Introduction
Colorectal cancer (CRC) is the third most common
cancer in the world. Although the morbidity of CRC shows a rising
trend in developing countries and a downward trend in developed
countries, it is still one of the most frequent malignancies in
people over the age of 60 (1,2).
During the past decades, increasing understanding regarding CRC
tumorigenesis and progression has been achieved in the laboratory
and the five-year survival of CRC has increased from 48.6 to 67.2%
(3–5). However, CRC remains to be a worldwide
problem because the molecular mechanism underlying CRC migration
and metastasis is largely unknown. It is most urgently needed to
explore the underling mechanisms of carcinogenesis in CRC, to
discover tumor-suppressor genes, oncogenes and cell signaling
pathways as diagnostic biomarkers and therapeutic targets at
various stages of CRC.
miRNAs are a group of evolutional highly conserved
small non-coding RNAs with a length of approximately 20–25
nucleotides (6–9). These nucleotides regulate gene
expression by imperfect base pairing with targeted mRNAs at the
3′-untranslated region (3′-UTR), leading to mRNA cleavage or
translational repression (10,11).
They have been detected in tissues as well as plasma, serum and
urine (12). miRNAs target a
wide-range of genes that are implicated in cell survival,
proliferation, differentiation and apoptosis, suggesting that
miRNAs are crucial for homeostasis and dysregulation of miRNAs
would lead to human diseases (13–15).
Previous studies have demonstrated that miRNAs have been regarded
as oncomiRs or tumor suppressor genes contributing to the
initiation and progression of CRC. miRNAs are globally deregulated
in cancer cells: several oncogenic miRNAs are upregulated, whereas
tumor suppressive miRNAs are often suppressed. Thus, therapeutic
delivery of tumor suppressive miRNAs and inhibition of oncogenic
miRNAs serve as a feasible way to cancer treatment (16–18).
Nuclear factor I/X (NFIX) is a member of NFI
transcription factor family that binds to nucleotide sequences and
regulates transcriptional activity (19). Several studies have reported that
NFIX may be implicated in cancer growth and metastasis. Heng et
al (20) demonstrated that
NFIX regulates proliferation and migration within the murine SVZ
neurogenic niche. NFIX is also targeted by miR-1290 and
upregulation of miR-1290 in esophageal squamous cell carcinoma
(ESCC) promotes disease progression (21). Closely related to our research is
that NFIX has been shown to be targeted by miR-1914 and miR-1915,
and loss of these two miRNAs leads to CRC chemotherapy resistance
through stabilization of NFIX (22). These results suggested that
supression of miRNAs that targeted NFIX may be a novel strategy to
inhibit cancer progression and metastasis. Its function and
expression in CRC and the association between the expression of
miR-1914 and its counterpart miR-647 has yet to be fully
elucidated.
In this study, we reported miR-647 and miR-1914 as
the top upregulated miRNAs in CRC samples and cell lines. miR-647
and miR-1914 acted as putative oncogenic miRNAs. Knockdown the
expression of both miRNAs suppressed CRC cell proliferation and
migration and overexpression of the two miRNAs promoted CRC cell
proliferation and migration. Bioinformatic analysis, isobaric tags
for relative and absolute quantification (iTRAQ) experiments and
luciferase reporter assay indicated NFIX was targeted by miR-647
and miR-1914 that mediated the oncogenic function. Taken together,
our study showed the involvement of miRNAs in CRC cells
proliferation and migration, targeting these oncogenic miRNAs may
be a novel strategy for the treatment of CRC.
Materials and methods
Cell lines and cell culture
The human colon cancer cell lines SW480, SW620 and
HT-29 were purchased from the American Type Culture Collection
(ATCC; Manassas, VA, USA). The human intestinal epithelial cell
(HIEC) line was preserved in our State Key Laboratory of Cancer
Biology (CBSKL, China). All cell lines were confirmed by STR
analysis. SW480 and SW620 cells were maintained in Leibovitz's L-15
(Macgene, Beijing, China) medium supplemented with 10% fetal bovine
serum (FBS; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA,
USA), HT-29 cells were maintained in MCCOY'S 5A (Macgene) medium
supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.),
and HIEC cells were maintained in Dulbecco's modified Eagle's
medium (DMEM; Macgene) supplemented with 10% FBS (Gibco; Thermo
Fisher Scientific, Inc.). All the cell lines were cultured in a
humidified incubator at 37°C and 5% CO2. The medium was
changed every 3 days, and the cells were trypsinized when 90%
confluence was reached.
Patients and clinicopathological data
collection
This study was approved by the hospital Ethics
Committee of Xijing Hospital (Xi'an, China). We collected 32 pairs
of matched samples from colon cancer patients, of which 10 were
used for the RNA-Seq analysis to seek the meaningful microRNAs,
while the other 22 pairs were used to verify the expression of
miR-647 and miR-1914 in CRC using RT-qPCR analysis. We obtained
written informed consent from all the patients before performing
operations and collecting tissues. Clinical examination and
histopathological analysis of the tissue specimens were also
performed to obtain an accurate diagnosis. The samples from the
surgery were collected and placed in liquid nitrogen to freeze the
tissues immediately. The 22 pairs of matched samples were then
stored at −80°C after 24 h in liquid nitrogen for later extraction
of total RNA. Clinical and pathological data, including sex, age,
pathological grade and lymph node metastasis, were obtained from
the medical records and are shown in Table I.
 | Table I.Relationship of miR-647 or miR-1914-5p
relative expression with the clinicopathological characteristics of
colorectal cancer patients. |
Table I.
Relationship of miR-647 or miR-1914-5p
relative expression with the clinicopathological characteristics of
colorectal cancer patients.
