involves
pathogenesis and interaction between circRNAs and their paring
miRNAs, aiming to explore circRNAs involved in RA and evaluate
possible miRNA targets through circRNAs MRE sequence analysis, and
provide a novel insight into the circRNAs in PBMC of RA patients in
this study.
Materials and methods
Sample collection
Totally twenty RA patients and healthy people age
and gender matched were selected at Xiangmihu People's Hospital,
the Fourth People's Hospital of Shenzhen. After selected, blood
specimens were collected in March-May in 2014. The experimental
study was approved by the hospital ethics committee. Besides, all
participants have signed the informed consents form. The PBMC of
the blood were immediately separated from 10 paired blood samples,
which were obtained from vein of RA patients and HCs.
RNA extraction
The samples were following placed in RNAlater
(Qiagen, Hilden, Germany) and stored at −80°C. After extraction of
total RNA with TRIzol (Thermo Fisher Scientific, Inc., Waltham, MA,
USA), the determination of RNA quantity and quality was conducted
according to the manufacturer's protocol by NanoDrop ND-1000
(NanoDrop; Thermo Fisher Scientific, Inc., Wilmington, DE, USA). In
addition, RNA integrity was assessed by standard denaturing agarose
gel electrophoresis.
RNA labeling and microarray
hybridization
Sample labeling and array hybridization were
performed according to the manufacturer's protocol (Arraystar Inc.,
Rockville, MD, USA). Briefly, linear RNAs were firstly removed by
Rnase R (Epicentre; Illumina, Inc., San Diego, CA, USA). Following,
a random priming method (Arraystar Super RNA Labeling kit;
Arraystar Inc.) was used to amplify and transcribed each sample
into fluorescent cRNA. Then, the labeled cRNAs were purified by
RNeasy Mini kit (Qiagen), and concentration and specific activity
of it (pmol Cy3/µg cRNA) were measured by NanoDrop ND-1000.
Subsequently, 1 µl 25X fragmentation buffer and 1 µl 25X
fragmentation buffer were added to 1 µl each labeled cRNA, then the
mixture were placed at 60°C for 30 min. After, 25 µl 2X
Hybridization buffer was used to dilute the labeled cRNA. The
microarray slides were incubated in an Agilent Hybridization Oven
at 65°C for 17 h. Finally, the hybridized array was fixed and
scanned using the Axon GenePix 4000B microarray scanner (Molecular
Devices, Inc., Sunnyvale, CA, USA) after washed.
Data collection
GenePix Pro 6.0 software (Axon; Molecular Devices,
LLC, Sunnyvale, CA, USA) was used for raw data extraction and grid
alignment from scanned images. The R software package was used for
quantile normalization of raw data and subsequent data processing.
After that, the filtering of low intensive substance was performed,
and at least 50% of the circRNAs samples considered flag
‘expressed’ (>2 times background standard deviation) were
conserved for further analyses.
The ‘fold-change’ of each circRNA was computed to
compare the profile differences between the samples of RA patients
and healthy controls. t-test was performed to evaluate the
statistical significance. P-values ≤0.05 with the ‘fold-changes’ of
circRNA ≥2.0 were considered significantly differentially expressed
ones. We filter the analysis outputs and rank the differentially
expressed circRNAs according to fold-change and P-value by using
Microsoft Excel's Data/Sort and Filter functionalities (Microsoft
Corporation, Redmond, WA, USA).
Data analysis and annotation for
circRNA/miRNA interaction
We selected the top 10 up- and downregulated
circRNAs in PBMCs and plasma for further analysis. PubMed (National
Center for Biotechnology Information, Bethesda, MD, USA) was used
to analyze differentially expressed circRNAs. circRNA/miRNA
interactions were predicted to facilitate our study by using
Arraystar miRNA target prediction software based on TargetScan and
miRanda. The differentially expressed circRNAs within all the
comparisons were annotated in detail with the circRNA/miRNA
interaction information. Additionally, we analyzed the sequences of
the MREs and the top 10 significantly differentially expressed
circRNAs and predicted miRNA targets were also analyzed for their
correlation with miRNAs previously described.
Validation of differentially expressed
circRNA by reverse transcription-quantitative polymerase chain
reation (RT-qPCR)
Five circRNAs exhibiting up- or downregulated
expression was chosen for results verification by RT-qPCR. The
primers are showed in Table I. In
order to reduce errors caused by potential variation in
concentration and transcription efficiency of RNA, evaluation of
the reference gene β-actin was served as an internal control.
Briefly, the cycle parameters for the PCR reaction were 95°C for 5
min, followed by 40 amplification cycles of a denaturing step at
the same temperature for 10 sec and an annealing/extension step of
60 sec at 60°C. Following, the amplification product concentration
for each sample was generated by Rotor-Gene Real-Time Analysis
Software 6.0 (Qiagen). Fold-changes were calculated using the
2−∆∆Cq method. The relative values of gene expression
were taken as the ratio of the sample to the internal control.
 | Table I.Reverse transcription-quantitative
polymerase chain reaction primer list. |
Table I.
Reverse transcription-quantitative
polymerase chain reaction primer list.
| Gene | Primer
sequence | Annealing temp
(°C) | Product size
(bp) |
|---|
| β-actin (H) | F:
5′-GTGGCCGAGGACTTTGATTG-3′ |
|
|
|
| R:
5′-CCTGTAACAACGCATCTCATATT-3′ | 60 | 73 |
|
hsa_circRNA_104593 | F:
5′-TGCCAGACACATACAAAGATAAGC-3′ |
|
|
|
(hsa_circ_0083964) | R:
5′-GGTATCTGGTGGCATCTCATCC-3′ | 60 | 160 |
|
hsa_circRNA_104194 | F:
5′-GATAAAAACTAACCACCATCTTGC-3′ |
|
|
|
(hsa_circ_0004712) | R:
5′-AAGGTAGTCTTCATCCAGCAGG-3′ | 60 | 177 |
|
hsa_circRNA_103334 | F:
5′-ATTTGGAGTCTGGGAGTGAT-3′ |
|
|
|
(hsa_circ_0064996) | R:
5′-ACAGTGTTTTGTTAGTTGCTTCT-3′ | 60 | 168 |
|
hsa_circRNA_101407 | F:
5′-CAGCACGACAACATTATTGCCTAC-3′ |
|
|
|
(hsa_circ_0032683) | R:
5′-ACGACGTTCCTTCTCAGACAGC-3′ | 60 | 142 |
|
hsa_circRNA_102594 | F:
5′-CACCATCAGCCATAGATCCTC-3′ |
|
|
|
(hsa_circ_0052012) | R:
5′-CTTGCCATCCATCCCACATA-3′ | 60 | 118 |
|
hsa_circRNA_102822 | F:
5′-ACTGCTGGATACTAAATGTGATGC-3′ |
|
|
|
(hsa_circ_0056536) | R:
5′-GTTCAAGTTTTCGAGCCTGTTC-3′ | 60 | 147 |
Results
Quality control
RNA quality and quantity were measured using the
NanoDrop ND-1000. RNA integrity was successfully tested by standard
denaturing agarose gel electrophoresis after RNA extraction and
prior to sample labeling. For spectrophotometer readings, the O.D.
