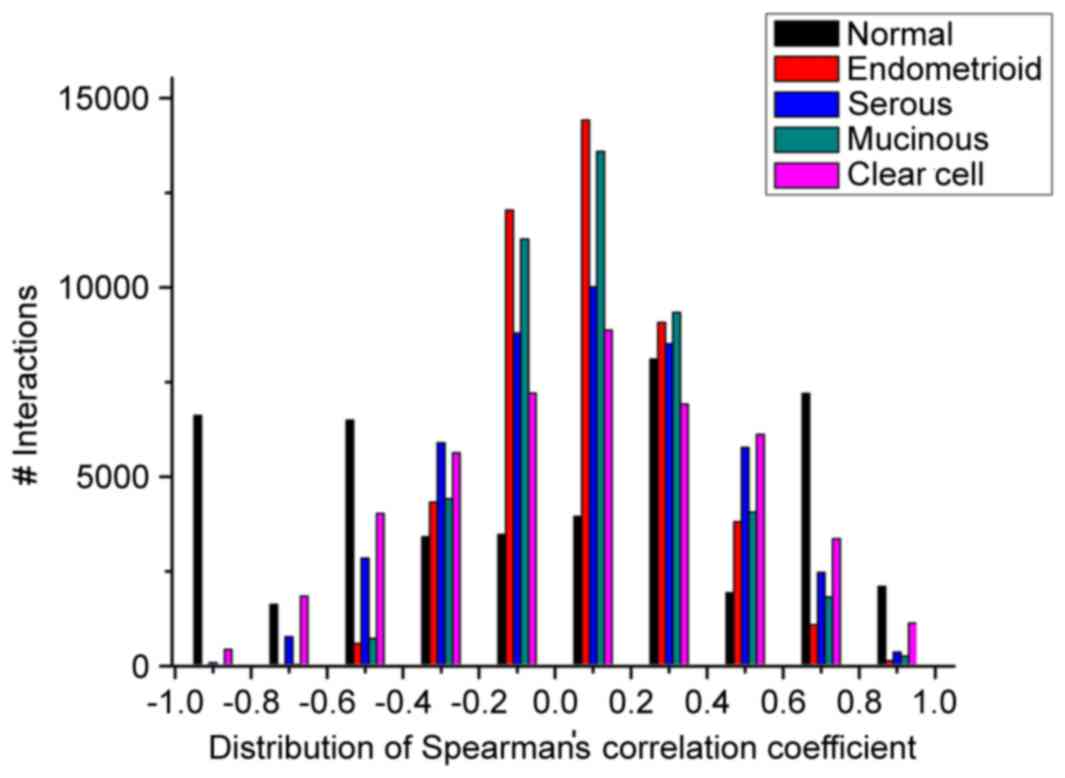
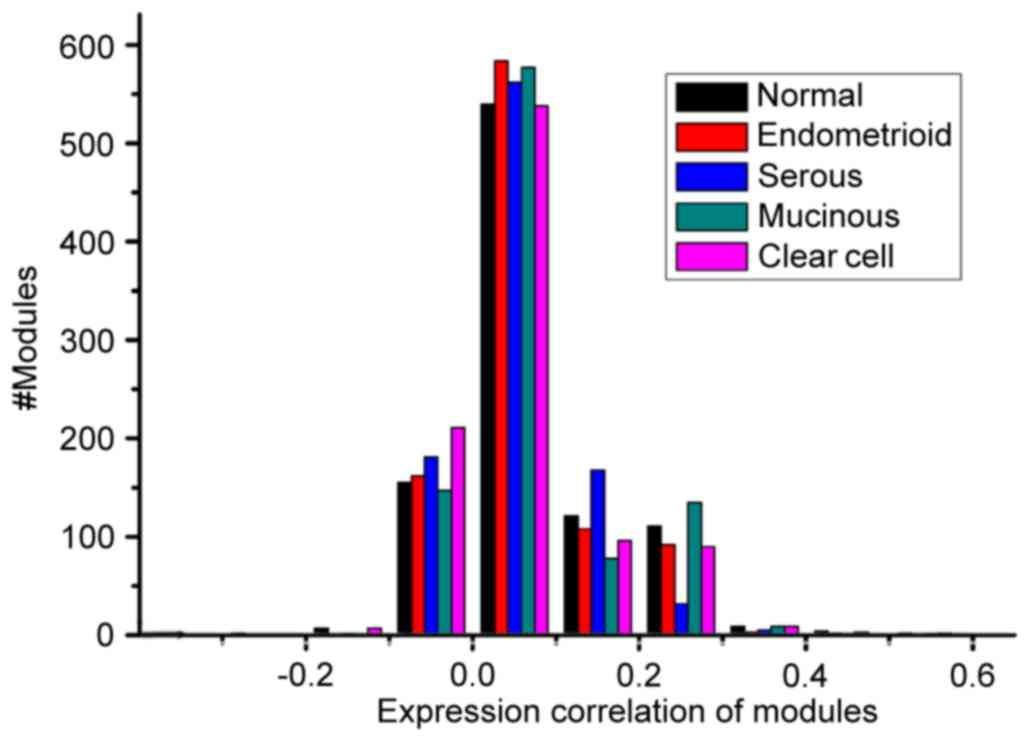
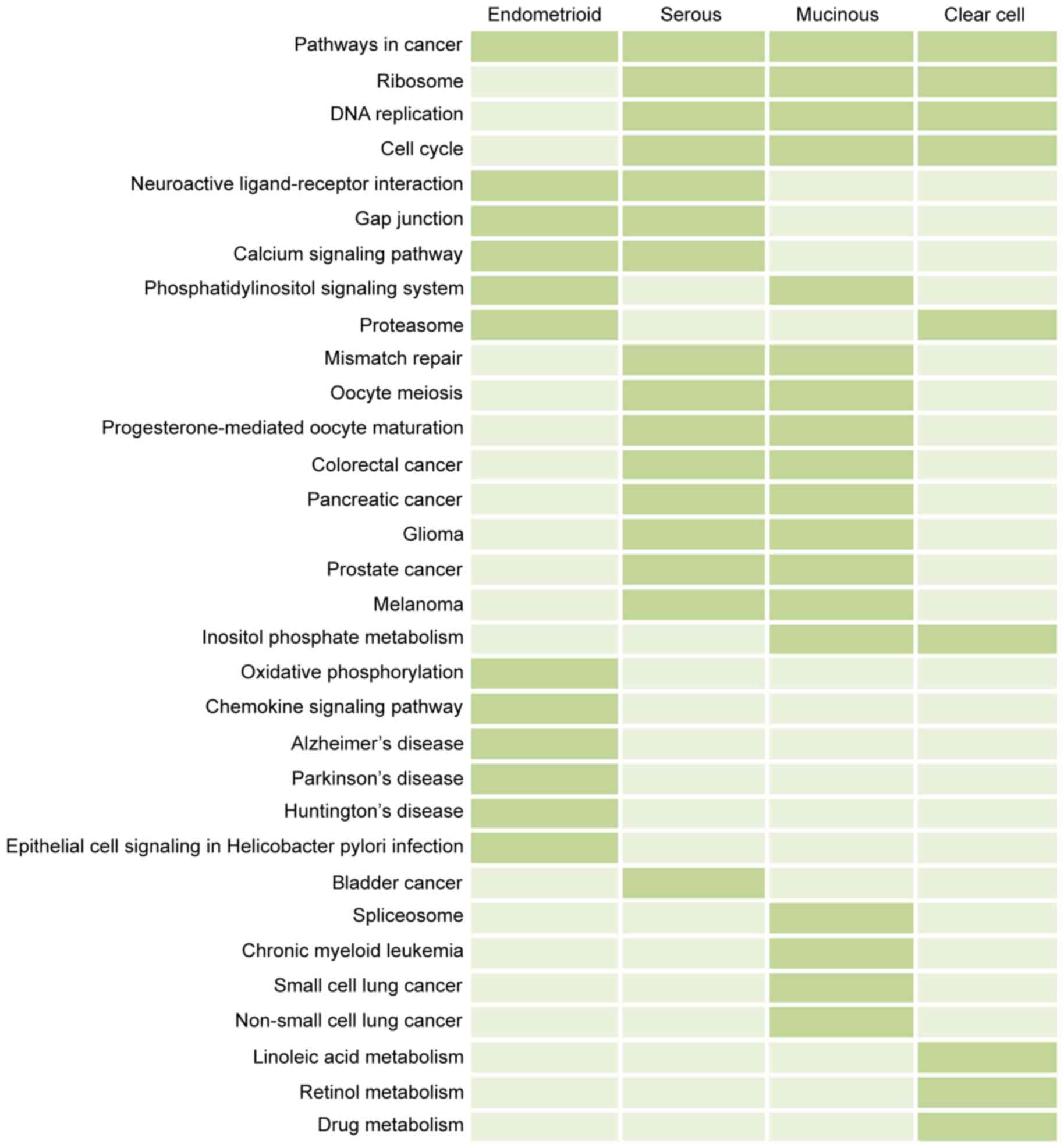
|
1
|
Coleman RL, Monk BJ, Sood AK and Herzog
TJ: Latest research and treatment of advanced-stage epithelial
ovarian cancer. Nat Rev Clin Oncol. 10:211–224. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fredrickson TN: Ovarian tumors of the hen.
