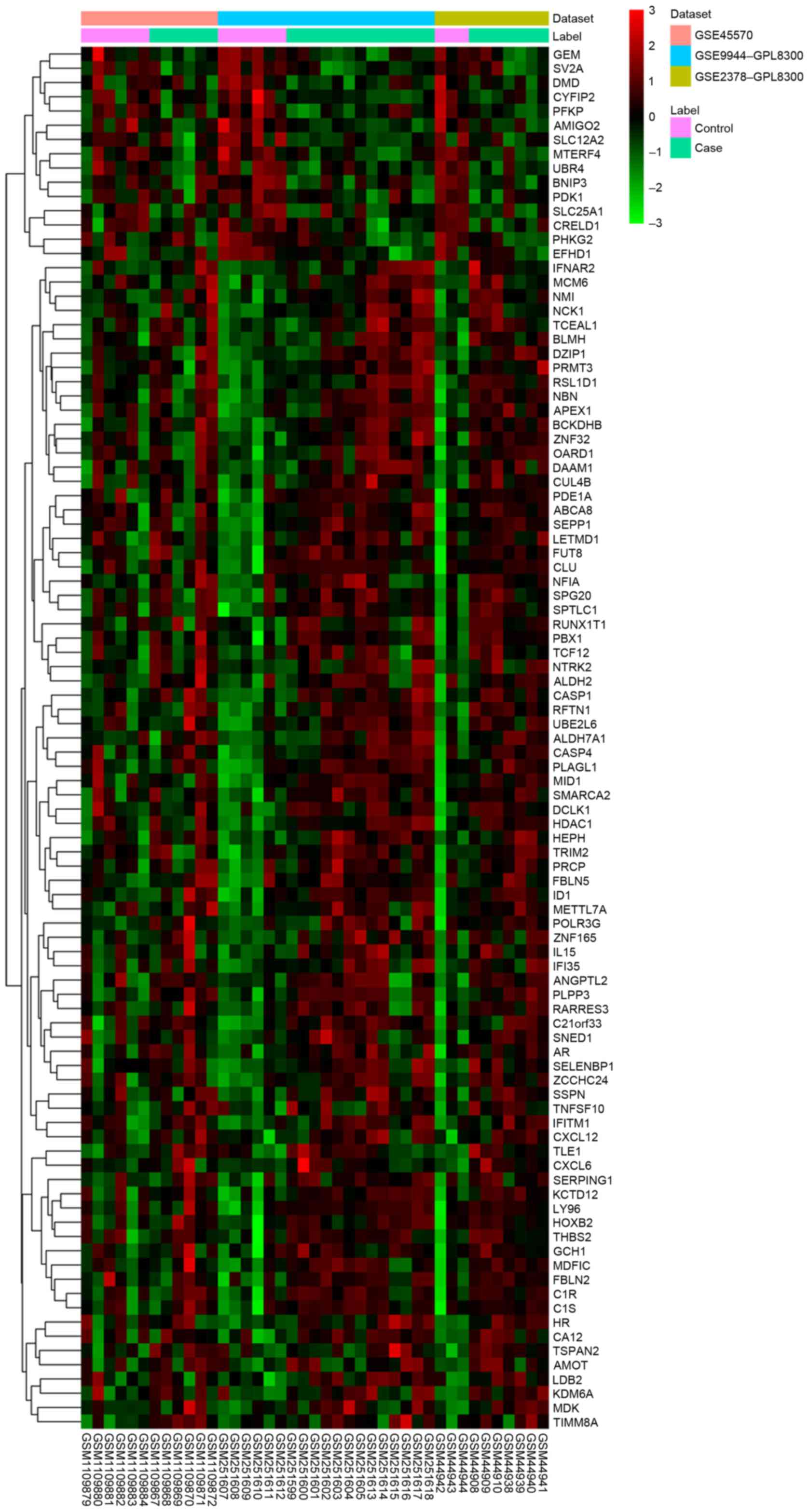
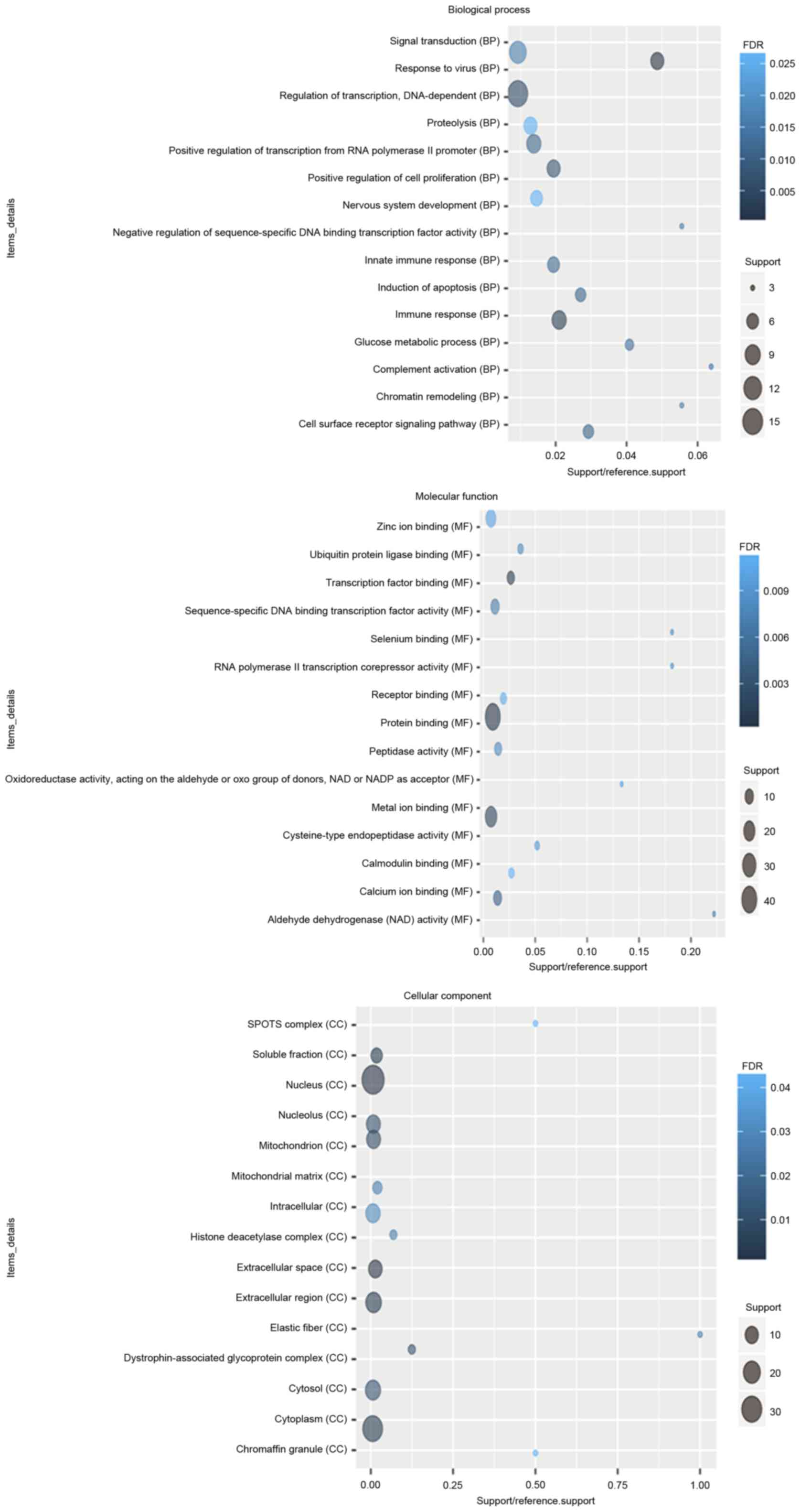
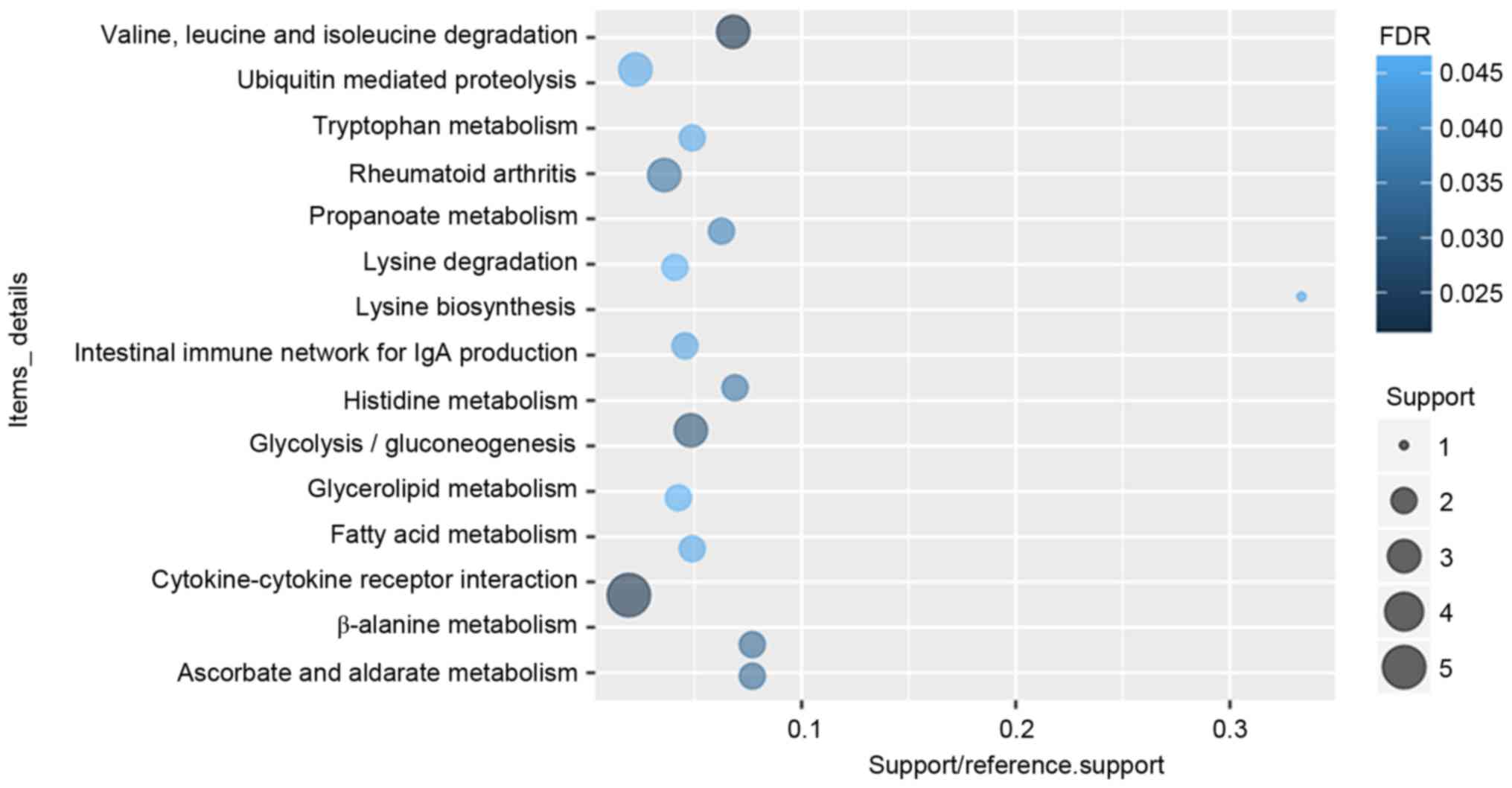
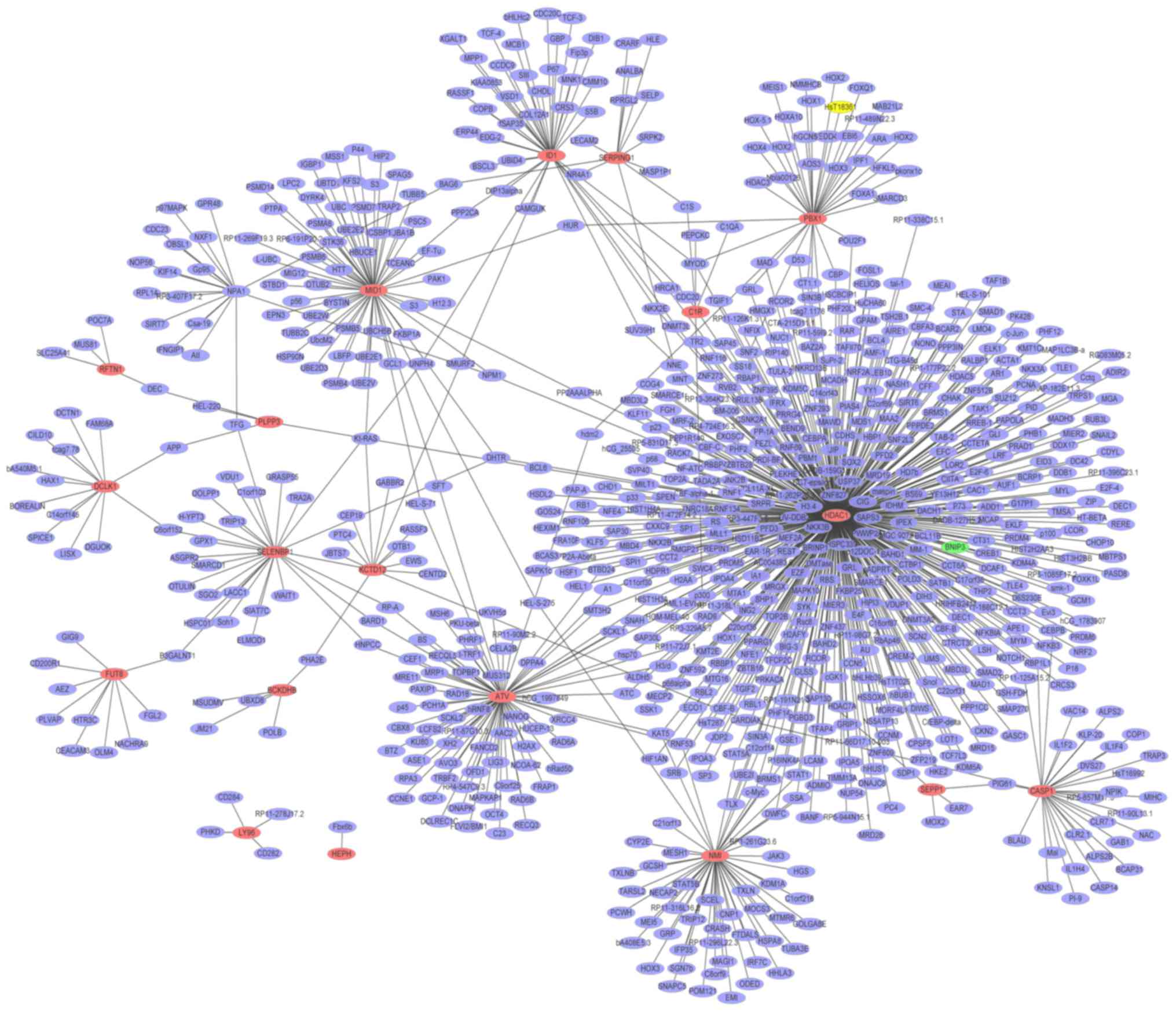
