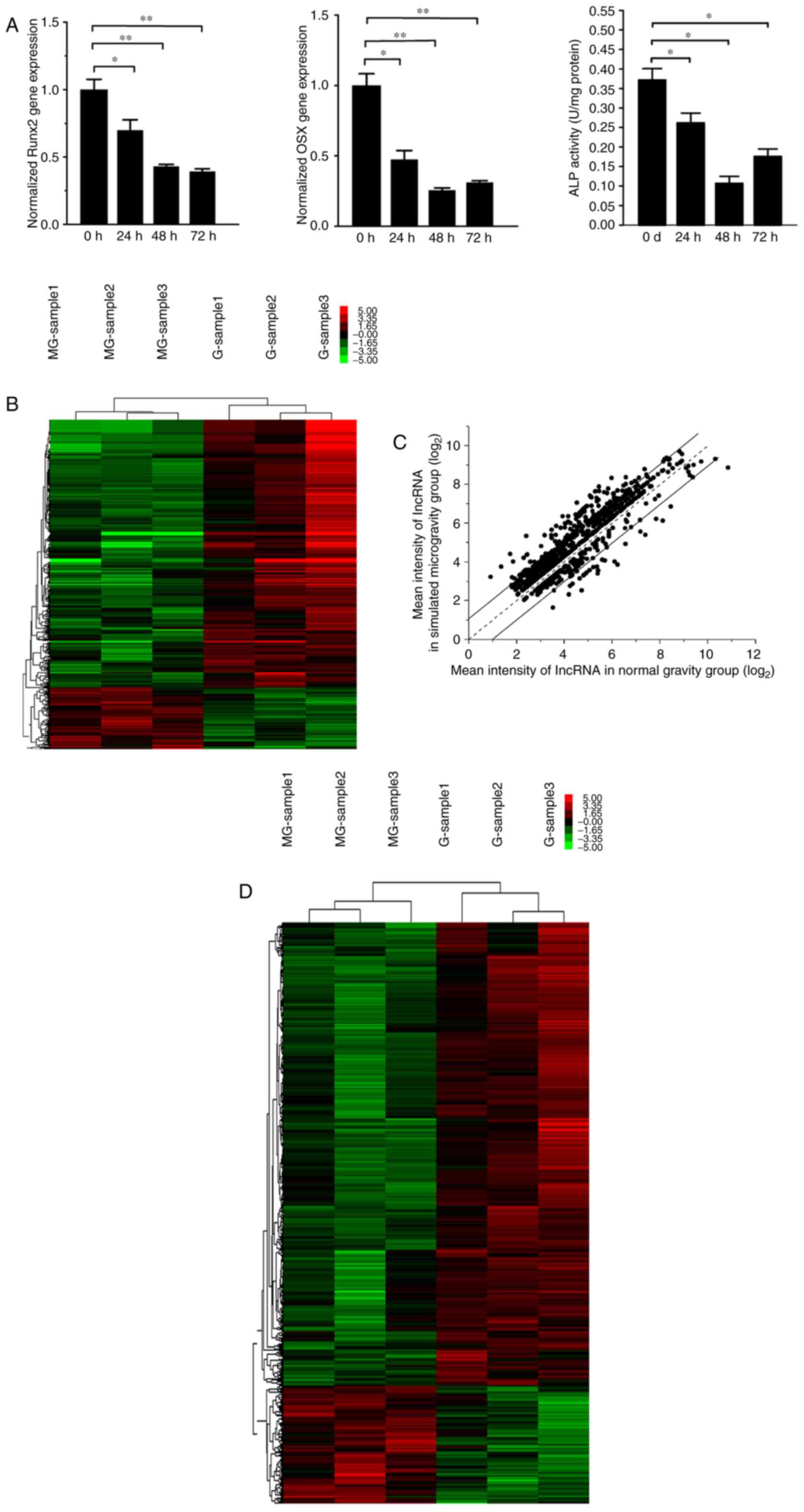
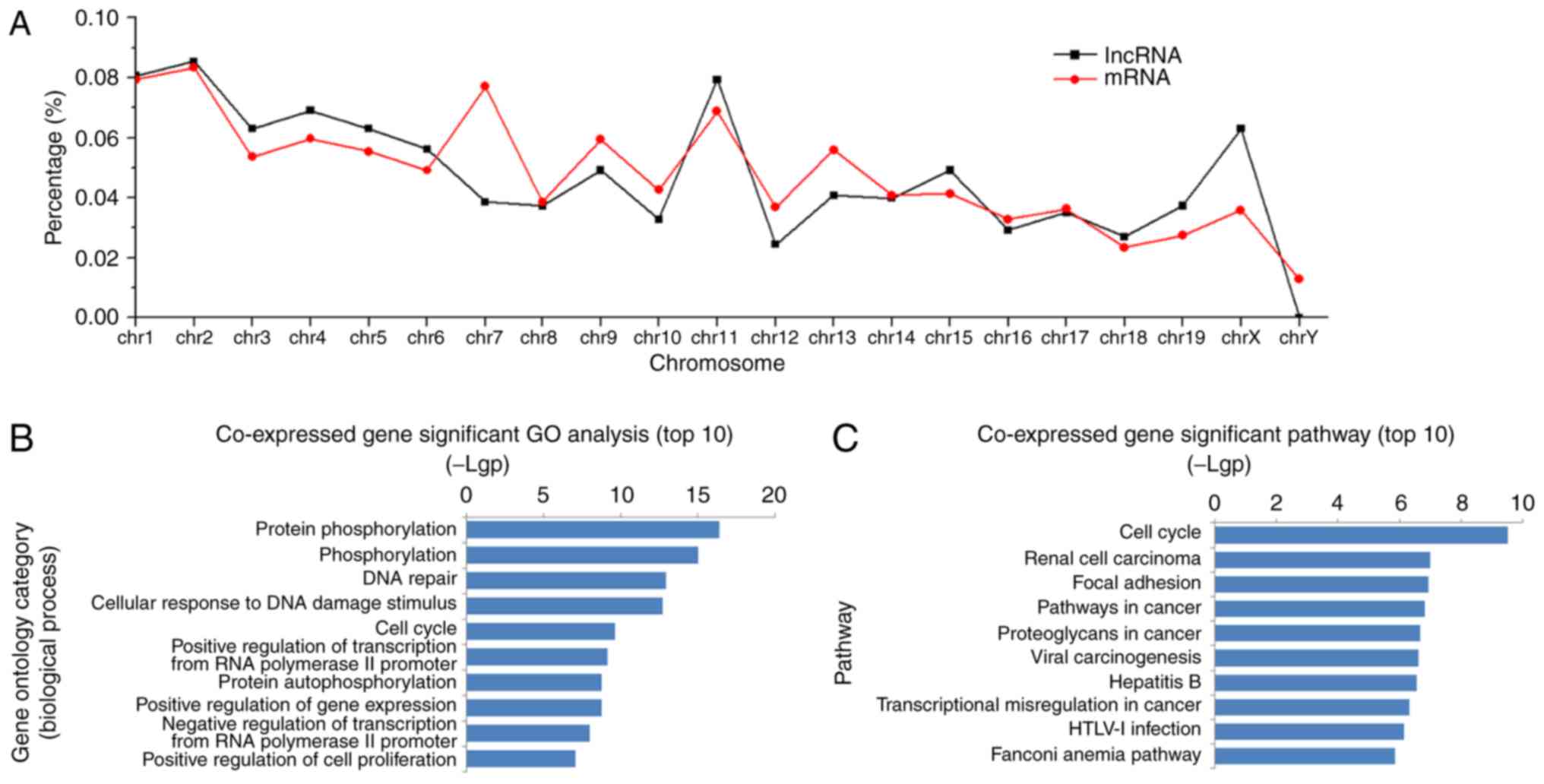
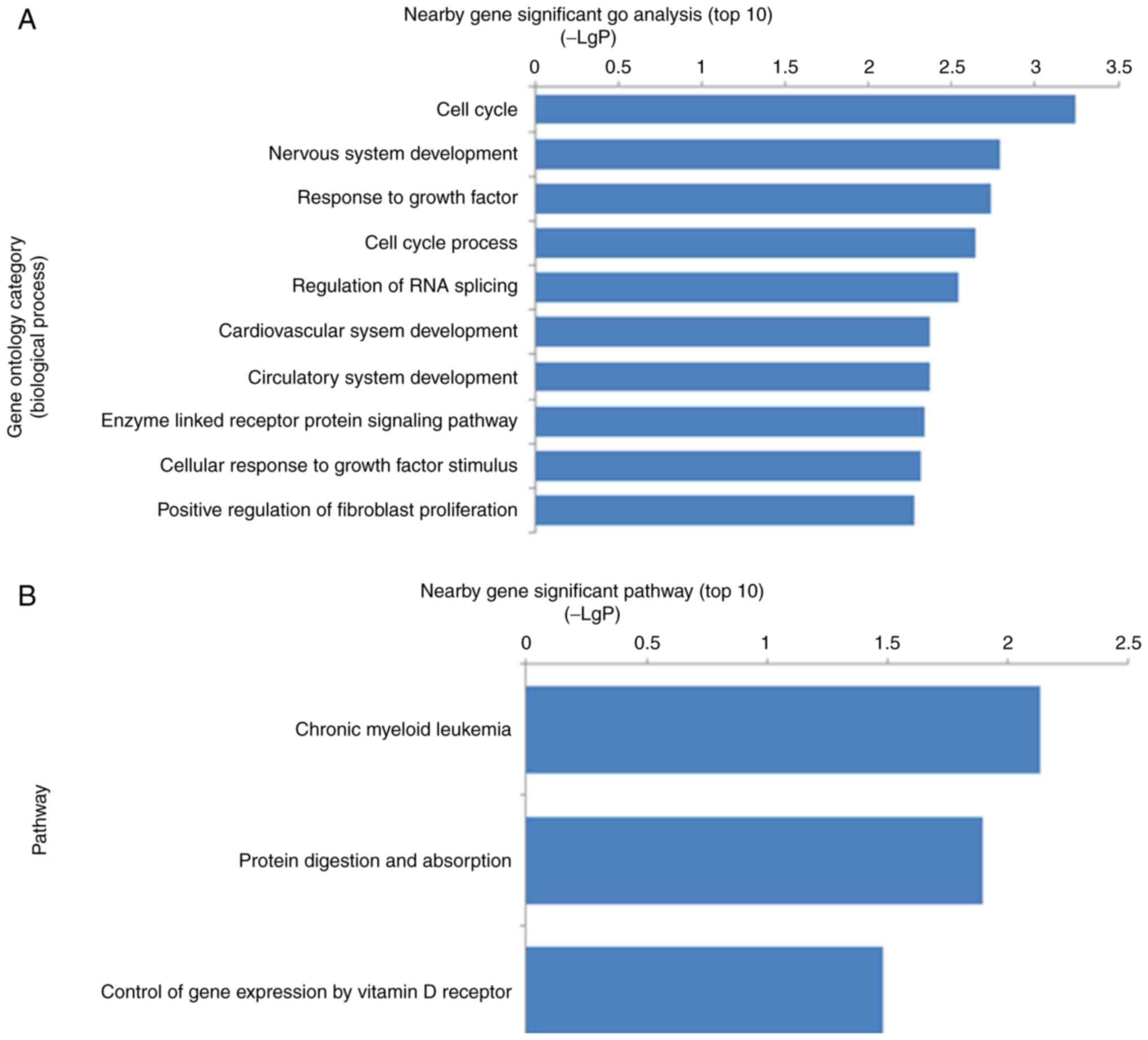
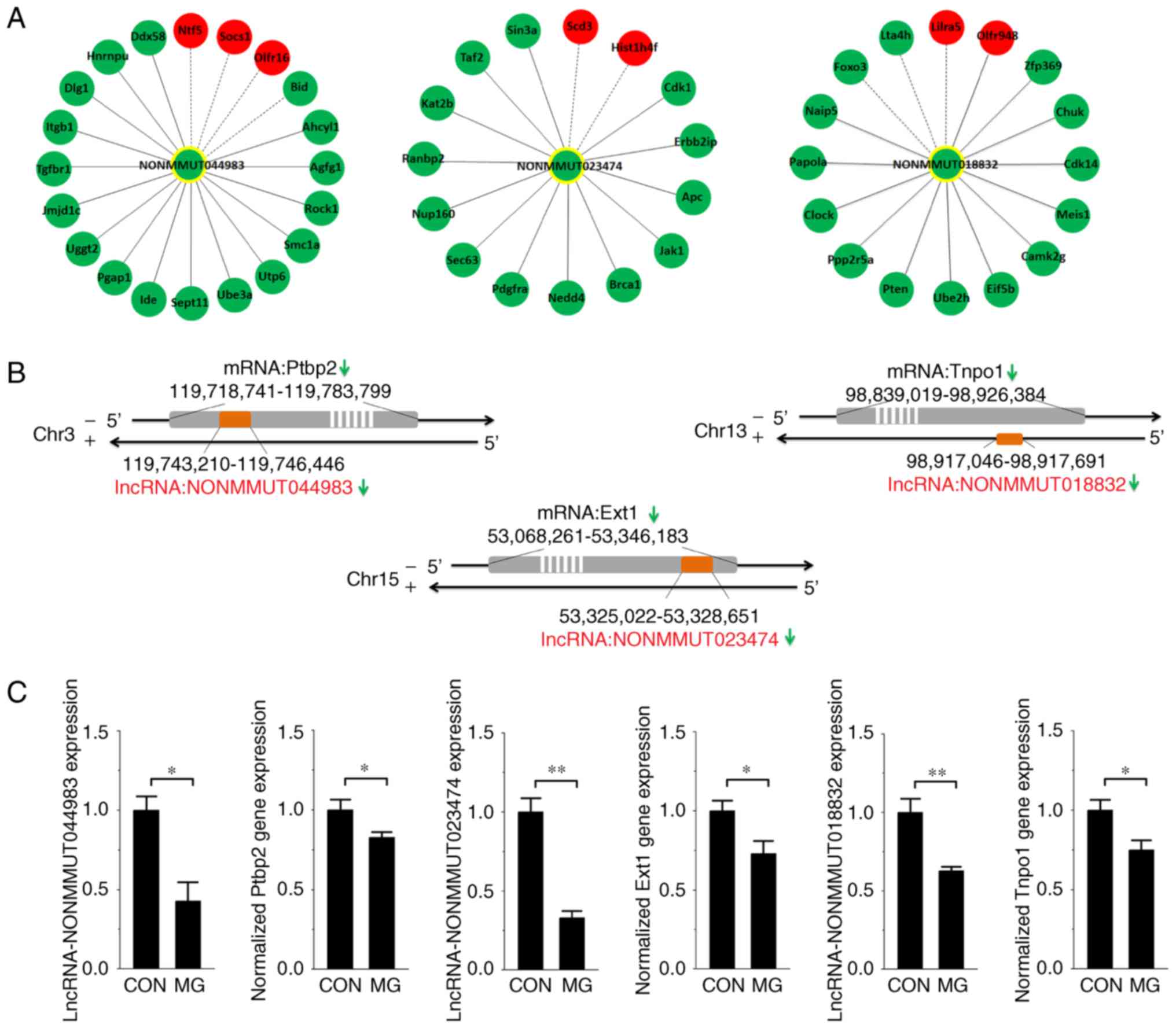
|
1
|
Bloomfield SA: Disuse osteopenia. Curr
Osteoporos Rep. 8:91–97. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ohshima H: Secondary osteoporosis UPDATE.
Bone loss due to bed rest and human space flight study. Clin
Calcium. 20:709–716. 2010.(In Japanese).
|
|
3
|
Morey ER and Baylink DJ: Inhibition of
bone formation during space flight. Science. 201:1138–1141. 1978.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zayzafoon M, Gathings WE and McDonald JM:
Modeled microgravity inhibits osteogenic differentiation of human
mesenchymal stem cells and increases adipogenesis. Endocrinology.
145:2421–2432. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rodionova NV: The dynamics of
proliferation and differentiation of osteogenic cells under
supportive unloading. Tsitol Genet. 45:22–27. 2011.PubMed/NCBI
|
|
6
|
Hu LF, Li JB, Qian AR, Wang F and Shang P:
Mineralization initiation of MC3T3-E1 preosteoblast is suppressed
under simulated microgravity condition. Cell Biol Int. 39:364–372.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Capulli M, Rufo A, Teti A and Rucci N:
Global transcriptome analysis in mouse calvarial osteoblasts
highlights sets of genes regulated by modeled microgravity and
identifies a ‘mechanoresponsive osteoblast gene signature’. J Cell
Biochem. 107:240–252. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Patel MJ, Liu W, Sykes MC, Ward NE, Risin
SA, Risin D and Jo H: Identification of mechanosensitive genes in
osteoblasts by comparative microarray studies using the rotating
wall vessel and the random positioning machine. J Cell Biochem.