|
|
| miR-647 relative
expression | miR-1914 relative
expression |
|---|
|
|
|
|
|
|---|
| Characteristics | Number | Low (%) | High (%) | Not detected (%) | Low (%) | High (%) |
|---|
| Age (years) |
|
|
|
|
|
|
|
<60 | 7 | 1 (14.29) | 5 (71.43) | 1 (14.29) | 1 (14.29) | 6 (85.71) |
|
≥60 | 15 | 2 (13.33) | 8 (53.33) | 5 (33.33) | 3 (20.00) | 12 (80.00) |
| Sex |
|
|
|
|
|
|
|
Male | 11 | 3 (27.27) | 3 (27.27) | 5 (45.45) | 4 (36.36) | 7 (63.64) |
|
Female | 11 | – | 10 (90.91) | 1 (9.09) | – | 11 (100) |
| Tumor location |
|
|
|
|
|
|
|
Colon | 13 | 1 (7.69) | 8 (61.54) | 4 (30.77) | 1 (7.69) | 12 (92.31) |
|
Rectum | 9 | 2 (22.22) | 5 (55.56) | 2 (22.22) | 2 (22.22) | 7 (77.78) |
| Histological
differentiation |
|
|
|
|
|
|
|
High/moderate | 15 | 3 (20.00) | 8 (53.33) | 4 (26.67) | 3 (20.00) | 12 (80.00) |
|
Low/undifferentiated | 1 | – | 1 (100.00) | – | – | 1 (100.00) |
| Lymphatic
metastasis |
|
|
|
|
|
|
|
Yes | 10 | 2 (20.00) | 7 (70.00) | 1 (10.00) | 2 (20.00) | 8 (80.00) |
| No | 12 | 1 (8.33) | 6 (50.00) | 5 (41.67) | 2 (16.67) | 10 (83.33) |
| TNM stage |
|
|
|
|
|
|
| I/II
stage | 12 | 1 (8.33) | 6 (50.00) | 5 (41.67) | 2 (16.67) | 10 (83.33) |
| III/IV
stage | 10 | 2 (20.00) | 7 (70.00) | 1 (10.00) | 2 (20.00) | 8 (80.00) |
RNA-Seq library preparation and
transcriptomic analysis
The significantly different expressed miRNAs were
predicted via using the mRNA-Seq by screening our laboratory
previous transcriptome data: The other 10 pairs clinical samples,
including CRC tissues and paired adjacent normal tissues. The
samples were used to predict the significantly different expressed
miRNAs for the first time and they were sent to Shanghai Novel
Bioinformatics Co., Ltd. (Shanghai, China) for further RNA-Seq,
cDNA libraries were generated using the TruSeq™ RNA
Sample Preparation kit (Illumina, San Diego, CA, USA) following the
manufacturer's instructions. To compare the differential expression
of genes among samples, the number of raw unique reads in each
sample was normalized to Reads Per Kilobases per Million mapped
sequence reads (RPKM) to obtain normalized gene expression levels.
DESeq was used to identify differentially expressed miRNAs or mRNA
by using a fold change of 2 and a nominal P-value of 0.05 as the
filtering criteria. Hierarchical clustering was performed using
average linkage clustering with Euclidean distances, treating
samples independently each other.
RNA extraction and quantitative
real-time PCR
Total RNA was extracted from the CRC tissues and
paired normal tissues (5 cm away from the colon cancer edge) and
maintained in −80°C after being washed with PBS. RNA was extracted
from the cultured cells using the same method. We followed the
TRIzol reagent (Takara Bio Inc., Dalian, China) protocol to extract
total RNA, dissolve the RNA after extraction in RNase-free
ddH2O, and quantify the RNA using an ultraviolet
spectrophotometer. To analyze the expression of NFIX mRNA, cDNA was
synthesized using PrimeScript™ RT Master Mix (Takara Bio Inc.). To
analyze the expression of miR-647 or miR-1914-5p, reverse
transcription (RT) was performed with a Mir-X™ miRNA
First-Strand Synthesis Kit (Clontech, Mountain View CA, USA), and
then, the cDNA was synthesized. Subsequently, quantitative
real-time PCR was performed on an ABI 7500 PCR system (Applied
Biosystems Life Technologies, Foster City, CA, USA) according to
the manufacturer's instructions. The results of mRNA and miRNA
real-time PCR were normalized to GAPDH or U6, respectively, as an
endogenous control. All real-time PCR reactions were performed in
triplicate, and average CT values and SD values were calculated.
The primer sequences were as follows: miR-647,
5′-GTGGCTGCACTCACTTCCTTC-3′; miR-1914-5p,
5′-CCCTGTGCCCGGCCCACTTCTG-3′; NFIX forward,
5′-TGTTCCCGACCGTTACTTTG-3′ and reverse,
5′-GTATTTCCCGCTATCTTCCTG-3′; U6 forward,
5′-GCTTCGGCAGCACATATACTAAAAT-3′ and reverse,
5′-CGCTTCACGAATTTGCGTGTCAT-3′; GAPDH forward,
5′-GGTGAAGGTCGGAGTCAACG-3′ and reverse,
5′-CAAAGTTGTCATGGATGHACC−3′.
Cell transfection
All the cells were seeded in 60 mm dishes and,
cultured in a humidified incubator at 37°C and 5% CO2.
When 50–60% confluence was reached, the cells were transfected
using Lipofectamine™ RNAi MAX (Invitrogen Life
Technologies, Carlsbad, CA, USA) according to the manufacturer's
instructions. After 8 h incubation at 37°C in FBS-free medium,
cells were incubated for the next 24 h at 37°C in 10% FBS medium.
For knockdown of miR-647 or miR-1914-5p, hsa-miR-647 antagomir or
hsa-miR-1914-5p antagomir, respectively, was used. For
overexpression of miR-647 or miR-1914-5p, hsa-miR-647 mimics or
hsa-miR-1914-5p mimics, respectively, were used. All the products
were chemically synthesized by GenePharma Co., Ltd. (Shanghai,
China). The antagomir and mimic sequences were as follows:
hsa-miR-647 antagomir, 5′-GAAGGAAGUGAGUGCAGCCAC−3′; hsa-miR-1914-5p
antagomir, 5′-CAGAAGUGGGCCGGGCACAGGG-3′; antagomir negative
control, 5′-UUGUACUACACAAAAGUACUG-3′; hsa-miR-647 mimics,
5′-GUGGCUGCACUCACUUCCUUC-3′; hsa-miR-1914-5p mimics,
5′-CCCUGUGCCCGGCCCACUUCUG-3′; mimics negative control,
5′-UUGUACUACACAAAAGUACUG−3′.
Plasmid construction and siRNA
efficiency assay
The 3′-UTR of human NFIX was synthesized and cloned
into a GV272 (SV40-Luc-MCS) firefly luciferase reporter vector by
Genechem (Shanghai, China). The wild type NFIX (NFIX WT) and
mutant-type NFIX (NFIX Mut) were synthesized by GeneChem Co., Ltd.
(Shanghai, China). siRNA for NFIX and the negative control were
chemi-cally synthesized by AuGCT (Beijing, China). The sequences
were as follows: hsa-miR-647 NFIX-WT,
5′-UGGAGCUGUGCACCAGCAGCCAA-3′; hsa-miR-647 NFIX-Mut,
5′-UGGAGCUGUGCACCACGUCGGUA-3′; NFIX-homo-1033 sense,
5′-GCCCGGCUUCUCUAAAGAATT-3′ and antisense,
5′-UUCUUUAGAGAAGCCGGGCTT-3′; negative control sense,
5′-UUCUCCGAACGUGUCACGUTT-3′ and antisense,
5′-ACGUGACACGUUCGGAGAATT−3′.
Cell proliferation assay
We used a
3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H tetrazolium bromide
(MTT) assay to test the cell proliferation ability. Transfected or
untransfected cells were re-seeded into 96-well plates at a density
of 3,000 cells/well, Then, the cells were incubated at 37°C in
humidified 5% CO2 for the next 4 days. At the same time
of day, MTT solution (5 mg/ml) was added to the medium and
incubated for the next 4 h. The MTT medium was removed, and then,
150 µl of dimethyl sulfoxide (DMSO) was added to each well. A
spectrophotometer was used to measure the absorbance of each well
at 490 nm (A490). All experiments were performed in
quadruplicate.
