Introduction
Lung cancer has always been the most dominant cancer
subtype. According to the latest statistics, the incidence and
mortality of lung cancer have consistently occupied the top spot
either in the country or worldwide and far outpaces other types of
malignant tumors (1). Because
there is no pertinent early diagnosis or optimal tissue-based
molecular diagnostic procedure, lung cancer is usually in the
advanced stages when diagnosed (2). Additionally, non-small cell lung
cancer (NSCLC) accounts for 80% of the total number of lung cancer
cases, including adenocarcinoma (ADC), squamous cell carcinoma
(SCC) and large cell carcinoma (LCC) (3). Despite great efforts in elucidating
the occurrence, development and prognosis of NSCLC in recent years,
researchers still have not illuminated the potential tumorigenesis
mechanism of NSCLC to date. As a consequence, studies are required
to further develop a novel molecular-target diagnosis and therapy
targeting NSCLC.
MicroRNAs (miRNAs), which are 20–25 nt in length,
are a type of endogenous non-coding small RNA that regulates gene
expression at either the messenger RNA (mRNA) or protein level
during the post-transcription period (4,5).
miRNA can either inhibit the translation of mRNA or directly induce
mRNA degradation by binding to the 3′ untranslated regions (3′UTRs)
of mRNA (6); these processes can
promote or restrain the proliferation, transformation,
differentiation, apoptosis and necrosis of cells (6–8).
Evidence has shown that abnormal miRNA expression is relative to
the tumorigenesis process of several types of cancer (9), including NSCLC (10). miRNA has become the valuable
biomarkers in a variety of diseases (11), especially cancer. miRNA can be used
not only for NSCLC subtype analysis but also for monitoring the
prognosis and recurrence of early stage NSCLC by identifying a
specific sequence (12–16).
Previous studies had contrary conclusions that
miR-146a acted to promote tumor growth in papillary thyroid
carcinoma (17,18) but also exerted tumor suppressor
activities in malignancies located in the following organs: Breast
(19), prostate (20–22),
pancreas (23) and stomach
(24–28). Regarding NSCLC, Wang et al
reported that miR-146a had higher expression levels in NSCLC cells
when compared with normal lung cells (29). Meanwhile, our previous study
(30) concluded that miR-146a had
low expression in NSCLC, but miR-146a mimic could inhibit cell
proliferation and metastasis as well as induce apoptosis through
the EGFR signaling pathway, which is in accordance with another
published study (31). Therefore,
we intended to detect the clinical significance and function
mechanism of miR-146a-5p (abbreviated as miR-146a) in NSCLC.
In vivo animal experiment models have become
an important means of NSCLC studies. To date, the chick embryo
chorioallantoic membrane (CAM) model has become an efficient
experimental animal model for studying tumors because it is cheap,
convenient, fast, simple and sensitive. Because of its natural
immunodeficiency and abundant formation of new blood vessels and an
arteriovenous network, the CAM model is suitable for researching
the mechanisms of angiogenesis, invasion and metastasis (32–39).
In this study, we intended to investigate the effect
and molecular mechanism by which miR-146a-5p affects NSCLC using a
constructed miR-146a-5p-expressing H460 NSCLC cell line and
transplanting the transduced cells into the CAM of chick embryos.
By establishing a CAM xenograft tumor model, we simulated the
tumorigenesis process of NSCLC and observed the resulting
angiogenesis. In addition, the local invasive and necrosis
conditions of tumor were measured using hematoxylin and eosin
(H&E) staining. In addition, the prediction of target genes in
silico also implicated the possible functional location and
pathways of miR-146a-5p.
Materials and methods
Ethics statement
This research project was conducted with the
permission of the Research Ethics Committee of the First Affiliated
Hospital of Guangxi Medical University (Nanning, China).
Cell cultivation and selection
NSCLC cell lines provided by Yangjie Experiment
Center of Guangxi Medical University consisted of two different
histological types: The human lung large cell cancer H460, and the
lung adenocarcinoma cancer cell lines A549, PC9 and H1299. The
NSCLC cell lines were cultured in either RPMI-1640 medium (H460,
A549 and H1299) or Dulbecco's modified Eagle's medium (DMEM) PC9 at
37°C in a humidified environment containing 5% CO2.