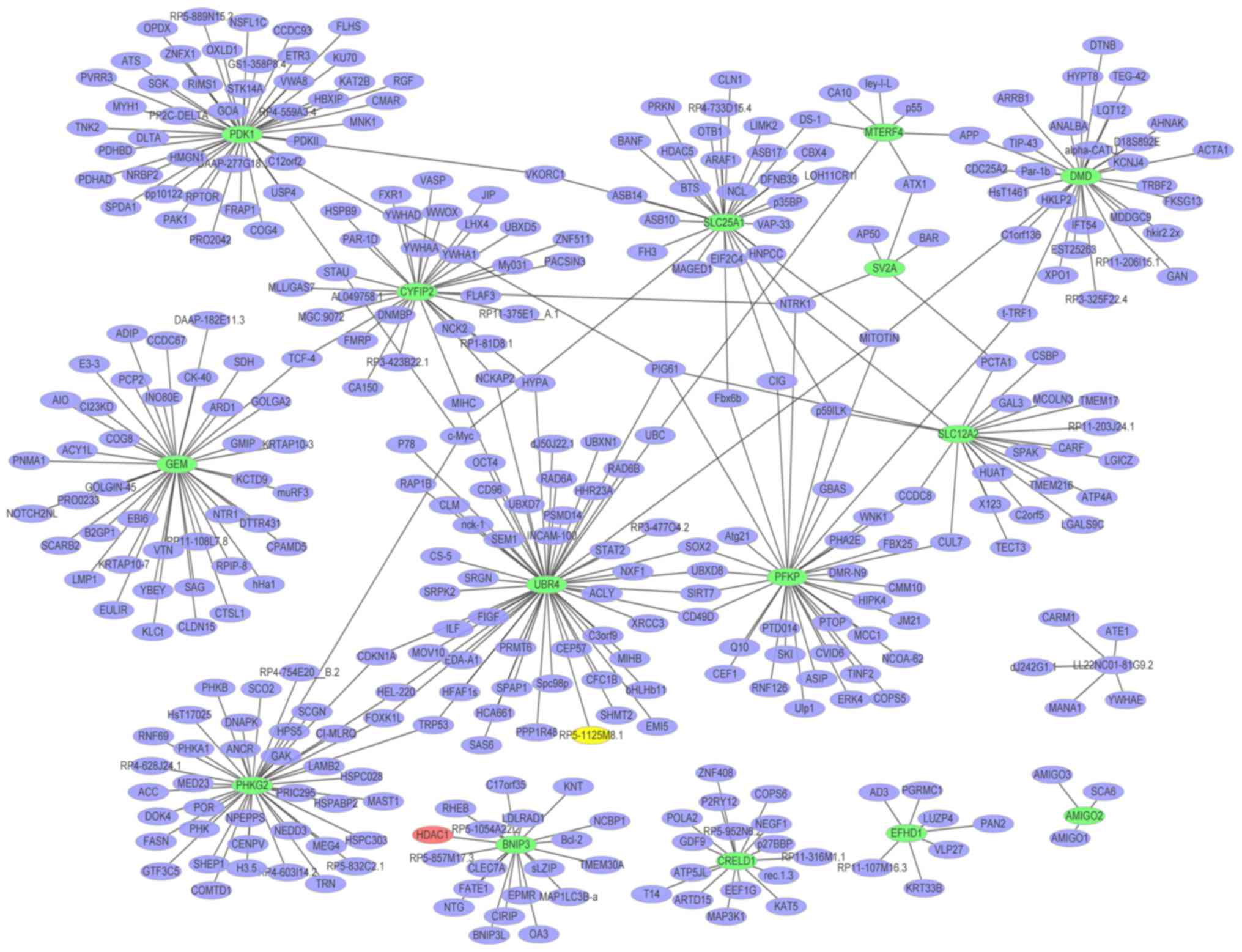
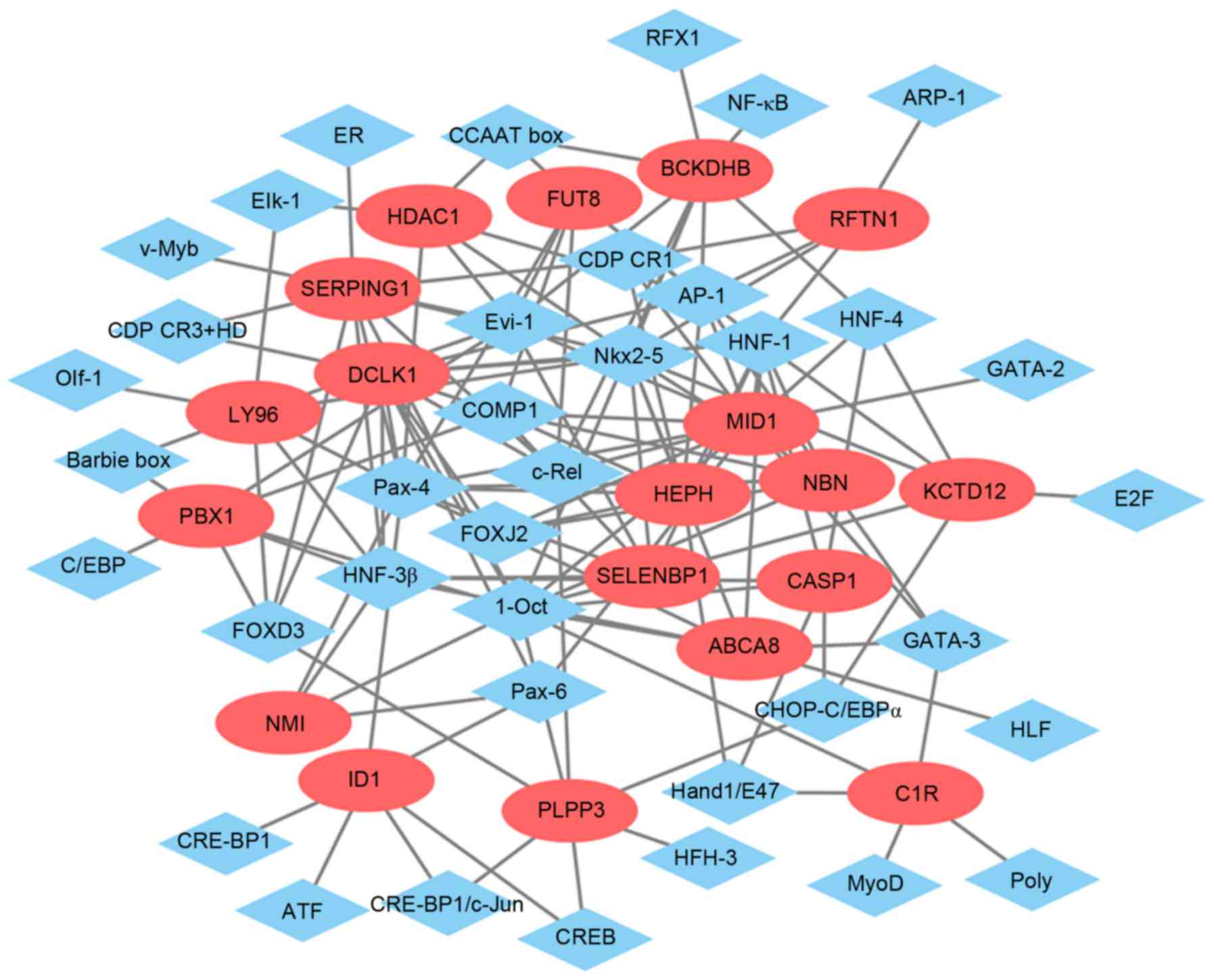
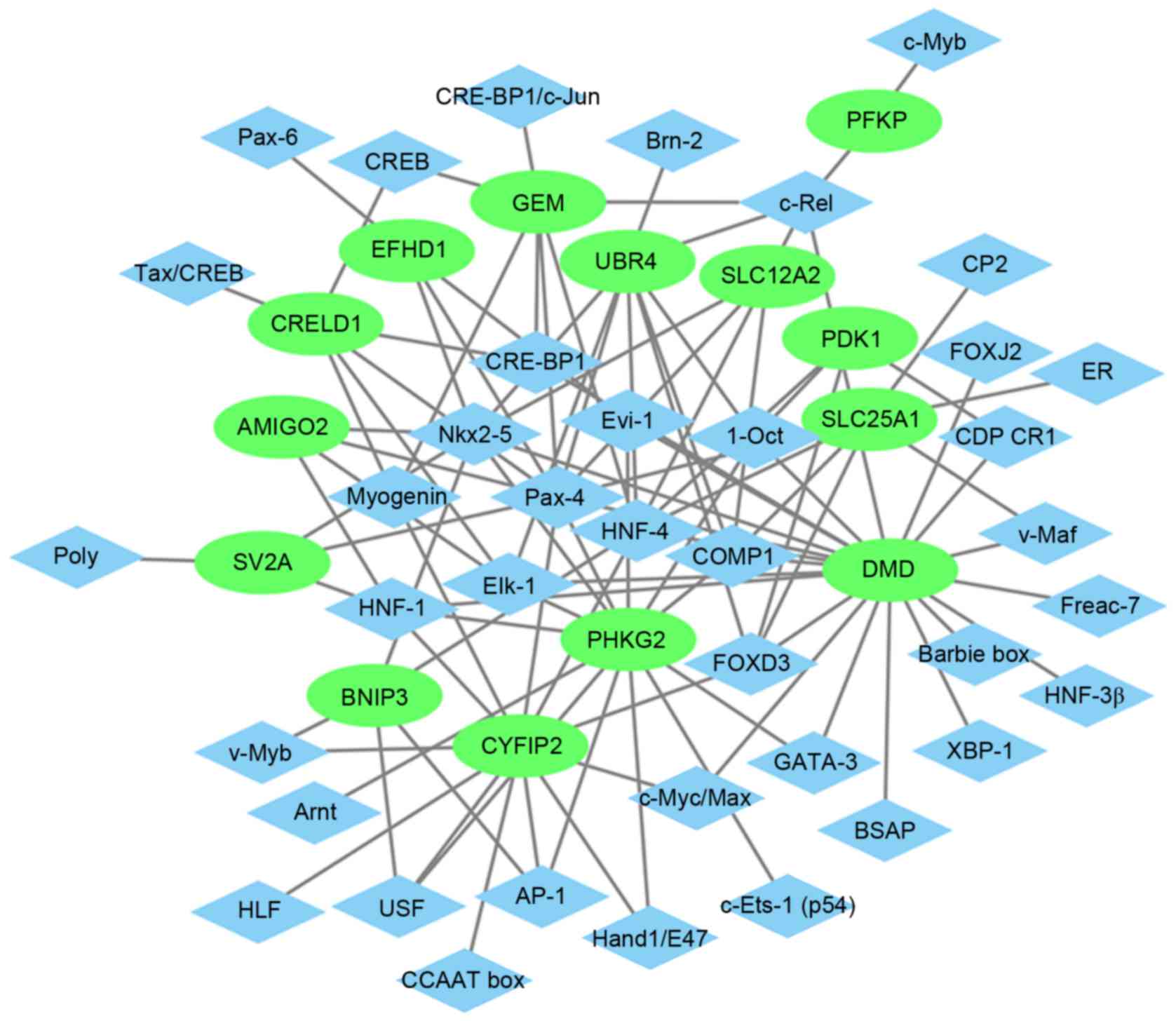
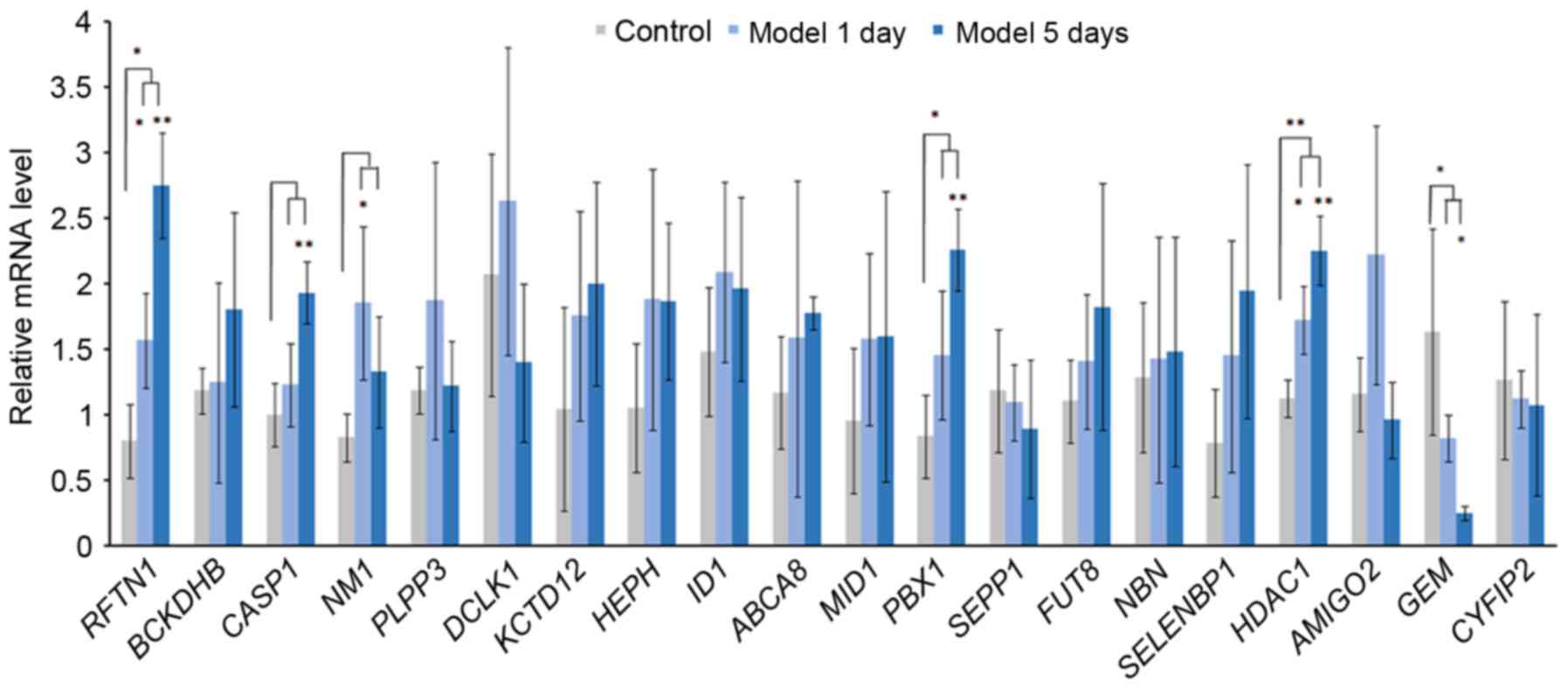
|
1
|
Yu-Wai-Man P, Stewart JD, Hudson G,
Andrews RM, Griffiths PG, Birch MK and Chinnery PF: OPA1 increases
the risk of normal but not high tension glaucoma. J Med Genet.
47:120–125. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Moreno MC, Sande P, Marcos HA, de Zavalía
N, Sarmiento MI Keller and Rosenstein RE: Effect of glaucoma on the
retinal glutamate/glutamine cycle activity. FASEB J. 19:1161–1162.
2005.PubMed/NCBI
|
|
3
|
Liu T, Xie L, Ye J and He X: Family-based
analysis identified CD2 as a susceptibility gene for primary open
angle glaucoma in Chinese Han population. J Cell Mol Med.