Cell wound scratch assay
The cells were seeded in 6-well plates and
transfected when 50–60% confluence was reached. After 8 h
transfection, cells were incubated for the next 24 h at 37°C in 10%
FBS medium. At the same time on the second day, a straight line was
made with a sterile pipette (200 µl) in each well. After the cells
were washed with PBS 3 times, they were incubated at 37°C in 1% FBS
medium for one day. We took images of each well from the same
perspective at the same time of day over the next 3 days.
iTRAQ experiments
miR-647 antagomir was transfected into SW620 cells
to downregulate the expression of miR-647 in SW620 cells, the
negative control was used to represent the cells with a normal
miR-647 expression. Then, the two protein samples were extracted
from each cell and subjected to a LC-shot gun analysis using the
iTRAQ method. Each peptide solution was labelled with one of the
iTRAQ reagents (iTRAQ reporter ions of 115 and 117 mass/charge
ratio) according to the manufacturer's instructions (AB Sciex,
Framingham, MA, USA). The labelled peptides were pooled and
fractionated via strong cation exchange (SCX) using a ChromXP
C18-CL column (Eksigent parts of AB Sciex, Dublin, CA, USA) and
analyzed with nano LC-MS/MS with a nano LC system (Eksigent parts
of AB Sciex). A total of 5929 effective proteins were got from
three independent experiments, among these 4,141 were
differentially expressed (2,078 were downregulated and 2,063 were
upregulated).
Dual luciferase reporter assays
The 293T cells were co-transfected with the GV272
(SV40-Luc-MCS) firefly luciferase reporter vector (NFIX WT/NFIX
Mut) and hsa-miR-647 mimics or negative control in 24-well plates.
After 8 h of transfection, the cells were incubated for the next 48
h at 37°C and 5% CO2 in 10% FBS medium. A luciferase
assay kit (Promega, Madison, WI, USA) was used to perform the
luciferase assays according to the manufacturer's directions.
Firefly luciferase activity was normalized to Renilla
luciferase activity for each well.
Statistical analysis
SPSS 19.0 software (SPSS, Inc., Chicago, IL, USA)
and GraphPad Prism software (version 5.0) were used for statistical
analyses. All data are expressed as the mean ± standard error. All
experiments were repeated three times to reduce the error. The
differences between the two groups were compared using Student's
t-test. ANOVA test was used when making comparisons in datasets
containing multiple groups (>2). P<0.05 was considered to
indicate a statistically significant difference.
Results
miR-647 and miR-1914-5p are both
upregulated in CRC tissues and cell lines, and they are correlated
with each other
To investigate the roles of miRNA in CRCs, we first
examined the miRNA expression profiles in 10 CRC samples and in
their adjacent normal tissues with RNA-Seq. After normalization
with the endogenous control, we found that 9 microRNAs were
upregulated and 4 microRNAs were downregulated in colon cancer
tissues compared with their adjacent normal tissues. The most
significant increase in these microRNAs are miR-1914, miR-1182 and
miR-198, meanwhile the most significant decreased in these
microRNAs are miR-147b, miR-378i and miR-650, they are shown in
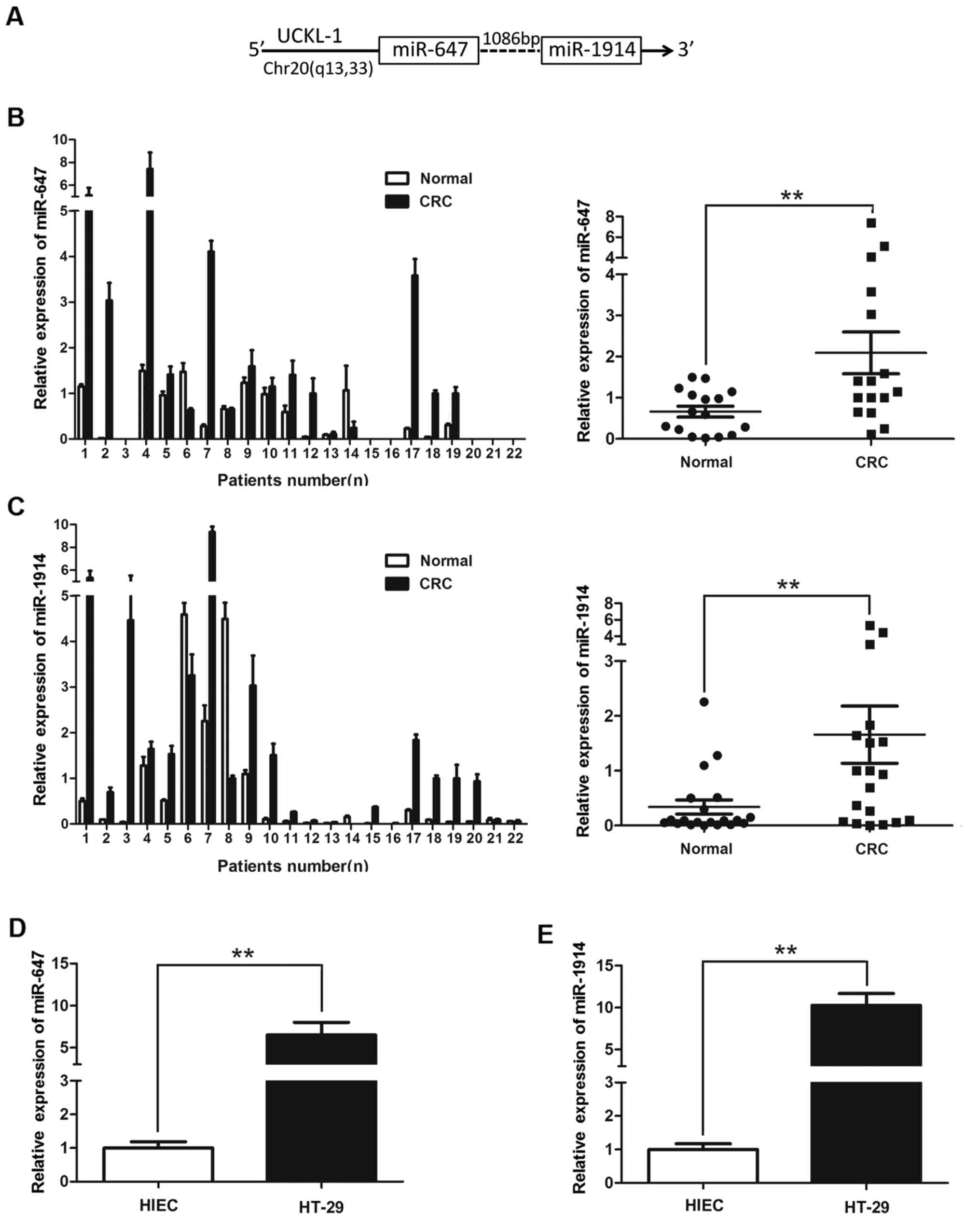
Table II. miR-647 is also
upregulated in CRC tissues as compared with normal tissues and
listed in Table II, miR-647 is
increased 3.85-fold in CRC, although the P-value is 0.07116, we
found that miR-1914 and miR-647 are positioned very close to each
other on chr20q13 and are only 1,086 bp from each other as shown in
Fig. 1A, indicating that they may
work as a miRNA cluster. miRNA cluster (miR-647 and miR-1914) that
is abnormal expression in CRC tissues. Whether they have the
synchronous expression and the same effects became interesting
questions to us. We therefore selected miR-647 and miR-1914 for
further investigation.
 | Table II.Significant dysregulated miRNAs in
CRCs. |
Table II.
