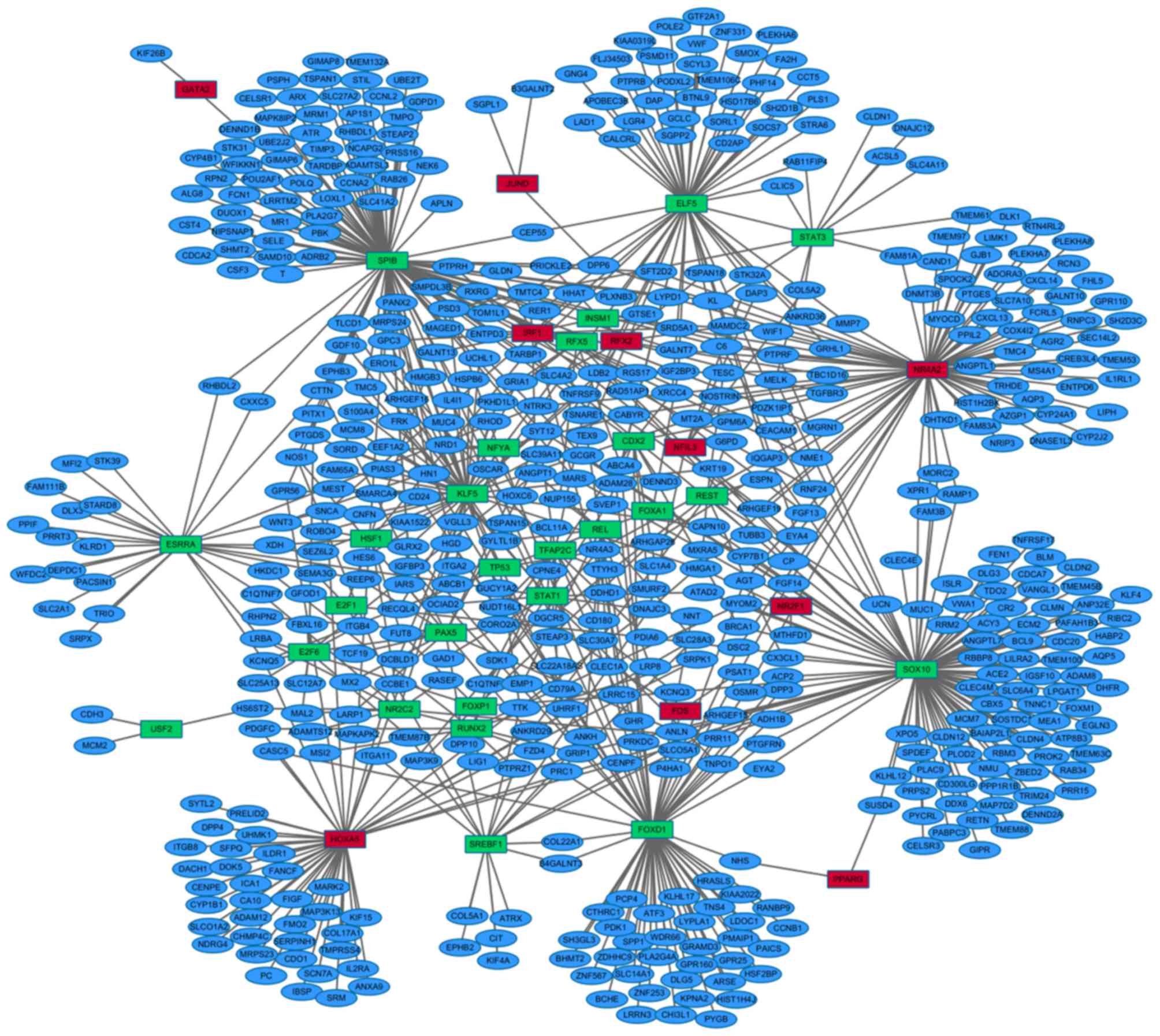
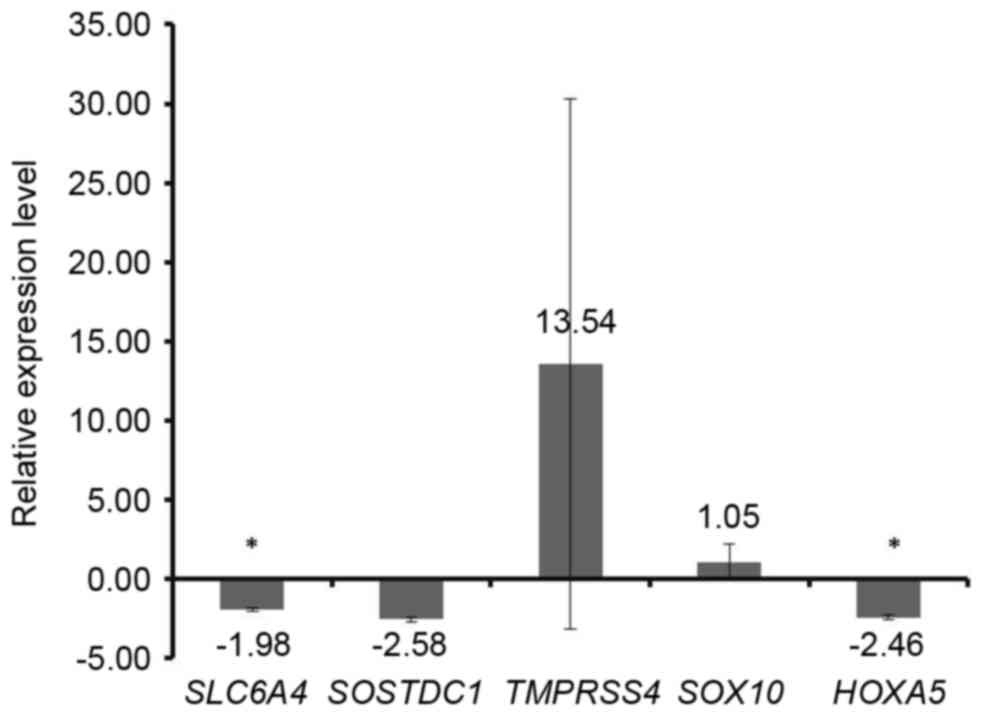
|
1
|
Minna JD, Roth JA and Gazdar AF: Focus on
lung cancer. Cancer cell. 1:49–52. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhao N, Liu Y, Chang Z, Li K, Zhang R,
Zhou Y, Qiu F, Han X and Xu Y: Identification of biomarker and
co-regulatory motifs in lung adenocarcinoma based on differential
interactions. PLoS One. 10:e01391652015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bandyopadhyay S, Mehta M, Kuo D, Sung MK,
Chuang R, Jaehnig EJ, Bodenmiller B, Licon K, Copeland W, Shales M,
et al: Rewiring of genetic networks in response to DNA damage.
Science. 330:1385–1389. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Liu X, Liu ZP, Zhao XM and Chen L:
Identifying disease genes and module biomarkers by differential
interactions. J Am Med Inform Assoc. 19:241–248. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Latchman DS: Transcription factors: An
overview. Int J Biochem Cell Biol. 29:1305–1312. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mitchell PJ and Tjian R: Transcriptional
regulation in mammalian cells by sequence-specific DNA binding
proteins. Science. 245:371–378. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Meng X, Lu P, Bai H, Xiao P and Fan Q:
Transcriptional regulatory networks in human lung adenocarcinoma.
Mol Med Rep. 6:961–966. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen CY, Chen ST, Fuh CS, Juan HF and
Huang HC: Coregulation of transcription factors and microRNAs in
human transcriptional regulatory network. BMC Bioinformatics. 12
Suppl 1:S412011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Garber ME, Troyanskaya OG, Schluens K,
Petersen S, Thaesler Z, Pacyna-Gengelbach M, van de Rijn M, Rosen
GD, Perou CM, Whyte RI, et al: Diversity of gene expression in
adenocarcinoma of the lung. Proc Natl Acad Sci USA. 98:pp.