A260/A280 ratio value was established as close to 2.0 for pure RNA
(ratios between 1.8 and 2.1 were acceptable), and the O.D.A260/A230
ratio were required to be >1.8. For agarose gels, the ribosomal
RNA bands of 28S and 18S were sharp and intensely stained whereas
low molecular weight RNAs such as tRNA and 5S ribosomal RNA
presented as diffuse bands. DNA contamination of the RNA
preparation was evident as a high molecular weight smear or a band
migrating above the 28S ribosomal RNA band. All quality criteria
established by the manufacturer were satisfied for each array to
ensure successful microarray analysis.
circRNAs expression profiling in
patients with RA
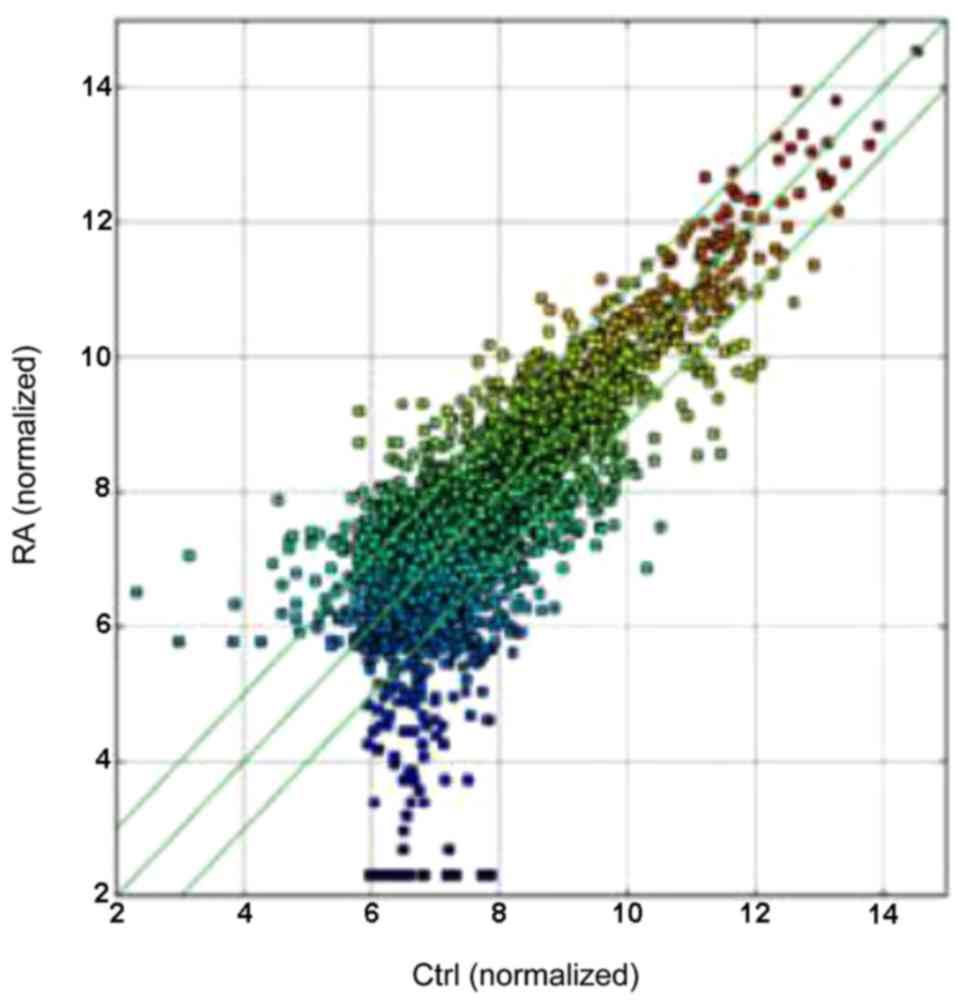
In this study, we identified 584 circRNAs that were
differentiallyexpressed in RA patients vs. HCs by circRNAs
microarray. A total of 255 circRNAs were significantly upregulated
and 329 were downregulated among the RA samples (Table II and Fig. 1), the differentiated circRNAs were
accounted for fold-change (FC≥2.0) and values (≤0.05) in the
upregulated and downregulated groups. The box plot view was to show
the distributions of expression values between RA and HCs.