Significant dysregulated miRNAs in
CRCs.
| Gene ID | miRNA sequence | miRNA | Style | Mean fold
change | P-value |
|---|
| MIR647 |
gtggctgcactcacttccttc | hsa-miR-647 | Up | 3.85420 | 0.07116 |
| MIR1914 |
ccctgtgcccggcccacttctg |
hsa-miR-1914-5p | Up | 3.56096 | 0.00280 |
| MIR1182 |
gagggtcttgggagggatgtgac | hsa-miR-1182 | Up | 3.09538 | 0.00297 |
| MIR198 |
ggtccagaggggagataggttc | hsa-miR-198 | Up | 2.83630 | 0.00259 |
| MIR147B |
gtgtgcggaaatgcttctgcta | hsa-miR-147b | Down | 0.13310 | 0.00069 |
| MIR378I |
actggactaggagtcagaagg | hsa-miR-378i | Down | 0.16201 | 0.00006 |
| MIR650 |
aggaggcagcgctctcaggac | hsa-miR-650 | Down | 0.20118 | 0.00005 |
qRT-PCR was initially performed to detect the
expression levels of miR-647 and miR-1914-5p in colon cancer
tissues and cells, and the result showed that the expression levels
of miR-647 and miR-1914-5p in CRC tissue were significantly
upregulated than in non-cancerous tissues (Fig. 1B and C). The expression levels of
miR-647 and miR-1914-5p in HT-29 were higher than in HIEC (Fig. 1D and E). There was a significant
difference when the two groups were compared (P<0.01). This
finding suggests that miR-647 and miR-1914-5p both play an
important role in the developmental processes of CRC.
miR-647 and miR-1914-5p promote the
growth of CRC cells
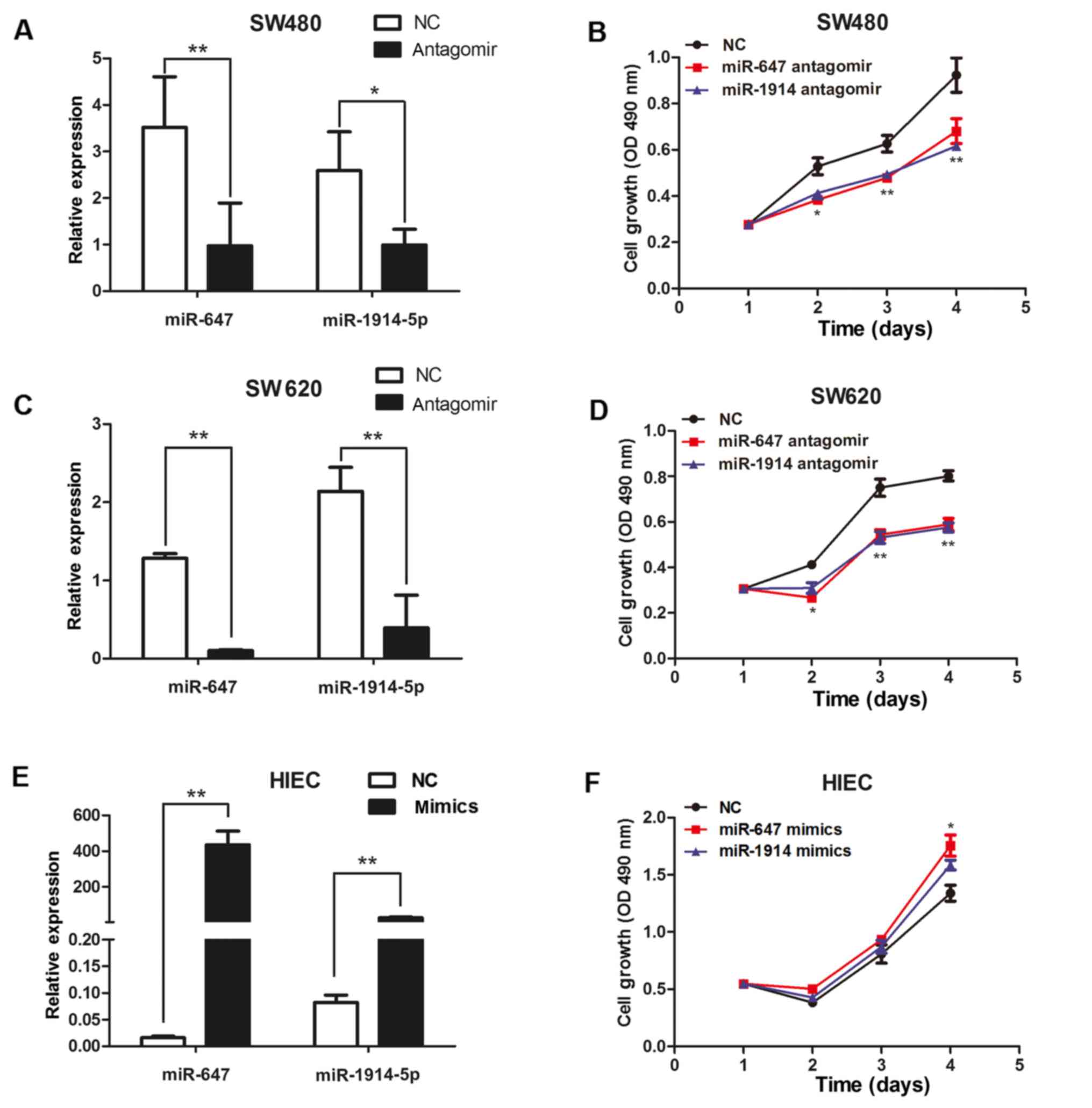
We use MTT to evaluate the function of miR-647 and
miR-1914-5p on the growth of CRC cells. We transfected miR-647
antagomir or miR-1914-5p antagomir into SW480 and SW620 cells to
reduce the expression of miR-647 or miR-1914-5p. Meanwhile, we
transfected either a miR-647 mimic or a miR-1914-5p mimic into
HIECs to induce the expression of miR-647 or miR-1914-5p. We use
q-PCR to detect the expression of miR-647 and miR-1914-5p (Fig. 2). The results of q-PCR analysis
revealed that the expression levels of miR-647 and miR-1914-5p in
the SW480 and SW620 cells transfected with antagomir were
significantly decreased, compared with the control cells (Fig. 2A and C). The expression levels of
miR-647 and miR-1914-5p in the HIEC cells transfected with mimics
were significantly increased, compared with the control cells
(Fig. 2E). We found that the
proliferation of SW480 and SW620 cells was suppressed after
transfection with miR-647 antagomir and miR-1914-5p antagomir
(Fig. 2B and D). The
overexpression of miR-647 and miR-1914-5p markedly promote the
proliferation of the HIEC cells (Fig.