13784–13789. 2001; View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Director's Challenge Consortium for the
Molecular Classification of Lung Adenocarcinoma, ; Shedden K,
Taylor JM, Enkemann SA, Tsao MS, Yeatman TJ, Gerald WL, Eschrich S,
Jurisica I, Giordano TJ, et al: Gene expression-based survival
prediction in lung adenocarcinoma: A multi-site, blinded validation
study. Nat Med. 14:822–827. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Stearman RS, Dwyer-Nield L, Zerbe L,
Blaine SA, Chan Z, Bunn PA Jr, Johnson GL, Hirsch FR, Merrick DT,
Franklin WA, et al: Analysis of orthologous gene expression between
human pulmonary adenocarcinoma and a carcinogen-induced murine
model. Am J Pathol. 167:1763–1775. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang W, Gong W, Ai H, Tang J and Shen C:
Gene expression analysis of lung adenocarcinoma and matched
adjacent non-tumor lung tissue. Tumori. 100:338–345.
2014.PubMed/NCBI
|
|
13
|
Jiang H, Deng Y, Chen HS, Tao L, Sha Q,
Chen J, Tsai CJ and Zhang S: Joint analysis of two microarray
gene-expression data sets to select lung adenocarcinoma marker
genes. BMC Bioinformatics. 5:812004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fu S, Pan X and Fang W: Differential
co-expression analysis of a microarray gene expression profiles of
pulmonary adenocarcinoma. Mol Med Rep. 10:713–718. 2014.PubMed/NCBI
|
|
15
|
Lin CC, Chen YJ, Chen CY, Oyang YJ, Juan
HF and Huang HC: Crosstalk between transcription factors and
microRNAs in human protein interaction network. BMC Syst Biol.
6:182012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li BQ, You J, Chen L, Zhang J, Zhang N, Li
HP, Huang T, Kong XY and Cai YD: Identification of
lung-cancer-related genes with the shortest path approach in a
protein-protein interaction network. Biomed Res Int.
2013:2673752013.PubMed/NCBI
|
|
17
|
Barrett T, Wilhite SE, Ledoux P,
Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH,
Sherman PM, Holko M, et al: NCBI GEO: Archive for functional
genomics data sets-update. Nucleic Acids Res. 41(Database Issue):
D991–D995. 2013.PubMed/NCBI
|
|
18
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hardcastle TJ: Generalized empirical
Bayesian methods for discovery of differential data in
high-throughput biology. Bioinformatics. 32:195–202.
2016.PubMed/NCBI
|
|
20
|
Eden E, Navon R, Steinfeld I, Lipson D and
Yakhini Z: GOrilla: A tool for discovery and visualization of
enriched GO terms in ranked gene lists. BMC Bioinformatics.
10:482009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Eden E, Lipson D, Yogev S and Yakhini Z:
Discovering motifs in ranked lists of DNA sequences. PLoS Comput
Biol. 3:e392007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The gene
ontology consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Nogales-Cadenas R, Carmona-Saez P, Vazquez
M, Vicente C, Yang X, Tirado F, Carazo JM and Pascual-Montano A:
GeneCodis: Interpreting gene lists through enrichment analysis and
integration of diverse biological information. Nucleic Acids Res.