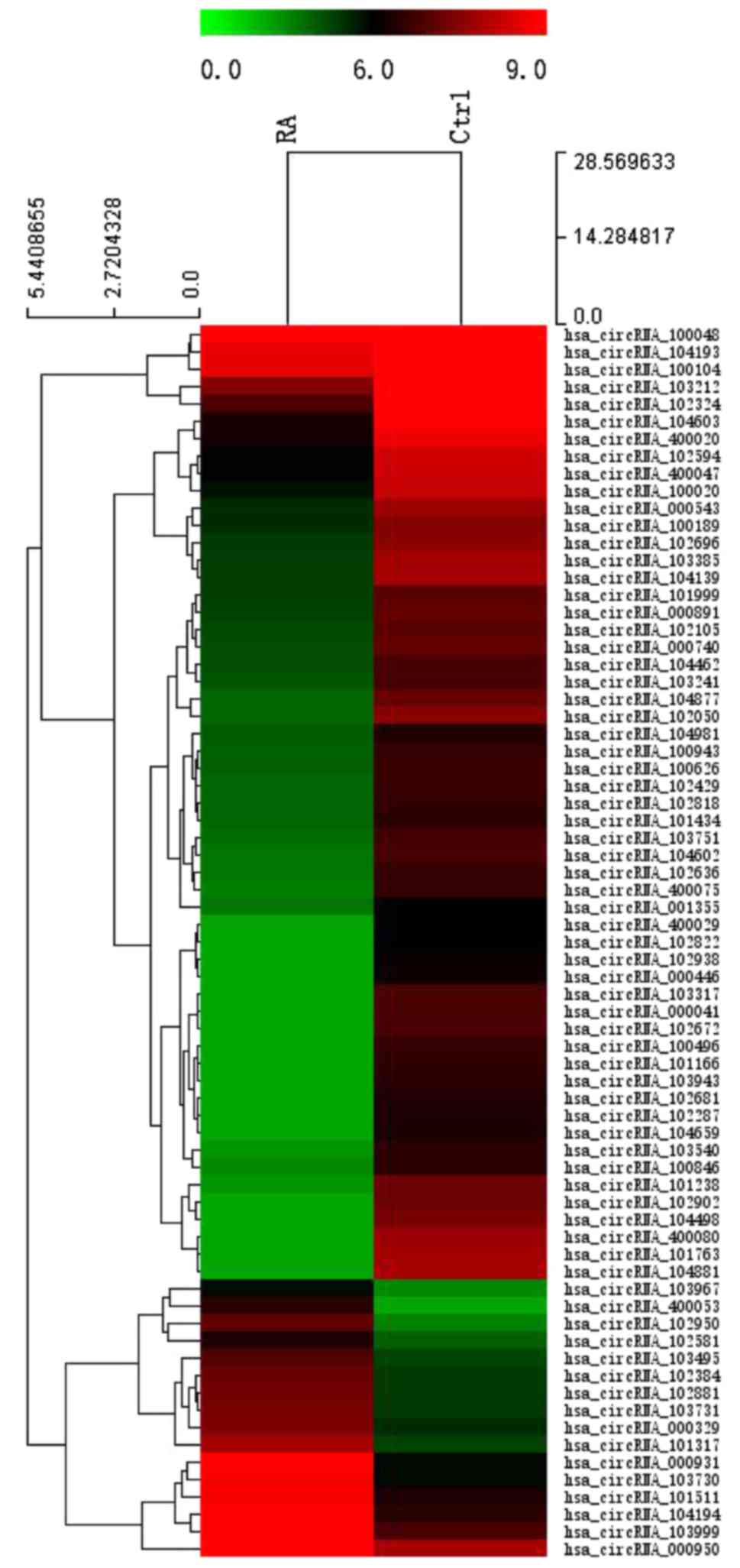
Moreover, on the basic of all the target circRNA, the 584
differentially expressed circRNAs were converted into a
Hierarchical clustering which revealed circRNA expression in normal
and RA samples to show distinguishable circRNA expression among
these samples (Fig. 2).
 | Table II.Comparison of RA vs. controls in
PBMCs. |
Table II.
Comparison of RA vs. controls in
PBMCs.
| circRNA | Alias | Fold-change | GeneSymbol | MRE1 | MRE2 | MRE3 | MRE4 | MRE5 |
|---|
|
hsa_circRNA_400053 |
hsa_circ_0092285 | 18.1669486 | PNKP | hsa-miR-627-3p | hsa-let-7g-5p | hsa-miR-488-5p |
hsa-miR-449c-5p |
hsa-miR-891a-3p |
|
hsa_circRNA_102950 |
hsa_circ_0058794 | 15.0354961 | AGAP1 | hsa-miR-431-3p |
hsa-miR-181c-5p | hsa-miR-619-3p | hsa-miR-15a-5p | hsa-miR-668-5p |
|
hsa_circRNA_000931 |
hsa_circ_0001045 | 10.4743877 | MRPS5 | hsa-miR-877-3p | hsa-miR-874-3p | hsa-miR-214-5p | hsa-miR-876-5p | hsa-miR-30a-5p |
|
hsa_circRNA_101317 |
hsa_circ_0004137 | 10.1073595 | RBM23 | hsa-miR-138-5p | hsa-miR-422a |
hsa-miR-378a-3p | hsa-miR-378d | hsa-miR-877-3p |
|
hsa_circRNA_103730 |
hsa_circ_0005654 |
7.6028184 | PRDM5 | hsa-miR-578 | hsa-miR-501-5p | hsa-miR-581 | hsa-miR-511-5p | hsa-miR-588 |
|
hsa_circRNA_104194 |
hsa_circ_0004712 |
7.0503716 | PDE7B | hsa-miR-455-3p | hsa-let-7g-5p | hsa-miR-876-3p | hsa-miR-661 |
hsa-miR-323a-5p |
|
hsa_circRNA_103967 |
hsa_circ_0074229 |
6.956375 | UBE2D2 | hsa-miR-512-3p | hsa-miR-215-3p | hsa-miR-105-5p | hsa-miR-377-3p | hsa-miR-876-3p |
|
hsa_circRNA_103731 |
hsa_circ_0070819 |
5.9690673 | PRDM5 | hsa-miR-578 | hsa-miR-511-5p | hsa-miR-588 | hsa-miR-619-3p | hsa-miR-183-5p |
|
hsa_circRNA_102881 |
hsa_circ_0057582 |
5.8787181 | HECW2 | hsa-miR-152-5p | hsa-miR-96-3p | hsa-miR-380-5p |
hsa-miR-365a-5p |
hsa-miR-103a-3p |
|
hsa_circRNA_103999 |
hsa_circ_0074854 |
5.6591339 | MAT2B | hsa-miR-646 | hsa-miR-367-3p | hsa-miR-32-5p | hsa-miR-338-3p | hsa-miR-25-3p |
|
hsa_circRNA_104881 |
hsa_circ_0088088 | −46.833615 | HSDL2 | hsa-miR-16-5p |
hsa-miR-135b-5p | hsa-miR-424-5p |
hsa-miR-135a-5p | hsa-miR-15a-5p |
|
hsa_circRNA_101763 |
hsa_circ_0038644 | −44.833615 | PRKCB |
hsa-miR-181b-5p |
hsa-miR-181c-5p |
hsa-miR-181d-5p |
hsa-miR-181a-5p | hsa-miR-329-5p |
|
hsa_circRNA_400080 |
hsa_circ_0092348 | −41.966949 | LARP7 |
hsa-miR-302b-3p |
hsa-miR-302a-3p |
hsa-miR-302d-3p |
hsa-miR-302c-3p |
hsa-miR-302b-5p |
|
hsa_circRNA_104498 |