2F). The results indicate that miR 647 and miR-1914-5p can
promote the proliferation of CRC cells.
miR-647 and miR-1914-5p promote the
migration of CRC cells
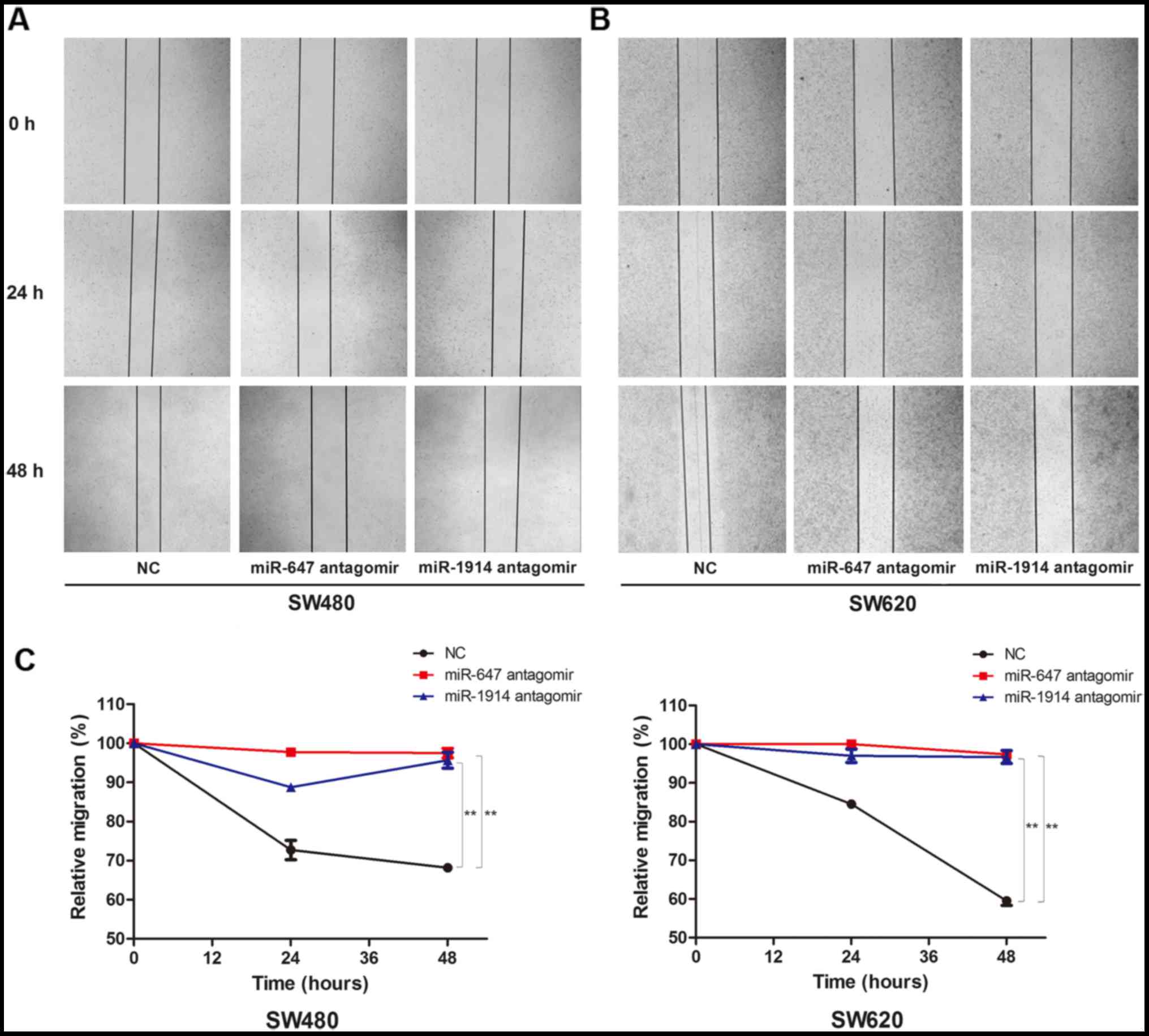
We subsequently investigated the effect of miR-647
and miR-1914 on the migration of CRC cells. First, we transfected
miR-647 antagomir or miR-1914-5p antagomir into SW480 and SW620
cells to reduce the expression of miR-647 or miR-1914-5p. After 8 h
of transfection and another 24 h of incubation, the cell wound
scratch assay was performed. The results showed that there was a
significant difference in migration ability between the NC group
and the miR-647 antagomir group, there also exists a significant
difference between the NC group and the miR-1914 antagomir group.
We found that both in the SW480 (Fig.
3A) and SW620 cells (Fig. 3B),
the migration capacity decreased visibly in the miR-647 antagomir
group and the miR-1914-5p antagomir group, in contrast to the NC
group. We use the relative migration percentage to quantify the
capacity of cell migration (Fig.
3C). The results show that the migration of SW480 and SW620 was
downregulated after the silence of the expression of miR-647 and
miR-1914-5p. This finding indicates that miR-647 and miR-1914-5p
can promote the migration of CRC cells.
NFIX is a common direct target of
miR-647 and miR-1914-5p
miR-1914 interacts with the 3′-UTR of NFIX and
reduces the level of NFIX in chemoresistant CRC cells, as reported
by Hu et al (22). First
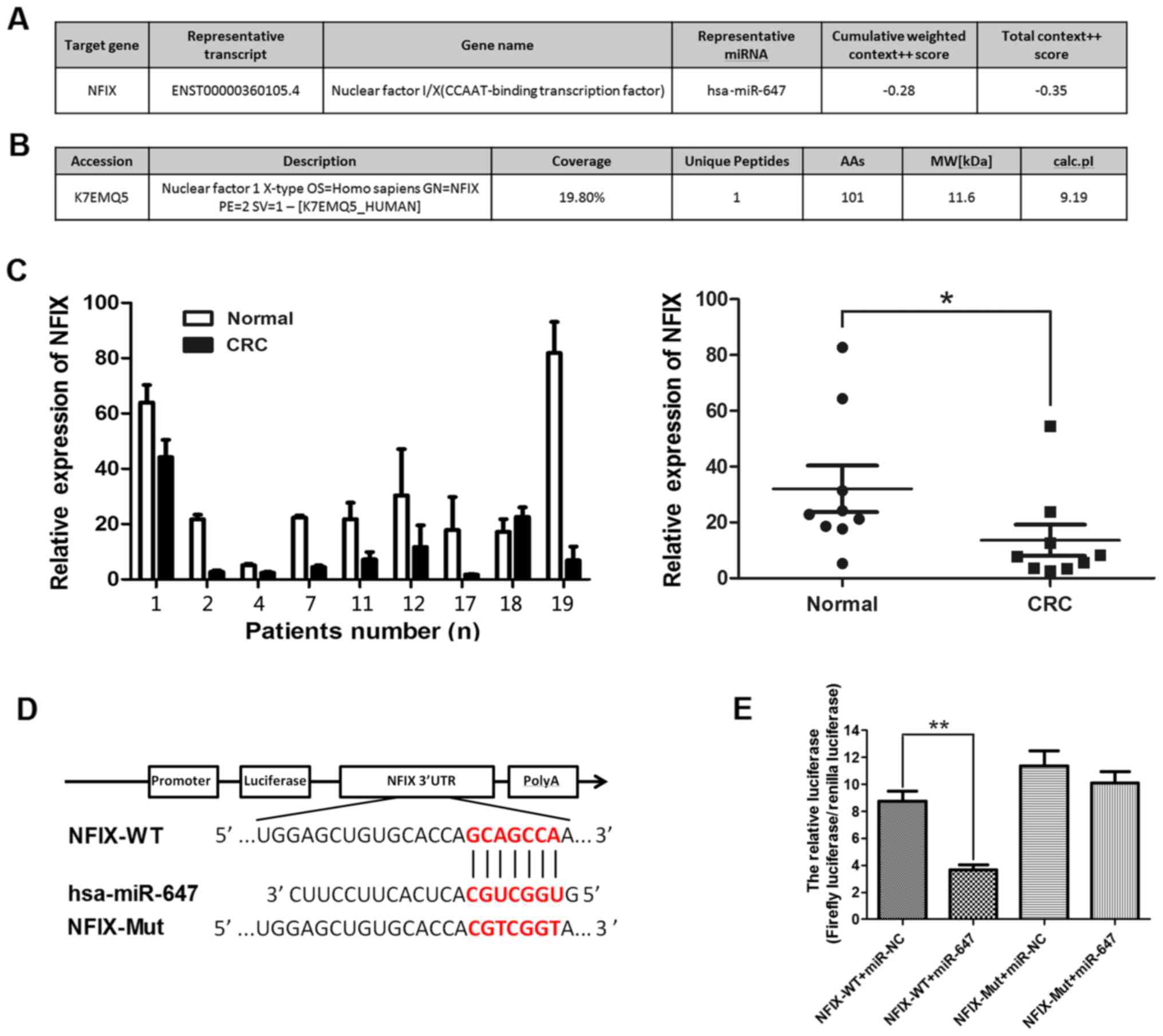
bioinformatics prediction was used to seek the target gene of
miR-647. TargetScan 7.0 was used to identify potential genes that
target miR-647. It predicted that NFIX may be a target gene and its
target site of miR-647 based on the putative target sequence in
base pairs 271–278 or 4,032–4,038 of the NFIX 3′UTR. The results
are illustrated in Fig. 4A. Then
iTRAQ was put into effect by the State Key Laboratory of Cancer
Biology (CBSKL). We found 4,141 differently expressed genes, NFIX
is the intersection of the bioinformatics prediction data and
i-TRAQ data (Fig. 4B). Taking into
consideration that miR-1914 and miR-647 are positioned very close
to each other, and NFIX is a target gene of miR-1914, we detected
the expression of NFIX in CRC samples for confirmation by side. We
detected the expression of NFIX in the samples in which the