18:600–609. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Guo MS, Wu YY and Liang ZB: Hyaluronic
acid increases MMP-2 and MMP-9 expressions in cultured trabecular
meshwork cells from patients with primary open-angle glaucoma. Mol
Vis. 18:1175–1181. 2012.PubMed/NCBI
|
|
5
|
Gazzard G, Foster PJ, Devereux JG, Oen F,
Chew P, Khaw PT and Seah S: Intraocular pressure and visual field
loss in primary angle closure and primary open angle glaucomas. Br
J Ophthalmol. 87:720–725. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rosenberg LF: Glaucoma: Early detection
and therapy for prevention of vision loss. Am Fam Physician.
52:2289–2298. 1995.PubMed/NCBI
|
|
7
|
Bikbova G, Oshitari T and Yamamoto S:
Diabetes mellitus and retinal vein occlusion as risk factors for
open angle glaucoma and neuroprotective therapies for retinal
ganglion cell neuropathy. J Clin Exper Ophthalmol. S3:2012.
|
|
8
|
McMenemy MG: Primary Open Angle Glaucoma.
Springer; New York, NY: pp. 1721. 2014
|
|
9
|
Girard MJA, Zimmo L, White ET, Mari JM,
Ethier CR and Strouthidis NG: Towards a biomechanically-based
diagnosis for glaucoma: In vivo deformation mapping of the human
optic nerve head. In: ASME 2012 Summer Bioengineering Conference.
Puerto Rico. pp. 423–424. 2012;
|
|
10
|
Wu Y, Zang WD and Jiang W: Functional
analysis of differentially expressed genes associated with glaucoma
from DNA microarray data. Genet Mol Res. 13:9421–9428. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lukas TJ, Miao H, Chen L, Riordan SM, Li
W, Crabb AM, Wise A, Du P, Lin SM and Hernandez MR: Susceptibility
to glaucoma: Differential comparison of the astrocyte transcriptome
from glaucomatous African American and Caucasian American donors.
Genome Biol. 9:R1112008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hernandez MR, Agapova OA, Yang P,
Salvador-Silva M, Ricard CS and Aoi S: Differential gene expression
in astrocytes from human normal and glaucomatous optic nerve head
analyzed by cDNA microarray. Glia. 38:45–64. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yan X, Yuan F, Chen X and Dong C:
Bioinformatics analysis to identify the differentially expressed
genes of glaucoma. Mol Med Rep. 12:4829–4836. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Rozsa FW, Scott KM, Pawar H, Samples JR,
Wirtz MK and Richards JE: Differential expression profile
prioritization of positional candidate glaucoma genes: The GLC1C
locus. Arch Ophthalmol. 125:117–127. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Fei Q, Lin J, Meng H, Wang B, Yang Y, Wang
Q, Su N, Li J and Li D: Identification of upstream regulators for
synovial expression signature genes in osteoarthritis. Joint Bone
Spine. 83:545–551. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Marot G, Foulley JL, Mayer CD and
Jaffrézic F: Moderated effect size and P-value combinations for
microarray meta-analyses. Bioinformatics. 25:2692–2696. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wei K, Pan S and Li Y: Functional
characterization of maize C2H2, zinc-finger
gene family. Plant Mol Biol Rep. 34:761–776. 2016. View Article : Google Scholar
|
|
18
|
Matys V, Fricke E, Geffers R, Gössling E,
Haubrock M, Hehl R, Hornischer K, Karas D, Kel AE, Kel-Margoulis
OV, et al: TRANSFAC: Transcriptional regulation, from patterns to
profiles. Nucleic Acids Res. 31:374–378. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Adachi M, Takahashi K, Nishikawa M, Miki H
and Uyama M: High intraocular pressure-induced ischemia and
reperfusion injury in the optic nerve and retina in rats. Graefes
Arch Clin Exp Ophthalmol. 234:445–451. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Adachi M, Takahashi K, Nishikawa M, Miki H
and Uyama M: High intraocular pressure-induced ischemia and
reperfusion injury in the optic nerve and retina in rats. Graefes
Arch Clin Exp Ophthalmol. 234:445–451. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Huang Y, Li Z, Wang N, van Rooijen N and
Cui Q: Roles of PI3K and JAK pathways in viability of retinal
ganglion cells after acute elevation of intraocular pressure in
rats with different autoimmune backgrounds. BMC Neurosci. 9:782008.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang S, Wang H, Lu Q, Qing G, Wang N,
Wang Y, Li S, Yang D and Yan F: Detection of early neuron
degeneration and accompanying glial responses in the visual pathway
in a rat model of acute intraocular hypertension. Brain Res.
1303:131–143. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sun D, Qu J and Jakobs TC: Reversible
reactivity by optic nerve astrocytes. Glia. 61:1218–1235. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lye-Barthel M, Sun D and Jakobs TC:
Morphology of astrocytes in a glaucomatous optic nerve. Invest
Ophthalmol Vis Sci. 54:909–917. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Morrison JC, Cepurna WO, Doser TA, Dyck JA
and Johnson EC: A short interval of controlled elevation of iop
(CEI) reproduces early chronic glaucoma model optic nerve head
(ONH) gene expression responses. Invest Ophthalmol Vis Sci.