Environ Health Perspect. 73:35–51. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Barua A, Bitterman P, Abramowicz JS, Dirks
AL, Bahr JM, Hales DB, Bradaric MJ, Edassery SL, Rotmensch J and
Luborsky JL: Histopathology of ovarian tumors in laying hens: A
preclinical model of human ovarian cancer. Int J Gynecol Cancer.
19:531–539. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Feeley KM and Wells M: Precursor lesions
of ovarian epithelial malignancy. Histopathology. 38:87–95. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bell DA: Origins and molecular pathology
of ovarian cancer. Mod Pathol. 18 Suppl 2:S19–S32. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Moorman PG, Palmieri RT, Akushevich L,
Berchuck A and Schildkraut JM: Ovarian cancer risk factors in
African-American and white women. Am J Epidemiol. 170:598–606.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sharifian A, Pourhoseingholi MA,
Norouzinia M and Vahedi M: Ovarian cancer in Iranian women, a trend
analysis of mortality and incidence. Asian Pac J Cancer Prev.
15:10787–10790. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Vaughan S, Coward JI, Bast RC Jr, Berchuck
A, Berek JS, Brenton JD, Coukos G, Crum CC, Drapkin R,
Etemadmoghadam D, et al: Rethinking ovarian cancer: Recommendations
for improving outcomes. Nat Rev Cancer. 11:719–725. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lengauer C, Kinzler KW and Vogelstein B:
Genetic instabilities in human cancers. Nature. 396:643–649. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Konstantinopoulos PA, Spentzos D and
Cannistra SA: Gene-expression profiling in epithelial ovarian
cancer. Nat Clin Pract Oncol. 5:577–587. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Meinhold-Heerlein I, Bauerschlag D, Zhou
Y, Sapinoso LM, Ching K, Frierson H Jr, Bräutigam K, Sehouli J,
Stickeler E, Könsgen D, et al: An integrated clinical-genomics
approach identifies a candidate multi-analyte blood test for serous
ovarian carcinoma. Clin Cancer Res. 13:458–466. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ansenberger K, Zhuge Y, Lagman JA,
Richards C, Barua A, Bahr JM and Hales DB: E-cadherin expression in
ovarian cancer in the laying hen, Gallus domesticus, compared to
human ovarian cancer. Gynecol Oncol. 113:362–369. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jackson E, Anderson K, Ashwell C, Petitte
J and Mozdziak PE: CA125 expression in spontaneous ovarian
adenocarcinomas from laying hens. Gynecol Oncol. 104:192–198. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhuge Y, Lagman JA, Ansenberger K, Mahon
CJ, Daikoku T, Dey SK, Bahr JM and Hales DB: CYP1B1 expression in
ovarian cancer in the laying hen Gallusdomesticus. Gynecol Oncol.
112:171–178. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zidar N, Odar K, Glavac D, Jerse M, Zupanc
T and Stajer D: Cyclooxygenase in normal human tissues-is COX-1
really a constitutive isoform and COX-2 an inducible isoform? J
Cell Mol Med. 13:3753–3763. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Spandidos DA, Dokianakis DN, Kallergi G
and Aggelakis E: Molecular basis of gynecological cancer. Ann NY
Acad Sci. 900:56–64. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cancer Genome Atlas Research Network, .
Integrated genomic analyses of ovarian carcinoma. Nature.
474:609–615. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lafky JM, Wilken JA, Baron AT and Maihle
NJ: Clinical implications of the ErbB/epidermal growth factor (EGF)
receptor family and its ligands in ovarian cancer. Biochim Biophys
Acta. 1785:232–265. 2008.PubMed/NCBI
|
|
19
|
Li Q, Zeng X, Cheng X, Zhang J, Ji J, Wang
J, Xiong K, Qi Q and Huang W: Diagnostic value of dual detection of
hepatocyte nuclear factor 1 beta (HNF-1beta) and napsin A for
diagnosing ovarian clear cell carcinoma. Int J Clin Exp Pathol.
8:8305–8310. 2015.PubMed/NCBI
|
|
20
|
Gavin AC, Aloy P, Grandi P, Krause R,
Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dümpelfeld B,
et al: Proteome survey reveals modularity of the yeast cell
machinery. Nature. 440:631–636. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jordán F, Nguyen TP and Liu WC: Studying
protein-protein interaction networks: A systems view on diseases.