expression of miR-647 is significantly upregulated compared with
their adjacent tissues (nos. 1, 2, 4, 7, 11, 12, 17, 18 and 19).
Fig. 4C showed that the expression
of NFIX was decreased in CRC tissues among these patients. To
further clearly determine the target gene of miR-647 and verify
whether miR-647 targets NFIX directly, NFIX-WT and NFIX-Mut
plasmids were constructed and cloned into the region downstream of
the GV272 (SV40-Luc-MCS) firefly luciferase reporter vector
(Fig. 4D). The subsequent
luciferase activity assay demonstrated that miR-647 significantly
decreased luciferase activity of NFIX-WT, but not NFIX-Mut in 293T
cells (Fig. 4E). Overall, our
results show that NFIX is a direct target of miR-647, in summary
NFIX is a common direct target of miR-647 and miR-1914-5p.
miR-647 and miR-1914-5p promote
proliferation and migration in CRC cells equivalently by targeting
NFIX
To clarify whether miR-647 and miR-1914-5p can
promote proliferation and migration in CRC cells by directly
targeting NFIX, NFIX siRNA (NFIX-homo-1033) were chemically
synthesized. The expression level of NFIX in the SW480 cells
transfected with NFIX-homo-1033 was significantly decreased
compared with the negative control (Fig. 5A). Then the NFIX siRNA were
co-transfected into SW480 cells with miR-647 antagomir or
miR-1914-5p antagomir separately. The MTT assay was carried out to
test the cell proliferation ability. The results showed that the
proliferation was greatly descend in miR-647 antagomir or
miR-1914-5p antagomir groups compared with the negative control
cells, in the meantime the proliferation ability was regained after
we transfect NFIX-homo-1033 in these two groups. The results were
illustrated in Fig. 5B. The
effects of miR-647 and miR-1914-5p on cell migration was detected
by the cell wound scratch assay. The results showed that the
migration ability was tremendously decline in miR-647 antagomir or
miR-1914-5p antagomir groups compared with the negative control
cells, and the migration capabilities was recovered after we
transfect NFIX-homo-1033 in these two groups (Fig. 5C). The detailed relative migration
percentage quantifying the capacity of cell migration was showed in
Fig. 5D. This result indicates
that miR-647 and miR-1914-5p promote proliferation and migration in
CRC cells by targeting NFIX directly.