37(Web Server Issue): W317–W322. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tabas-Madrid D, Nogales-Cadenas R and
Pascual-Montano A: GeneCodis3:A non-redundant and modular
enrichment analysis tool for functional genomics. Nucleic Acids
Res. 40(Web Server Issue): W478–W483. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Carmona-Saez P, Chagoyen M, Tirado F,
Carazo JM and Pascual-Montano A: GENECODIS: A web-based tool for
finding significant concurrent annotations in gene lists. Genome
Biol. 8:R32007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Matys V, Fricke E, Geffers R, Gössling E,
Haubrock M, Hehl R, Hornischer K, Karas D, Kel AE, Kel-Margoulis
OV, et al: TRANSFAC: Transcriptional regulation, from patterns to
profiles. Nucleic Acids Res. 31:374–378. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Smoot ME, Ono K, Ruscheinski J, Wang PL
and Ideker T: Cytoscape 2.8: New features for data integration and
network visualization. Bioinformatics. 27:431–432. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang KK, Liu N, Radulovich N, Wigle DA,
Johnston MR, Shepherd FA, Minden MD and Tsao MS: Novel candidate
tumor marker genes for lung adenocarcinoma. Oncogene. 21:7598–7604.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Backen AC, Cole CL, Lau SC, Clamp AR,
McVey R, Gallagher JT and Jayson GC: Heparan sulphate synthetic and
editing enzymes in ovarian cancer. Br J Cancer. 96:1544–1548. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chikaishi Y, Uramoto H, Koyanagi Y, Yamada
S, Yano S and Tanaka F: TMPRSS4 expression as a marker of
recurrence in patients with lung cancer. Anticancer Res.
36:121–127. 2016.PubMed/NCBI
|
|
33
|
Oegema K, Savoian MS, Mitchison TJ and
Field CM: Functional analysis of a human homologue of the
Drosophila actin binding protein anillin suggests a role in
cytokinesis. J Cell Biol. 150:539–552. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Suzuki C, Daigo Y, Ishikawa N, Kato T,
Hayama S, Ito T, Tsuchiya E and Nakamura Y: ANLN plays a critical
role in human lung carcinogenesis through the activation of RHOA
and by involvement in the phosphoinositide 3-kinase/AKT pathway.
Cancer Res. 65:11314–11325. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Han SS, Kim WJ, Hong Y, Hong SH, Lee SJ,
Ryu DR, Lee W, Cho YH, Lee S, Ryu YJ, et al: RNA sequencing
identifies novel markers of non-small cell lung cancer. Lung
Cancer. 84:229–235. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Cai X, Luo J, Yang X, Deng H, Zhang J, Li
S, Wei H, Yang C, Xu L, Jin R, et al: In vivo selection for
spine-derived highly metastatic lung cancer cells is associated
with increased migration, inflammation and decreased adhesion.
Oncotarget. 6:22905–22917. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chen L, Zhuo D, Chen J and Yuan H:
Screening feature genes of lung carcinoma with DNA microarray
analysis. Int J Clin Exp Med. 8:12161–12171. 2015.PubMed/NCBI
|
|
38
|
Lim MY and Thomas PS: Biomarkers in
exhaled breath condensate and serum of chronic obstructive
pulmonary disease and non-small-cell lung cancer. Int J Chronic
Dis. 2013:5786132013.PubMed/NCBI
|
|
39
|
Brooks GD, McLeod L, Alhayyani S, Miller
A, Russell PA, Ferlin W, Rose-John S, Ruwanpura S and Jenkins BJ:
IL6 Trans-signaling Promotes KRAS-Driven lung carcinogenesis.
Cancer Res. 76:866–876. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Katoh M and Katoh M: Human FOX gene family
(Review). Int J Oncol. 25:1495–1500. 2004.PubMed/NCBI
|
|
41
|
Nakayama S, Soejima K, Yasuda H, Yoda S,
Satomi R, Ikemura S, Terai H, Sato T, Yamaguchi N, Hamamoto J, et
al: FOXD1 expression is associated with poor prognosis in non-small
cell lung cancer. Anticancer Res. 35:261–268. 2015.PubMed/NCBI
|
|
42
|
Ciribilli Y, Singh P, Spanel R, Inga A and
Borlak J: Decoding c-Myc networks of cell cycle and apoptosis
regulated genes in a transgenic mouse model of papillary lung
adenocarcinomas. Oncotarget. 6:31569–31592. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wang CC, Su KY, Chen HY, Chang SY, Shen
CF, Hsieh CH, Hong QS, Chiang CC, Chang GC, Yu SL and Chen JJ:
HOXA5 inhibits metastasis via regulating cytoskeletal remodelling
and associates with prolonged survival in non-small-cell lung
carcinoma. PLoS One. 10:e01241912015. View Article : Google Scholar : PubMed/NCBI
|