Brief Funct Genomics. 11:497–504. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wu C, Zhu J and Zhang X: Integrating gene
expression and protein-protein interaction network to prioritize
cancer-associated genes. BMC Bioinformatics. 13:1822012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Srihari S and Ragan MA: Systematic
tracking of dysregulated modules identifies novel genes in cancer.
Bioinformatics. 29:1553–1561. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hendrix ND, Wu R, Kuick R, Schwartz DR,
Fearon ER and Cho KR: Fibroblast growth factor 9 has oncogenic
activity and is a downstream target of Wnt signaling in ovarian
endometrioid adenocarcinomas. Cancer Res. 66:1354–1362. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Irizarry RA, Hobbs B, Collin F,
Beazer-Barclay YD, Antonellis KJ, Scherf U and Speed TP:
Exploration, normalization, and summaries of high density
oligonucleotide array probe level data. Biostatistics. 4:249–264.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Nguyen PV, Srihari S and Leong HW:
Identifying conserved protein complexes between species by
constructing interolog networks. BMC Bioinformatics. 14 Suppl
16:S82013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liu G, Wong L and Chua HN: Complex
discovery from weighted PPI networks. Bioinformatics. 25:1891–1897.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tomita E, Tanaka A and Takahashi H: The
worst-case time complexity for generating all maximal cliques and
computational experiments. Theoret Comput Sci. 363:28–42. 2006.
View Article : Google Scholar
|
|
30
|
Benjamini Y, Drai D, Elmer G, Kafkafi N
and Golani I: Controlling the false discovery rate in behavior
genetics research. Behav Brain Res. 125:279–284. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
da W Huang, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009.PubMed/NCBI
|
|
32
|
Chen J, Bai M, Ning C, Xie B, Zhang J,
Liao H, Xiong J, Tao X, Yan D, Xi X, et al: Gankyrin facilitates
follicle-stimulating hormone-driven ovarian cancer cell
proliferation through the PI3K/AKT/HIF-1alpha/cyclin D1 pathway.
Oncogene. 35:2506–2517. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kim YS, Choi KC and Hwang KA: Genistein
suppressed epithelial-mesenchymal transition and migration
efficacies of BG-1 ovarian cancer cells activated by estrogenic
chemicals via estrogen receptor pathway and downregulation of TGF-β
signaling pathway. Phytomedicine. 22:993–999. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tong X, Barbour M, Hou K, Gao C, Cao S,
Zheng J, Zhao Y, Mu R and Jiang HR: Interleukin-33 predicts poor
prognosis and promotes ovarian cancer cell growth and metastasis
through regulating ERK and JNK signaling pathways. Mol Oncol.
10:113–125. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang Q, Tang Y, Yu H, Yin Q, Li M, Shi L,
Zhang W, Li D and Li L: CCL18 from tumor-cells promotes epithelial
ovarian cancer metastasis via mTOR signaling pathway. Mol Carcinog.
55:1688–1699. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Su YQ, Wigglesworth K, Pendola FL, O'Brien
MJ and Eppig JJ: Mitogen-activated protein kinase activity in
cumulus cells is essential for gonadotropin-induced oocyte meiotic
resumption and cumulus expansion in the mouse. Endocrinology.
143:2221–2232. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Guadagno TM and Ferrell JE Jr: Requirement
for MAPK activation for normal mitotic progression in Xenopus egg
extracts. Science. 282:1312–1315. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Gao Y, Ren J, Zhang L, Zhang Y, Wu X,
Jiang H, Xu F, Yuan B, Yu X and Zhang J: The effects of
demecolcine, alone or in combination with sucrose on bovine oocyte
protrusion rate, MAPK1 protein level and c-mos gene expression
level. Cell Physiol Biochem. 34:1974–1982. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Posada J, Yew N, Ahn NG, Woude GF Vande
and Cooper JA: Mos stimulates MAP kinase in Xenopus oocytes and
activates a MAP kinase kinase in vitro. Mol Cell Biol.
13:2546–2553. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Shibuya EK and Ruderman JV: Mos induces
the in vitro activation of mitogen-activated protein kinases in
lysates of frog oocytes and mammalian somatic cells. Mol Biol Cell.