hsa_circ_0082452 | −32.166949 | EXOC4 | hsa-miR-493-5p | hsa-miR-20a-3p |
hsa-miR-1185-5p | hsa-miR-762 | hsa-miR-433-5p |
|
hsa_circRNA_102902 |
hsa_circ_0057980 | −28.966949 | PIKFYVE | hsa-miR-455-5p |
hsa-miR-181a-3p | hsa-miR-589-5p | hsa-miR-320b | hsa-miR-320a |
|
hsa_circRNA_101238 |
hsa_circ_0004372 | −22.841417 | PAN3 | hsa-miR-320b | hsa-miR-320a | hsa-miR-138-5p | hsa-miR-593-5p | hsa-miR-320c |
|
hsa_circRNA_000041 |
hsa_circ_0000035 | −22.833615 | WASF2 | hsa-miR-372-5p | hsa-miR-345-3p | hsa-miR-491-3p | hsa-miR-204-3p | hsa-miR-558 |
|
hsa_circRNA_103317 |
hsa_circ_0064735 | −22.833615 | UBP1 | hsa-miR-136-5p | hsa-miR-138-5p | hsa-miR-134-3p | hsa-miR-30b-3p | hsa-miR-223-5p |
|
hsa_circRNA_102672 |
hsa_circ_0003195 | −22.133615 | TTC27 | hsa-miR-589-3p |
hsa-miR-518f-5p |
hsa-miR-520c-5p |
hsa-miR-518d-5p | hsa-miR-526a |
|
hsa_circRNA_100496 |
hsa_circ_0017076 | −19.366949 | GNG4 | hsa-miR-30b-3p | hsa-miR-650 | hsa-miR-485-5p |
hsa-let-7f-2-3p | hsa-miR-370-3p |
Interaction between differential
expression of circRNAs and miRNA
With the Arraystar's homemade miRNA target
prediction software based on miRanda, microRNAs paring to circRNAs
was predicted and the the top 5 predicted miRNA targets of circRNA
were selected. The differentially expressed circRNAs is listed with
their MREs as supplementary data. The top ten up and downregulated
circRNA and their target microRNAs are also shown in the Table II. As we and others have
previously observed the differential expression of miRNAs which
shown to be associated with physiopathologicmachanisms in RA. All
the differentially expressed circRNAs in our study are were notated
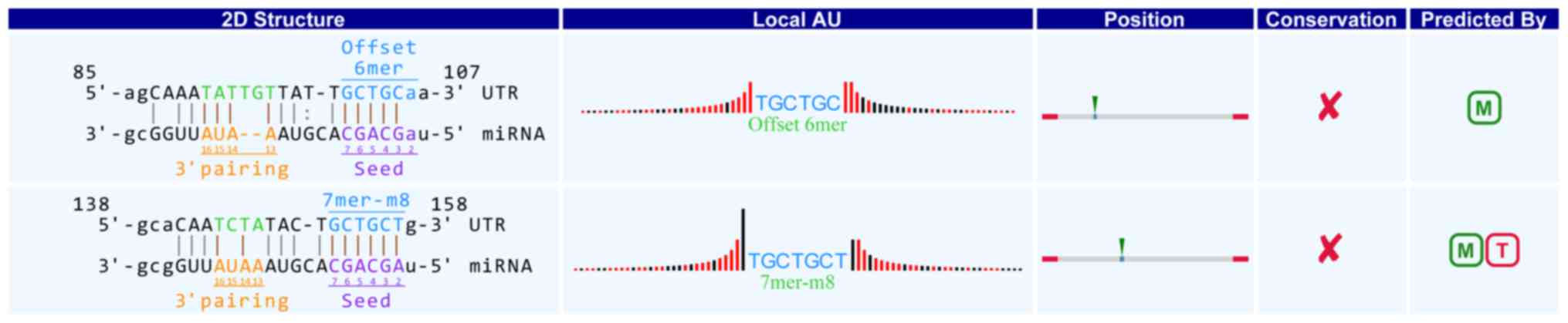
in detail. For example, in hsa_circRNA_104881, the 101st-106th and
152nd-157th nucleotides are beginning from the 5′ terminal region
in the higher expressed gene HSDL2. The nucleotides were completely
and represent a complete matched to the seed region of miR-16-5p in
the binding mode of Offset 6mer and 7mer-m8 respectively (positions
2–8) (Fig. 3). Through target gene
prediction, target genes of the 5 aforementioned miRNAs were
collected. The results of Gene symbol analysis on the up and
downregulated circRNAs with identified target genes were shown in
Table II. Gene symbol analysis
revealed that some target genes were involved in some biological
processes. These processes were associated with pathogenesis of
this disorder.