51:52162010.
|
|
27
|
Morrison JC, Cepurna WO, Tehrani S, Choe
TE, Jayaram H, Lozano DC, Fortune B and Johnson EC: A period of
controlled elevation of IOP (CEI) produces the specific gene
expression responses and focal injury pattern of experimental rat
glaucoma. Invest Ophthalmol Vis Sci. 57:6700–6711. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Downs JC, Burgoyne CF, Seigfreid WP,
Reynaud JF, Strouthidis NG and Sallee V: 24-hour IOP telemetry in
the nonhuman primate: Implant system performance and initial
characterization of IOP at multiple timescales. Invest Ophthalmol
Vis Sci. 52:7365–7375. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Crowston JG, Kong YX, Trounce IA, Dang TM,
Fahy ET, Bui BV, Morrison JC and Chrysostomou V: An acute
intraocular pressure challenge to assess retinal ganglion cell
injury and recovery in the mouse. Exp Eye Res. 141:3–8. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Briggs EL Ashworth, Toh T, Eri R, Hewitt
AW and Cook AL: TIMP1, TIMP2 and TIMP4 are increased in aqueous
humor from primary open angle glaucoma patients. Mol Vis.
21:1162–1172. 2015.PubMed/NCBI
|
|
31
|
Janssen SF, Gorgels TG, Van der Spek PJ,
Jansonius NM and Bergen AA: In silico analysis of the molecular
machinery underlying aqueous humor production: Potential
implications for glaucoma. J Clin Bioinforma. 3:212013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chen JH, Wang D, Huang C, Zheng Y, Chen H,
Pang CP and Zhang M: Interactive effects of ATOH7 and RFTN1 in
association with adult-onset primary open-angle glaucoma. Invest
Ophthalmol Vis Sci. 53:779–785. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Macgregor S, Hewitt AW, Hysi PG, Ruddle
JB, Medland SE, Henders AK, Gordon SD, Andrew T, McEvoy B,
Sanfilippo PG, et al: Genome-wide association identifies ATOH7 as a
major gene determining human optic disc size. Hum Mol Genet.
19:2716–2724. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Xue W, Du P, Lin S, Dudley VJ, Hernandez
MR and Sarthy VP: Gene expression changes in retinal Müller (glial)
cells exposed to elevated pressure. Curr Eye Res. 36:754–767. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Futterweit W, Ritch R, Teekhasaenee C and
Nelson ES: Coexistence of Prader-Willi syndrome, congenital
ectropion uveae with glaucoma, and factor XI deficiency. JAMA.
255:3280–3282. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Schmitt HM, Schlamp CL and Nickells RW:
Role of HDACs in optic nerve damage-induced nuclear atrophy of
retinal ganglion cells. Neurosci Lett. 625:11–15. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ueki Y and Reh TA: Activation of
BMP-Smad1/5/8 signaling promotes survival of retinal ganglion cells
after damage in vivo. PLoS One. 7:e386902012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Biermann J, Boyle J, Pielen A and Lagrèze
WA: Histone deacetylase inhibitors sodium butyrate and valproic
acid delay spontaneous cell death in purified rat retinal ganglion
cells. Mol Vis. 17:395–403. 2011.PubMed/NCBI
|
|
39
|
Chindasub P, Lindsey JD, Duong-Polk K,
Leung CK and Weinreb RN: Inhibition of histone deacetylases 1 and 3
protects injured retinal ganglion cells. Invest Ophthalmol Vis Sci.
54:96–102. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Messina-Baas OM, González-Huerta LM,
Chima-Galán C, Kofman-Alfaro SH, Rivera-Vega MR, Babayán-Mena I and
Cuevas-Covarrubias SA: Molecular analysis of the CYP1B1 gene:
Identification of novel truncating mutations in patients with
primary congenital glaucoma. Ophthalmic Res. 39:17–23. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Mansergh FC, Kenna PF, Ayuso C, Kiang AS,
Humphries P and Farrar GJ: Novel mutations in the TIGR gene in
early and late onset open angle glaucoma. Hum Mutat. 11:244–251.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Nakamura M: New insights into the
pathogenesis of glaucomatous optic neuropathy and refinement of the
objective assessment of its functional damage. Nippon Ganka Gakkai
Zasshi. 116:298–344. 2012.PubMed/NCBI
|
|
43
|
Sowden JC: Molecular and developmental
mechanisms of anterior segment dysgenesis. Eye. 21:1310–1318. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Kloss BA, Reis LM, Brémond-Gignac D,
Glaser T and Semina EV: Analysis of FOXD3 sequence variation in
human ocular disease. Mol Vis. 18:1740–1749. 2012.PubMed/NCBI
|
|
45
|
Lehmann OJ, Ebenezer ND, Jordan T, Fox M,
Ocaka L, Payne A, Leroy BP, Clark BJ, Hitchings RA, Povey S, et al:
Chromosomal duplication involving the forkhead transcription factor
gene FOXC1 causes iris hypoplasia and glaucoma. Am J Hum Genet.
67:1129–1135. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Chakrabarti S, Kaur K, Rao KN, Mandal AK,
Kaur I, Parikh RS and Thomas R: The transcription factor gene FOXC1
exhibits a limited role in primary congenital glaucoma. Invest
Ophthalmol Vis Sci. 50:75–83. 2009. View Article : Google Scholar : PubMed/NCBI
|