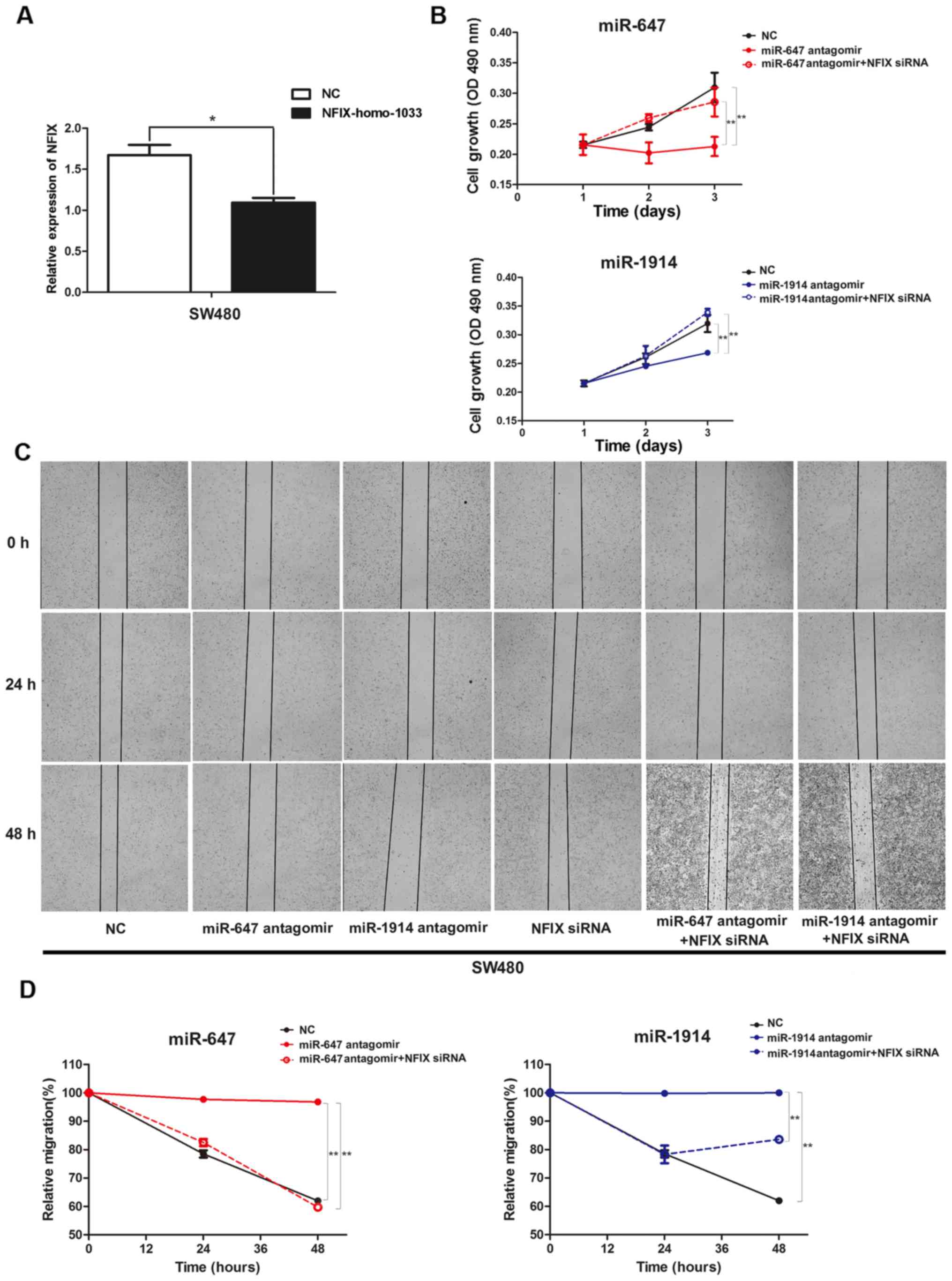 | Figure 5.(A) Expression levels of NFIX in SW480
cells transfected with NFIX siRNA (NFIX-homo-1033), determined
using qPCR. (B) Cell proliferation was analyzed using an MTT assay
in SW480 cells, which were co-transfected with microRNA antagomir
and NFIX-homo-1033. (C) Wound closure of SW480 cells, which were
co-transfected with microRNA antagomir and NFIX-homo-1033, in
contrast with NC after 0, 24 and 48 h. (D) The relative migration
percentage to quantify the capacity of cell migration among the NC
group, miR-647 antagomir group, miR-1914 antagomir group, NFIX
siRNA group, miR-647 antagomir + NFIX siRNA group and miR-1914
antagomir + NFIX siRNA group in SW480 cells. *P<0.05 and
**P<0.01, by ANOVA. NFIX, nuclear factor I/X. |
Discussion
In this study, we investigated the differential
expression of miRNAs between patients with CRC and their adjacent
normal tissues through screening with RNA-Seq. We identified 9
upregulated microRNAs and 4 downregulated microRNAs. The most
significant increase in these microRNAs is miR-1914 as well as its
counterpart miR-647. We discovered that the expression of the miRNA
cluster (miR-647 and miR-1914) were elevated in CRC specimens and
cell lines. More importantly, the expression of the two microRNAs
varied simultaneously. By detecting with iTRAQ, bioinformatics
algorithm and dual luciferase reporter assays, NFIX was
demonstrated to be their co-target gene. The expression of miR-647
was negatively associated with the levels of NFIX. The microRNA
cluster can promote CRC cells proliferation and migration by
downregulation the expression of NFIX. miR-647/1914 cluster act as
an oncogene mediate the tumorigenesis in CRC. By exploring and
understanding the detailed mechanisms of these microRNAs that
regulate colon cancer, miR-647/1914 may serve as therapeutic
molecules against CRC.
Hu et al indicated that plasma miR-1914* and
miR-1915 interact with NFIX RNA and reduce the level of NFIX in CRC
cells that are resistant to first-line chemotherapy. Elevated
levels of miR-1914* and miR-1915 in vivo not only reduced
the expression of NFIX but also increased the sensitivity of
HCT116/5-Fu/OXA cells to 5-FU and L-OHP, they came to the
conclusion that miR-1914* and miR-1915 can affect the cell response
to drug-induced apoptosis, tumor growth, invasion and tumor
suppressor genes (22). The
conclusion miR-1914 interacts with the 3′-UTR of NFIX and reduces
the level of NFIX in chemoresistant CRC cells in part confirms our
results. We found in subsequent experiments that miR-647 as its
counterpart is negatively related to the expression of NFIX and
NFIX is a direct target of miR-647 too. Increasingly, researchers
are aware of the opinion that there is a wide variety of
alterations and modifications in genetics and epigenetics that
occur during the development of CRC at the molecular level
(23,24), such as oncogenes that are activated
and anti-oncogenes that are inactivated (25,26).
Aberrantly activated miRNAs can be silenced using antagomirs
(27). Re-expression of miRNAs
that are lost in cancers can be achieved by the induction of miRNA
mimics (28). The present study
demonstrated that downregulation the expression of miR-647/1914
cluster with microRNA antagomir notably decreased the proliferation
and migration of SW480 and SW620 cells, and overexpress
miR-647/1914 cluster with microRNA mimics increased the
proliferation of HIEC cells.
To our knowledge, this is the first study to explore
the expression and the function of miR-647/1914 cluster in CRC.
miR-647 and miR-1914 are newly identified molecules, there have
been few studies on miR-647 or miR-1914-5p so far. Yang et
al (29) reported that several
lymphangiogenesis-related miRNAs including miR-647 are upregulated
significantly altered during lymphatic metastasis of gastric
cancer. On the contrary, Cao et al (30) reparted that miR-647 was markedly
downregulated in gastric cancer (GC) and GC cell lines and miR-647
exerts powerful antitumorigenic effects in vitro and in
vivo, and may represent a promising therapeutic agent against
GC. Hu et al (22) reported
the function of miR-1914 in colon cancer for the first time. Our
results were consistent with the previous study. Because of the
limited study about the expression and function of miR-647/miR-1914
in CRC, there are still some questions facing us. Whether the
cluster is regulated by a common transcriptional factor still
remains unknown. It is extremely essential to determine the
upstream regulator of miR-647 and miR-1914 in CRC.
In conclusion, we demonstrate that miR-647 and
miR-1914 promote the proliferation and migration equivalently by
downregulating NFIX in CRC cells in vitro. The miR-647 and
miR-1914 cluster may serve as a clinical indicator and a molecular
therapeutic for patients with CRC. Although the specific molecular
regulatory mechanism and signal transduction pathways still remain
unclear, more research is needed to increase the five-year survival
rate of patients with CRC and improve the prognosis.
Acknowledgements
This study was supported by the National Natural
Science Foundation of China (grant nos. 81572816, 31571437,
81230043 and 81421003) and the Science and Technology Co-ordination
Innovation Project of Shaanxi Province (grant no.
2015KTCQ03-02).
References
|
1
|
Chakradhar S: Colorectal cancer: 5 big
questions. Nature. 521:S162015. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Brody H: Colorectal cancer. Nature.