Validation of circRNAs using
RT-qPCR
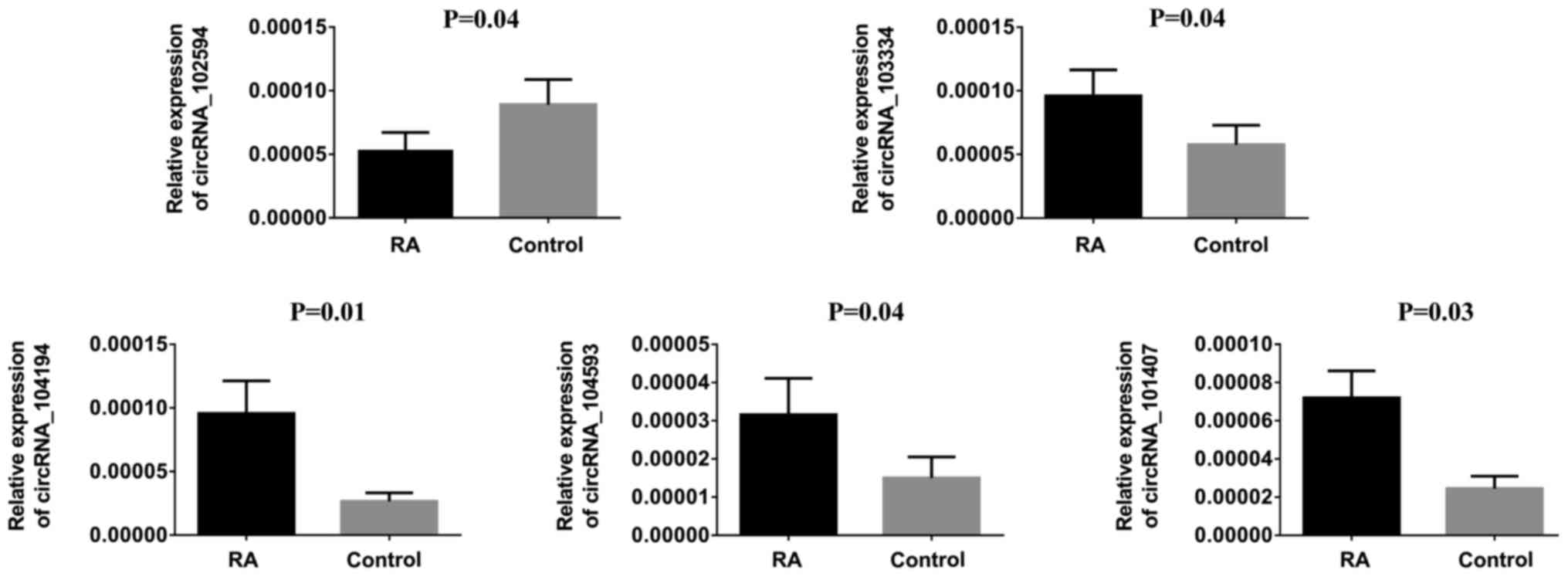
To verify our observations, hsa_circRNA_104194,
hsa_circRNA_104593, hsa_circRNA_103334, hsa_circRNA_101407 and
hsa_circRNA_102594 were selected to be further verifled using
RT-qPCR. The results are shown in Fig.
4. For example, the expression of hsa_circRNA_104194 was
upregulated 1.64-fold in the RA group compared to the control group
(P<0.010), which was consistent with the microarray results.
β-actin was used as the internal control; The calculated ratios of
other genes expression are given in Fig. 4.
Discussion
circRNAs are ubiquitous and widely expressed, shows
it is functionally important. The most well known circRNAs is
CDR1as, located in cytoplasm (12), and owned 74 miR-7 seed binding
sites. It can effectively reduce exerted miR-7 by the function as
miRNA sponge in body tissuse and affect disease such as cancers
(18). circular Sry is also a
demonstration of miRNA sponge (7)
and 16 miRNA-138 target sites were found in circular Sry. Besides,
there are evidences shows the decrease in polymorphisms at miRNA
target sites in circRNAs (19,20).
circRNAs could potentially regulate mechanism of pathology in
diseases.
Although lack of 5′cap and 3′poly (A) tail, circRNA
have protein-coding sequences, cap-independent translation of
certain linear mRNA and open reading frames, which illustrated that
circRNA have potential to translation (21,22),
However, no evident was supported the existence of translatable
circRNAs (23).
The most in-depth study of the relationship between
circRNAs and diseases is cancer. Researchers found their potential
of biomarkers to diagnose tumors. For example, cir-ITCH
downregulated in esophageal squamous cell carcinoma and colorectal
cancer by increasing ITCH expression by suppressing the
Wnt/β-catenin pathway (15,24).
Decreasing Has_circ_002059 can be consider as biomarker in gastric
cancer tissues (25) and
Has_-circ_0001649 low expressed in hepatocellular carcinoma (HCC)
tissues (26). CircRNAs has
abundant miRNA attachment sites and more than 800 miRNAs regulate
one-third of the genes, especially immune-related genes (27). CircRNAs have high degree of
stability in mammalian cells that we speculate that its change can
really reflect relationship with the disease compared with other
noncoding RNA.
We revealed that circRNAs were differentially
expressed in PBMC of RA patients compared with healthy controls in
this study. The maximum ratio of up and downregulated is 18.17 and
46.83. Several cicRNAs aberrant distinctly, these circRNAs may play
an important role in the pathogenesis of RA.
RA is immune system disorders involved in a variety
of factors of immune system disorders. It affects the activities of
the joints as well as the functions of many other organs. Although
the treatment and diagnosis of RA was improved, the specific
mechanism of etiology is still remains unclear. The function
circRNAs of in disorders gives us a new direction to understand the
RA. The circRNAs can regulate the expression of the mRNA by
reducing the inhibitory effect of miRNAs (28).
In this study, we found that hsa_circ_0092285,
hsa_circ_0058794 were significantly higher in PBMC of RA patients
and hsa_circ_0088088, hsa_circ_0038644 were lower.
hsa_circ_0092285 is aligned from the PNKP gene.
Polynucleotide phosphatase/kinase in humans is encoded by the PNKP
gene (29). Oxidative
stress-induced free radicals can be used as oxidants and
inflammatory mediators involving in the pathogenesis of RA
(30,31). Polynucleotide phosphatase/kinase
plays a major role in the restoration of correct DNA termini
following strand cleavage by ROS (32). which induces mutations in
itochondrial DNA (mtDNA) lesions (33) and sensitizes cells to hydrogen
peroxide (34). Downregulated PNKP
gene are the major source of accumulation of strand breaks in mtDNA
of hydrogen peroxide-treated cells and H2O2-induced cells (35). Therefore, we hypothesized that PNKP
is associated with the development of oxidative stress and
inflammation in RA.
hsa_circ_0058794 is spliced from the AGAP1 gene.
AGAP1 is a prototype of the ArfGAP protein with the GTPase-like
domain and a phosphoinositide-dependent Arf GAP and affects the
dynamics of the actin cytoskeleton (36). One of the characteristics of RA
synoviocytes is the invasion and migration of cartilage (37). Migrating to the unaffected joints
is a manifestation of disease progression. Invasion and migration
require the integration of signals from the extracellular
environment and remodeling the actin cytoskeleton (38,39).