101:587–599. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Claverie JM: Fewer genes, more noncoding
RNA. Science. 309:1529–1530. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kapranov P, Cheng J, Dike S, Nix DA,
Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermüller J,
Hofacker IL, et al: RNA maps reveal new RNA classes and a possible
function for pervasive transcription. Science. 316:1484–1488. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mercer TR, Dinger ME and Mattick JS: Long
noncoding RNAs: Insights into functions. Nat Rev Genet. 10:155–159.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Brosnan CA and Voinnet O: The long and the
short of noncoding RNAs. Curr Opin Cell Biol. 21:416–425. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhuang W, Ge X, Yang S, Huang M, Zhuang W,
Chen P, Zhang X, Fu J, Qu J and Li B: Upregulation of lncRNA MEG3
promotes osteogenic differentiation of mesenchymal stem cells from
multiple myeloma patients by targeting BMP4 transcription. Stem
Cells. 33:1985–1997. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hu Z, Wang Y, Sun Z, Wang H, Zhou H, Zhang
L, Zhang S and Cao X: miRNA-132-3p inhibits osteoblast
differentiation by targeting Ep300 in simulated microgravity. Sci
Rep. 5:186552015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The gene
ontology consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Draghici S, Khatri P, Tarca AL, Amin K,
Done A, Voichita C, Georgescu C and Romero R: A systems biology
approach for pathway level analysis. Genome Res. 17:1537–1545.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
da W Huang, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009.PubMed/NCBI
|
|
20
|
Stuart JM, Segal E, Koller D and Kim SK: A
gene-coexpression network for global discovery of conserved genetic
modules. Science. 302:249–255. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Prieto C, Risueño A, Fontanillo C and De
las Rivas J: Human gene coexpression landscape: Confident network
derived from tissue transcriptomic profiles. PLoS One. 3:e39112008.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Barabási AL and Oltvai ZN: Network
biology: Understanding the cell's functional organization. Nat Rev
Genet. 5:101–113. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hong D, Chen HX, Yu HQ, Liang Y, Wang C,
Lian QQ, Deng HT and Ge RS: Morphological and proteomic analysis of
early stage of osteoblast differentiation in osteoblastic
progenitor cells. Exp Cell Res. 316:2291–2300. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Verlinden L, Kriebitzsch C, Beullens I,
Tan BK, Carmeliet G and Verstuyf A: Nrp2 deficiency leads to
trabecular bone loss and is accompanied by enhanced osteoclast and
reduced osteoblast numbers. Bone. 55:465–475. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tang ZZ, Sharma S, Zheng S, Chawla G,
Nikolic J and Black DL: Regulation of the mutually exclusive exons
8a and 8 in the CaV1.2 calcium channel transcript by polypyrimidine
tract-binding protein. J Biol Chem. 286:10007–10016. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lekva T, Ueland T, Bøyum H, Evang JA,
Godang K and Bollerslev J: TXNIP is highly regulated in bone
biopsies from patients with endogenous Cushing's syndrome and
related to bone turnover. Eur J Endocrinol. 166:1039–1048. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ogasawara T, Kawaguchi H, Jinno S, Hoshi
K, Itaka K, Takato T, Nakamura K and Okayama H: Bone morphogenetic
protein 2-induced osteoblast differentiation requires Smad-mediated
down-regulation of Cdk6. Mol Cell Biol. 24:6560–6568. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Fan D, Liu S, Jiang S, Li Z, Mo X, Ruan H,
Zou GM and Fan C: The use of SHP-2 gene transduced bone marrow
mesenchymal stem cells to promote osteogenic differentiation and
bone defect repair in rat. J Biomed Mater Res A. 104:1871–1881.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sun Z, Cao X, Hu Z, Zhang L, Wang H, Zhou
H, Li D, Zhang S and Xie M: miR-103 inhibits osteoblast
proliferation mainly through suppressing Cav1.2 expression in
simulated microgravity. Bone. 76:121–128. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chang SF, Chang TK, Peng HH, Yeh YT, Lee
DY, Yeh CR, Zhou J, Cheng CK, Chang CA and Chiu JJ: BMP-4 induction
of arrest and differentiation of osteoblast-like cells via p21 CIP1
and p27 KIP1 regulation. Mol Endocrinol. 23:1827–1838. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yano H, Hamanaka R, Nakamura-Ota M, Adachi
S, Zhang JJ, Matsuo N and Yoshioka H: Sp7/Osterix induces the mouse
pro-α2 (I) collagen gene (Col1a2) expression via the proximal
promoter in osteoblastic cells. Biochem Biophys Res Commun.