521:S12015. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Van Cutsem E, Verheul HM, Flamen P,
Rougier P, Beets-Tan R, Glynne-Jones R and Seufferlein T: Imaging
in colorectal cancer: Progress and challenges for the clinicians.
Cancers (Basel). 8:E812016.doi: 10.3390/cancers8090081. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Baratti D, Kusamura S, Pietrantonio F,
Guaglio M, Niger M and Deraco M: Progress in treatments for
colorectal cancer peritoneal metastases during the years 2010–2015.
A systematic review. Crit Rev Oncol Hematol. 100:209–222. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hardingham JE, Grover P, Winter M, Hewett
PJ, Price TJ and Thierry B: Detection and clinical significance of
circulating tumor cells in colorectal cancer-20 years of progress.
Mol Med. 21 Suppl 1:S25–S31. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cummins JM, He Y, Leary RJ, Pagliarini R,
Diaz LA Jr, Sjoblom T, Barad O, Bentwich Z, Szafranska AE,
Labourier E, et al: The colorectal microRNAome. Proc Natl Acad Sci
USA. 103:pp. 3687–3692. 2006; View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Landi D, Gemignani F, Pardini B, Naccarati
A, Garritano S, Vodicka P, Vodickova L, Canzian F, Novotny J,
Barale R and Landi S: Identification of candidate genes carrying
polymorphisms associated with the risk of colorectal cancer by
analyzing the colorectal mutome and microRNAome. Cancer.
118:4670–4680. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li L and Ma HQ: MicroRNA-216a inhibits the
growth and metastasis of oral squamous cell carcinoma by targeting
eukaryotic translation initiation factor 4B. Mol Med Rep.
12:3156–3162. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mohammadi A, Mansoori B and Baradaran B:
The role of microRNAs in colorectal cancer. Biomed Pharmacother.
84:705–713. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chatterjee V, Beard RS Jr, Reynolds JJ,
Haines R, Guo M, Rubin M, Guido J, Wu MH and Yuan SY: MicroRNA-147b
regulates vascular endothelial barrier function by targeting ADAM15
expression. PLoS One. 9:e1102862014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang Y, Wang Y, Wei Y, Li M, Yu S, Ye M,
Zhang H, Chen S, Liu W and Zhang J: miR-129-3p promotes docetaxel
resistance of breast cancer cells via CP110 inhibition. Sci Rep.
5:154242015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Marí-Alexandre J, Sánchez-Izquierdo D,
Gilabert-Estellés J, Barceló-Molina M, Braza-Boïls A and Sandoval
J: miRNAs regulation and its role as biomarkers in endometriosis.
Int J Mol Sci. 17:E932016.doi: 10.3390/ijms17010093. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang LL, Wang L, Wang XY, Shang D, Yin SJ,
Sun LL and Ji HB: MicroRNA-218 inhibits the proliferation,
migration and invasion and promotes apoptosis of gastric cancer
cells by targeting LASP1. Tumour Biol. 37:15241–15252. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tan S, Li R, Ding K, Lobie PE and Zhu T:
miR-198 inhibits migration and invasion of hepatocellular carcinoma
cells by targeting the HGF/c-MET pathway. FEBS Lett. 585:2229–2234.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wu M, Zhang Y, Tang A and Tian L: miR-506
inhibits cell proliferation and invasion by targeting TET family in
colorectal cancer. Iran J Basic Med Sci. 19:316–322.
2016.PubMed/NCBI
|
|
16
|
Sun XF, Sun JP, Hou HT, Li K, Liu X and Ge
QX: MicroRNA-27b exerts an oncogenic function by targeting Fbxw7 in
human hepatocellular carcinoma. Tumour Biol. 37:15325–15332. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu Y, Uzair-Ur-Rehman, Guo Y, Liang H,
Cheng R, Yang F, Hong Y, Zhao C, Liu M, Yu M, et al: miR-181b
functions as an oncomiR in colorectal cancer by targeting PDCD4.
Protein Cell. 7:722–734. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Rawlings-Goss RA, Campbell MC and Tishkoff
SA: Global population-specific variation in miRNA associated with
cancer risk and clinical biomarkers. BMC Med Genomics. 7:532014.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Rossi G, Antonini S, Bonfanti C,
Monteverde S, Vezzali C, Tajbakhsh S, Cossu G and Messina G: Nfix
regulates temporal progression of muscle regeneration through
modulation of myostatin expression. Cell Rep. 14:2238–2249. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Heng YH, Zhou B, Harris L, Harvey T, Smith
A, Horne E, Martynoga B, Andersen J, Achimastou A, Cato K, et al:
NFIX regulates proliferation and migration within the murine SVZ
neurogenic niche. Cereb Cortex. 25:3758–3778. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mao Y, Liu J, Zhang D and Li B: miR-1290
promotes cancer progression by targeting nuclear factor I/X(NFIX)
in esophageal squamous cell carcinoma (ESCC). Biomed Pharmacother.
76:82–93. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hu J, Cai G, Xu Y and Cai S: The plasma
microRNA miR-1914* and −1915 suppresses chemoresistant in
colorectal cancer patients by down-regulating NFIX. Curr Mol Med.
16:70–82. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bertero T, Grosso S, Robbe-Sermesant K,
Lebrigand K, Hénaoui IS, Puisségur MP, Fourre S, Zaragosi LE,
Mazure NM, Ponzio G, et al: ‘Seed-Milarity’ confers to hsa-miR-210
and hsa-miR-147b similar functional activity. PLoS One.
7:e449192012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Feng L, Xie Y, Zhang H and Wu Y:
Down-regulation of NDRG2 gene expression in human colorectal cancer
involves promoter methylation and microRNA-650. Biochem Biophys Res
Commun. 406:534–538. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Omrane I and Benammar-Elgaaied A: The
immune microenvironment of the colorectal tumor: Involvement of
immunity genes and microRNAs belonging to the TH17 pathway. Biochim
Biophys Acta. 1856:28–38. 2015.PubMed/NCBI
|
|
26
|
Zhao XD, Lu YY, Guo H, Xie HH, He LJ, Shen
GF, Zhou JF, Li T, Hu SJ, Zhou L, et al: MicroRNA-7/NF-kB signaling
regulatory feedback circuit regulates gastric carcinogenesis. J
Cell Biol. 210:613–627. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Krützfeldt J, Rajewsky N, Braich R, Rajeev
KG, Tuschl T, Manoharan M and Stoffel M: Silencing of microRNAs in
vivo with ‘antagomirs’. Nature. 438:685–689. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sokilde R, Newie I, Persson H, Borg Å and
Rovira C: Passenger strand loading in overexpression experiments
using microRNA mimics. RNA Biol. 12:787–791. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yang B, Jing C, Wang J, Guo X, Chen Y, Xu
R, Peng L, Liu J and Li L: Identification of microRNAs associated
with lymphangiogenesis in human gastric cancer. Clin Transl Oncol.
16:374–379. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cao W, Wei W, Zhan Z, Xie D, Xie Y and
Xiao Q: Role of miR-647 in human gastric cancer suppression. Oncol
Rep. 37:1401–1411. 2017. View Article : Google Scholar : PubMed/NCBI
|