AGAP1 could induce and localizes to Rab4/AP1 containing a putative
endosome structure and specifically altering the stress fiber.
AGAP1 is a possible link between endocytic traffic anthe actin
cytoskeleton (36).
hsa_circ_0088088 is the most pronounced circRNA
downregulated in RA and spliced from hydroxysteroid
dehydrogenase-like 2 genes. HSDL2 is one of the members of
short-chain dehydrogenases/reductase (SDR) superfamily containing a
peroxisomal targeting signal (40). Human HSDL2 was found in peroxisomes
or mitochondria, some researches assumed the involvement of HSDL2
in fatty acid and cholesterol metabolism and homeostasis for its
localization (41). Existence of
dyslipidemia in the early RA and RA progression has been confirmed
(42). Cardiovascular risk is the
main cause of early death in patients with RA (43). Inflammatory cytokines promote lipid
breakdown and increase free fatty acid levels, causing endothelial
dysfunction, thereby increasing the risk of atherosclerosis in RA
(44).
hsa_circ_0038644 is encoded within PRKCB.PRKCB is
indicated as a candidate gene associated with the LPS immune
response, which involves B cell viability and antigen response
(45). Lipopolysaccharide (LPS) is
an inflammatory factor, which is a common ligand for TLR2 and TLR4.
When LPS binds to TLR2 and TLR4, it activates TLR signal
transduction pathway and participates in the immune regulation
function (46). Immune disorder is
the main pathogenesis of RA. TLRs and LPS considered members of the
innate immune system inducing proinflammatory cytokine production
has an important role in this disorder. However, TLRs, especially
TLR2 and TLR4, play an important role in the pathogenesis of RA
(47). PRKCB gene could recruit
IKK into lipid rafts and is involved in B-cell receptor
(BCR)-mediated NF-κΒ activation (48).
miRNAs are tissue-specific and stage-specific of
disease progression. It involves cancer, cell apoptosis,
inflammation, even autoimmune arthritis (49). miRNAs participates in generation of
MMPs, leukocyte activation, inflammatory responses, and cell
proliferation of synoviocytes in rheumatoid joints (50,51).
Previous studies have found that many miRNAs were associated with
RA.miR-146a, miR-155, miR-132, and miR-16 are increased in PBMCs in
RA patients compared to HCs (52).
Recently, miR-24-3p, and miR125a-5p were reported to
be helpful in the diagnosis of RA (53) and generation of chemokine and
inflammatory cytokines. Many studies have shown that some potential
effects between cicrRNAson miRNAs, however, the role of circRNAs
and miRNAs on the impaction of RA is not very clear. Theoretically,
as a sponge of miRNAs, the circRNA can competitively bind and
inhibit the corresponding miRNAs, increasing the target of the
corresponding miRNA. hsa_circ_0057980 is significantly
downregulated in this study. miR-181d is one of its MREs. Our
findings are consistent with a recent study in which miR-181d is
significantly increased in RA patients compared with HCs (54). Likewise, hsa-miR-16-5p, one of the
hsa_circ_0088088 downregulated in our study with the largest times,
means increase distinctly. miR-16 is mainly correlated with the
degree of activity of the disease such erythrocyte sedimentation
rate (ESR), C-reactive protein (CRP) value, tender joint count
(TJC) and TNF-α in RA patients (52). It also parallel to DAS28 index of
RA (55). Therefore, we could
conclude that miR-16 paly important role in pathogenesis of RA. On
the contrary, miR-30a was reported to have dropped significantly in
RA, which has a consistent result with our research that
hsa_circ_0001045 is significantly upregulated. Low levels of
miR-30a have been shown to be associated with reduced cell
apoptosis in RA synovialis (56).
Although many miRNAs above have relation with RA
described by other researchers, there are still many of them are
not clear or not studied so far. Some of them may have certain
correlation with the occurrence and development of RA. Studies have
found that hsa-let-7g-5p is significantly higher in patients by
affecting many metabolic pathways, such as lipid metabolism
(57). miR-488 was significantly
decreased in osteoarthritis (OA) chondrocytes by regulating zinc
transporter SLC39A8/ZIP8. Notably, OA involves musculoskeletal
disorder and also leads to joint injury (58).
Even though we have analyzed and discussed the role
of circRNA and miRNA in RA, there are also some limitations in this
study. First of all, the sample size is too small to get an
affirmative conclusion. Secondly, all specimens come from one
hospital, resulting in the possibility of regional characteristics.
Thirdly, detailed functional analysis is insufficient, so further
research needs to improve this deficiency. Maybe, we could
investigate the circRNA profile of regulatory T cells or some other
cell kinds which have been demonstrated to play key roles in RA
instead of peripheral blood mononuclear cells. Although our study
did not select and validate a specific circRNA for the
pathophysiology and molecular mechanisms of RA, nevertheless our
study may provide a new perspective for the early diagnosis and
pathogenesis of RA.
Overall, our study indicated that the expression of
circRNAs is significantly different between RA patients and HCs.
The circRNA as a miRNA sponge may play an important functional role
in RA. We should make good use of their roles in sponging miRNA to
prevent the harms caused by these overexpressed or low expressed
miRNAs in RA patients for treatment. There is no doubt that the
deep study is needed to reveal the role of molecular mechanism of
circRNA in the development and progression of RA.
Acknowledgements
This study was supported by Shenzhen Science and
Technology Projector of Guangdong Province (no.
JCYJ20160422164313440). We thank all subjects who participated in
this study. We also thank the Department of Nephrology, Shenzhen
Key Subject of Medicine of Guangdong Province.
References
|
1
|
Firestein GS: Evolving concepts of
rheumatoid arthritis. Nature. 423:356–361. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Salemi S, Biondo MI, Fiorentino C, Argento
G, Paolantonio M, Di Murro C, Malagnino VA, Canzoni M, Diamanti AP
and D'Amelio R: Could early rheumatoid arthritis resolve after
periodontitis treatment only?: Case report and review of the
literature. Medicine (Baltimore). 93:e1952014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ursini F, Russo E, Hribal M Letizia, Mauro
D, Savarino F, Bruno C, Tripolino C, Rubino M, Naty S and Grembiale
RD: Abatacept improves whole-body insulin sensitivity in rheumatoid
arthritis: An observational study. Medicine (Baltimore).