4:781–790. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Huang N, Wang J, Xie W, Lyu Q, Wu J, He J,
Qiu W, Xu N and Zhang Y: MiR-378a-3p enhances adipogenesis by
targeting mitogen-activated protein kinase 1. Biochem Biophys Res
Commun. 457:37–42. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yao R and Cooper GM: Requirement for
phosphatidylinositol-3 kinase in the prevention of apoptosis by
nerve growth factor. Science. 267:2003–2006. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Liang J and Slingerland JM: Multiple roles
of the PI3K/PKB (Akt) pathway in cell cycle progression. Cell
Cycle. 2:339–345. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Lee CM, Fuhrman CB, Planelles V, Peltier
MR, Gaffney DK, Soisson AP, Dodson MK, Tolley HD, Green CL and
Zempolich KA: Phosphatidylinositol 3-kinase inhibition by LY294002
radiosensitizes human cervical cancer cell lines. Clin Cancer Res.
12:250–256. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Keysar SB, Astling DP, Anderson RT, Vogler
BW, Bowles DW, Morton JJ, Paylor JJ, Glogowska MJ, Le PN,
Eagles-Soukup JR, et al: A patient tumor transplant model of
squamous cell cancer identifies PI3K inhibitors as candidate
therapeutics in defined molecular bins. Mol Oncol. 7:776–790. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Bertelsen BI, Steine SJ, Sandvei R, Molven
A and Laerum OD: Molecular analysis of the PI3K-AKT pathway in
uterine cervical neoplasia: Frequent PIK3CA amplification and AKT
phosphorylation. Int J Cancer. 118:1877–1883. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Datta SR, Brunet A and Greenberg ME:
Cellular survival: A play in three Akts. Genes Dev. 13:2905–2927.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Won HS, Jung CK, Chun SH, Kang JH, Kim YS,
Sun DI and Kim MS: Difference in expression of EGFR, pAkt, and PTEN
between oropharyngeal and oral cavity squamous cell carcinoma. Oral
Oncol. 48:985–990. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Chalhoub N and Baker SJ: PTEN and the
PI3-kinase pathway in cancer. Annu Rev Pathol. 4:127–150. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zhang S and Yu D: PI(3)king apart PTEN's
role in cancer. Clin Cancer Res. 16:4325–4330. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Arnold A and Papanikolaou A: Cyclin D1 in
breast cancer pathogenesis. J Clin Oncol. 23:4215–4224. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Motokura T, Bloom T, Kim HG, Jüppner H,
Ruderman JV, Kronenberg HM and Arnold A: A novel cyclin encoded by
a bcl1-linked candidate oncogene. Nature. 350:512–515. 1991.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Wang TC, Cardiff RD, Zukerberg L, Lees E,
Arnold A and Schmidt EV: Mammary hyperplasia and carcinoma in
MMTV-cyclin D1 transgenic mice. Nature. 369:669–671. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Worsley SD, Ponder BA and Davies BR:
Overexpression of cyclin D1 in epithelial ovarian cancers. Gynecol
Oncol. 64:189–195. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Sui L, Tokuda M, Ohno M, Hatase O and
Hando T: The concurrent expression of p27(kip1) and cyclin D1 in
epithelial ovarian tumors. Gynecol Oncol. 73:202–209. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Vousden KH and Prives C: Blinded by the
light: The growing complexity of p53. Cell. 137:413–431. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Vogelstein B, Lane D and Levine AJ:
Surfing the p53 network. Nature. 408:307–310. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Lynch HT, Casey MJ, Snyder CL, Bewtra C,
Lynch JF, Butts M and Godwin AK: Hereditary ovarian carcinoma:
Heterogeneity, molecular genetics, pathology, and management. Mol
Oncol. 3:97–137. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Reles A, Wen WH, Schmider A, Gee C,
Runnebaum IB, Kilian U, Jones LA, El-Naggar A, Minguillon C,
Schönborn I, et al: Correlation of p53 mutations with resistance to
platinum-based chemotherapy and shortened survival in ovarian
cancer. Clin Cancer Res. 7:2984–2997. 2001.PubMed/NCBI
|
|
60
|
Piek JM, van Diest PJ, Zweemer RP, Jansen
JW, Poort-Keesom RJ, Menko FH, Gille JJ, Jongsma AP, Pals G,
Kenemans P and Verheijen RH: Dysplastic changes in prophylactically
removed Fallopian tubes of women predisposed to developing ovarian
cancer. J Pathol. 195:451–456. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Lu D, Kuhn E, Bristow RE, Giuntoli RL II,
Kjaer SK, Shih IeM and Roden RB: Comparison of candidate serologic
markers for type I and type II ovarian cancer. Gynecol Oncol.
122:560–566. 2011. View Article : Google Scholar : PubMed/NCBI
|