452:531–536. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Haasper C, Jagodzinski M, Drescher M,
Meller R, Wehmeier M, Krettek C and Hesse E: Cyclic strain induces
FosB and initiates osteogenic differentiation of mesenchymal cells.
Exp Toxicol Pathol. 59:355–363. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Choi YH, Han Y, Lee SH, Jin YH, Bahn M,
Hur KC, Yeo CY and Lee KY: Cbl-b and c-Cbl negatively regulate
osteoblast differentiation by enhancing ubiquitination and
degradation of Osterix. Bone. 75:201–209. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yi S, Yu M, Yang S, Miron RJ and Zhang Y:
Tcf12, A member of basic helix-loop-helix transcription factors,
mediates bone marrow mesenchymal stem cell osteogenic
differentiation in vitro and in vivo. Stem Cells. 35:386–397. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Di Benedetto A, Sun L, Zambonin CG, Tamma
R, Nico B, Calvano CD, Colaianni G, Ji Y, Mori G, Grano M, et al:
Osteoblast regulation via ligand-activated nuclear trafficking of
the oxytocin receptor. Proc Natl Acad Sci USA. 111:pp. 16502–16507.
2014; View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Matsumoto Y, Matsumoto K, Irie F, Fukushi
J, Stallcup WB and Yamaguchi Y: Conditional ablation of the heparan
sulfate-synthesizing enzyme Ext1 leads to dysregulation of bone
morphogenic protein signaling and severe skeletal defects. J Biol
Chem. 285:19227–19234. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lu J, Qu S, Yao B, Xu Y, Jin Y, Shi K,
Shui Y, Pan S, Chen L and Ma C: Osterix acetylation at K307 and
K312 enhances its transcriptional activity and is required for
osteoblast differentiation. Oncotarget. 7:37471–37486. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kacena MA, Todd P and Landis WJ:
Osteoblasts subjected to spaceflight and simulated space shuttle
launch conditions. In Vitro Cell Dev Biol Anim. 39:454–459. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Bucaro MA, Fertala J, Adams CS, Steinbeck
M, Ayyaswamy P, Mukundakrishnan K, Shapiro IM and Risbud MV: Bone
cell survival in microgravity: Evidence that modeled microgravity
increases osteoblast sensitivity to apoptogens. Ann N Y Acad Sci.
1027:64–73. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Necsulea A, Soumillon M, Warnefors M,
Liechti A, Daish T, Zeller U, Baker JC, Grützner F and Kaessmann H:
The evolution of lncRNA repertoires and expression patterns in
tetrapods. Nature. 505:635–640. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Matthew T Weirauch: Gene coexpression
networks for the analysis of DNA microarray dataApplied Statistics
for Network Biology: Methods in Systems Biology. Dehmer M,
Emmert-Streib F, Graber A and Salvador A: Wiley-VCH Verlag GmbH
& Co. KGaA; Weinheim, Germany: 2011
|
|
42
|
Martianov I, Ramadass A, Barros A Serra,
Chow N and Akoulitchev A: Repression of the human dihydrofolate
reductase gene by a non-coding interfering transcript. Nature.
445:666–670. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Feng J, Bi C, Clark BS, Mady R, Shah P and
Kohtz JD: The Evf-2 noncoding RNA is transcribed from the Dlx-5/6
ultraconserved region and functions as a Dlx-2 transcriptional
coactivator. Genes Dev. 20:1470–1484. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Olivares-Navarrete R, Sutha K, Hyzy SL,
Hutton DL, Schwartz Z, McDevitt T and Boyan BD: Osteogenic
differentiation of stem cells alters vitamin D receptor expression.
Stem Cells Dev. 21:1726–1735. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Duncan RL, Akanbi KA and Farach-Carson MC:
Calcium signals and calcium channels in osteoblastic cells. Semin
Nephrol. 18:178–190. 1998.PubMed/NCBI
|
|
46
|
Iqbal J and Zaidi M: Molecular regulation
of mechanotransduction. Biochem Biophys Res Commun. 328:751–755.
2005. View Article : Google Scholar : PubMed/NCBI
|