94:e8882015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Aletaha D, Neogi T, Silman AJ, Funovits J,
Felson DT, Bingham CO III, Birnbaum NS, Burmester GR, Bykerk VP,
Cohen MD, et al: 2010 Rheumatoid arthritis classification criteria:
An American College of Rheumatology/European League Against
Rheumatism collaborative initiative. Arthritis Rheum. 62:2569–2581.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Goekoop-Ruiterman YP, de Vries-Bouwstra
JK, Allaart CF, van Zeben D, Kerstens PJ, Hazes JM, Zwinderman AH,
Ronday HK, Han KH, Westedt ML, et al: Clinical and radiographic
outcomes of four different treatment strategies in patients with
early rheumatoid arthritis (the BeSt study): A randomized,
controlled trial. Arthritis Rheum. 52:3381–3390. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chen XM, Huang QC, Yang SL, Chu YL, Yan
YH, Han L, Huang Y and Huang RY: Role of Micro RNAs in the
pathogenesis of rheumatoid arthritis: Novel perspectives based on
review of the literature. Medicine (Baltimore). 94:e13262015.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Qu S, Yang X, Li X, Wang J, Gao Y, Shang
R, Sun W, Dou K and Li H: Circular RNA: A new star of noncoding
RNAs. Cancer Lett. 365:141–148. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Conn SJ, Pillman KA, Toubia J, Conn VM,
Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA and
Goodall GJ: The RNA binding protein quaking regulates formation of
circRNAs. Cell. 160:1125–1134. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Qu S, Song W, Yang X, Wang J, Zhang R,
Zhang Z, Zhang H and Li H: Microarray expression profile of
circular RNAs in human pancreatic ductal adenocarcinoma. Genom
Data. 5:385–387. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sanger HL, Klotz G, Riesner D, Gross HJ
and Kleinschmidt AK: Viroids are single-stranded covalently closed
circular RNA molecules existing as highly base-paired rod-like
structures. Proc Natl Acad Sci USA. 73:pp. 3852–3856. 1976;
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hansen TB, Kjems J and Damgaard CK:
Circular RNA and miR-7 in cancer. Cancer Res. 73:5609–5612. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kulcheski FR, Christoff AP and Margis R:
Circular RNAs are miRNA sponges and can be used as a new class of
biomarker. J Biotechnol. 238:42–51. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li F, Zhang L, Li W, Deng J, Zheng J, An
M, Lu J and Zhou Y: Circular RNA ITCH has inhibitory effect on ESCC
by suppressing the Wnt/β-catenin pathway. Oncotarget. 6:6001–6013.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Nagpal JK, Dasgupta S, Jadallah S, Chae
YK, Ratovitski EA, Toubaji A, Netto GJ, Eagle T, Nissan A,
Sidransky D and Trink B: Profiling the expression pattern of GPI
transamidase complex subunits in human cancer. Mod Pathol.
21:979–991. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mendell JT, ap Rhys CM and Dietz HC:
Separable roles for rent1/hUpf1 in altered splicing and decay of
nonsense transcripts. Science. 298:419–422. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Peng L, Yuan XQ and Li GC: The emerging
landscape of circular RNA ciRS-7 in cancer (Review). Oncol Rep.
33:2669–2674. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Thomas LF and Saetrom P: Circular RNAs are
depleted of polymorphisms at microRNA binding sites.
Bioinformatics. 30:2243–2246. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang Y and Wang Z: Efficient backsplicing
produces translatable circular mRNAs. RNA. 21:172–179. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gilbert WV: Alternative ways to think
about cellular internal ribosome entry. J Biol Chem.
285:29033–29038. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Guo JU, Agarwal V, Guo H and Bartel DP:
Expanded identification and characterization of mammalian circular
RNAs. Genome Biol. 15:4092014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jeck WR, Sorrentino JA, Wang K, Slevin MK,
Burd CE, Liu J, Marzluff WF and Sharpless NE: Circular RNAs are
abundant, conserved, and associated with ALU repeats. RNA.
19:141–157. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Huang G, Zhu H, Shi Y, Wu W, Cai H and
Chen X: cir-ITCH plays an inhibitory role in colorectal cancer by
regulating the Wnt/β-catenin pathway. PLoS One. 10:e01312252015.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhao ZJ and Shen J: Circular RNA
participates in the carcinogenesis and the malignant behavior of
cancer. RNA Biol. 14:514–521. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Qin M, Liu G, Huo X, Tao X, Sun X, Ge Z,
Yang J, Fan J, Liu L and Qin W: Hsa_circ_0001649: A circular RNA
and potential novel biomarker for hepatocellular carcinoma. Cancer
Biomark. 16:161–169. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bentwich I, Avniel A, Karov Y, Aharonov R,
Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, et al:
Identification of hundreds of conserved and nonconserved human
microRNAs. Nat Genet. 37:766–770. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cheng DL, Xiang YY, Ji LJ and Lu XJ:
Competing endogenous RNA interplay in cancer: Mechanism,
methodology, and perspectives. Tumour Biol. 36:479–488. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Weinfeld M, Mani RS, Abdou I, Aceytuno RD
and Glover JN: Tidying up loose ends: The role of polynucleotide
kinase/phosphatase in DNA strand break repair. Trends Biochem Sci.
36:262–271. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hitchon CA and El-Gabalawy HS: Oxidation
in rheumatoid arthritis. Arthritis Res Ther. 6:265–278. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Phaniendra A, Jestadi DB and Periyasamy L:
Free radicals: Properties, sources, targets, and their implication
in various diseases. Indian J Clin Biochem. 30:11–26. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Das A, Wiederhold L, Leppard JB, Kedar P,
Prasad R, Wang H, Boldogh I, Karimi-Busheri F, Weinfeld M,
Tomkinson AE, et al: NEIL2-initiated, APE-independent repair of
oxidized bases in DNA: Evidence for a repair complex in human
cells. DNA Repair (Amst). 5:1439–1448. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ralph SJ, Rodriguez-Enriquez S, Neuzil J,
Saavedra E and Moreno-Sánchez R: The causes of cancer revisited:
‘Mitochondrial malignancy’ and ROS-induced oncogenic
transformation-why mitochondria are targets for cancer therapy. Mol
Aspects Med. 31:145–170. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Rasouli-Nia A, Karimi-Busheri F and
Weinfeld M: Stable down-regulation of human polynucleotide kinase
enhances spontaneous mutation frequency and sensitizes cells to
genotoxic agents. Proc Natl Acad Sci USA. 101:pp. 6905–6910. 2004;
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Tahbaz N, Subedi S and Weinfeld M: Role of
polynucleotide kinase/phosphatase in mitochondrial DNA repair.
Nucleic Acids Res. 40:3484–3495. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Nie Z, Stanley KT, Stauffer S, Jacques KM,
Hirsch DS, Takei J and Randazzo PA: AGAP1, an endosome-associated,
phosphoinositide-dependent ADP-ribosylation factor
GTPase-activating protein that affects actin cytoskeleton. J Biol
Chem. 277:48965–48975. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Bartok B, Hammaker D and Firestein GS:
Phosphoinositide 3-kinase δ regulates migration and invasion of
synoviocytes in rheumatoid arthritis. J Immunol. 192:2063–2070.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ridley AJ, Schwartz MA, Burridge K, Firtel
RA, Ginsberg MH, Borisy G, Parsons JT and Horwitz AR: Cell
migration: Integrating signals from front to back. Science.
302:1704–1709. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Parsons JT, Horwitz AR and Schwartz MA:
Cell adhesion: Integrating cytoskeletal dynamics and cellular
tension. Nat Rev Mol Cell Biol. 11:633–643. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Dai J, Xie Y, Wu Q, Wang L, Yin G, Ye X,
Zeng L, Xu J, Ji C, Gu S, et al: Molecular cloning and
characterization of a novel human hydroxysteroid dehydrogenase-like
2 (HSDL2) cDNA from fetal brain. Biochem Genet. 41:165–174. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Skogsberg J, Lundström J, Kovacs A,
Nilsson R, Noori P, Maleki S, Köhler M, Hamsten A, Tegnér J and
Björkegren J: Transcriptional profiling uncovers a network of
cholesterol-responsive atherosclerosis target genes. PLoS Genet.
4:e10000362008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Peters MJ, Vis M, van Halm VP, Wolbink GJ,
Voskuyl AE, Lems WF, Dijkmans BA, Twisk JW, de Koning MH, van de
Stadt RJ and Nurmohamed MT: Changes in lipid profile during
infliximab and corticosteroid treatment in rheumatoid arthritis.
Ann Rheum Dis. 66:958–961. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Pincus T, Sokka T and Wolfe F: Premature
mortality in patients with rheumatoid arthritis: Evolving concepts.
Arthritis Rheum. 44:1234–1236. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Boden G: Obesity and free fatty acids.
Endocrinol Metab Clin North Am. 37:635–646, viii-ix. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Siwek M, Slawinska A, Rydzanicz M, Wesoly
J, Fraszczak M, Suchocki T, Skiba J, Skiba K and Szyda J:
Identification of candidate genes and mutations in QTL regions for
immune responses in chicken. Anim Genet. 46:247–254. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Gierut A, Perlman H and Pope RM: Innate
immunity and rheumatoid arthritis. Rheum Dis Clin North Am.
36:271–296. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Roelofs MF, Wenink MH, Brentano F,
Abdollahi-Roodsaz S, Oppers-Walgreen B, Barrera P, van Riel PL,
Joosten LA, Kyburz D, van den Berg WB and Radstake TR: Type I
interferons might form the link between Toll-like receptor (TLR)
3/7 and TLR4-mediated synovial inflammation in rheumatoid arthritis
(RA). Ann Rheum Dis. 68:1486–1493. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Sheng YJ, Gao JP, Li J, Han JW, Xu Q, Hu
WL, Pan TM, Cheng YL, Yu ZY, Ni C, et al: Follow-up study
identifies two novel susceptibility loci PRKCB and 8p11.21 for
systemic lupus erythematosus. Rheumatology (Oxford). 50:682–688.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Nagata Y, Nakasa T, Mochizuki Y, Ishikawa
M, Miyaki S, Shibuya H, Yamasaki K, Adachi N, Asahara H and Ochi M:
Induction of apoptosis in the synovium of mice with
autoantibody-mediated arthritis by the intraarticular injection of
double-stranded MicroRNA-15a. Arthritis Rheum. 60:2677–2683. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Nakasa T, Nagata Y, Yamasaki K and Ochi M:
A mini-review: microRNA in arthritis. Physiol Genomics. 43:566–570.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Baxter D, McInnes IB and
Kurowska-Stolarska M: Novel regulatory mechanisms in inflammatory
arthritis: A role for microRNA. Immunol Cell Biol. 90:288–292.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Pauley KM, Satoh M, Chan AL, Bubb MR,
Reeves WH and Chan EK: Upregulated miR-146a expression in
peripheral blood mononuclear cells from rheumatoid arthritis
patients. Arthritis Res Ther. 10:R1012008. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Liu Y, Dong J, Mu R, Gao Y, Tan X, Li Y,
Li Z and Yang G: MicroRNA-30a promotes B cell hyperactivity in
patients with systemic lupus erythematosus by direct interaction
with Lyn. Arthritis Rheum. 65:1603–1611. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Wang W, Zhang Y, Zhu B, Duan T, Xu Q, Wang
R, Lu L and Jiao Z: Plasma microRNA expression profiles in Chinese
patients with rheumatoid arthritis. Oncotarget. 6:42557–42568.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Murata K, Yoshitomi H, Tanida S, Ishikawa
M, Nishitani K, Ito H and Nakamura T: Plasma and synovial fluid
microRNAs as potential biomarkers of rheumatoid arthritis and
osteoarthritis. Arthritis Res Ther. 12:R862010. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Xu K, Xu P, Yao JF, Zhang YG, Hou WK and
Lu SM: Reduced apoptosis correlates with enhanced autophagy in
synovial tissues of rheumatoid arthritis. Inflamm Res. 62:229–237.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Zafari S, Backes C, Meese E and Keller A:
Circulating biomarker panels in Alzheimer's disease. Gerontology.
61:497–503. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Egloff C, Hugle T and Valderrabano V:
Biomechanics and pathomechanisms of osteoarthritis. Swiss Med Wkly.
142:w135832012.PubMed/NCBI
|