Detection of the miR-146a-5p expression levels in these lung cancer
cell lines was performed to identify the cell line with the lowest
expression levels of miR-146a-5p; this cell line (H460) was set
aside for lentivirus transduction.
RNA extraction and qRT-PCR
Operative steps for determining miR-146a-5p
expression level were as follows. Total RNA was extracted using a
total RNA extraction kit (no. 9767; Takara Biotechnology Co., Ltd.,
Dailan, China) according to the manufacturer's instructions. After
determining the concentration and purity of the RNA by measuring
the absorbance at 260 and 280 nm, cDNA was generated by applying
Takara Mir-X™ miRNA First-Strand Synthesis. The reactions were
prepared using the SYBR® qRT-PCR User Manual. Primers
were designed by using Online Prime 3.0, and then Invitrogen
synthesized the primers.
Lentivirus transduction and
interference efficiency verification
hsa-miR-146a lentivirus (LV-hsa-miR-146a) and a
negative control lentivirus (LV-no load, LVCON238) were both
purchased from the Shanghai Jikai Gene Chemical Co., Ltd.
(Shanghai, China) and stored at −80°C. The target sequence of
miR-146a-5p (miRBase accession, MI0000477) was
TGAGAACTGAATTCCATGGGTT, and the vector was GV369, which contained
the component order Ubi-MCS-SV40-EGFP-IRES-puromycin. The sequence
of the negative control was TTCTCCGAACGTGTCACGT. During the
transduction process, the selected cell line H460 was divided into
three groups: Blank control group (untreated non-transduced H460
lung cancer cell line), the experimental group (LV-hsa-miR-146a
group) and the negative control group (LV-no load, LVCON238). Cell
morphology and fluorescence intensity were monitored under the
fluorescence microscope at 48 and 72 h after transduction,
respectively. Finally, total RNA was extracted from the cell lines,
and qRT-PCR was applied to quantify the relative miR-146a-5p
expression.
Chick embryo preparation
Fertilized chick embryos were purchased from Nanning
Liangfeng Agriculture and Farming Company Limited. After it was
sterilized in 75% alcohol, the chick embryo was placed in an
incubation box (which was also sterilized by 75% alcohol) at
37.6–38°C with 70–80% humidity. The chick embryos were incubated
until they were 8-day-old for experimental use.
CAM tumor xenograft assay
i) When the chick embryos were 7 days old, the
contours of the embryoid body, gas chamber and large blood vessels
attached to the chick chorioallantoic membrane on the egg shells
were sketched under the exposure of an egg tester on sterile
platform. ii) When the chick embryos were 8 day-old, they were
removed and sterilized with 75% alcohol. A 2-mm diameter hole was
drilled into the gas chamber side of every egg, and the embryo egg
membrane was pricked with a fine needle. Next, an approximately 2×1
cm window was ground out on the egg shell near the embryoid body
and large blood vessels to expose the white membrane. Afterwards,
air was repeatedly suctioned through the previously drilled hole
into the gas chamber to isolate the white membrane and chick
chorioallantoic membrane. When the white membrane was removed to
expose the vessels, a 5 mm silicon rubber ring was gently placed
above the vessel rich area on the CAM. Finally, the small hole in
the gas chamber was sealed up with sterile transparent materials.
The above procedure was conducted on a sterile platform. iii) A
cell suspension solution from each group was gently added to the
silicon rubber ring, and the window was sealed using sterile
transparent materials. After incubating for 24 h in the incubation
box, the silicon rubber ring was removed, and the model system was
then placed back into the incubation box to incubate for 120 h.
Throughout this duration, the tumorigenesis condition was monitored
and recorded every 24 h. iv) After 120 h, the chick embryo and
sterile transparent materials were discarded, and the xenograft
tumors were removed from the CAM. These tumors were photographed
and measured to record their size. v) The vessel ratio and window's
area were estimated by using Image-Pro Plus. vi) After embedding
the tumors with paraffin, the samples were sliced and stained with
HE-staining to observe the cellular morphology of tumor cell as
well as the metastasis conditions.
Identifying potential target genes in
silico and bioinformatics analysis
The predicted and validated microRNA gene targets
were obtained from miRWalk2.0 (http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/).
The twelve prediction databases were miRWalk, MicroT4, miRanda,
mirbridge, miRDB, miRMap, miRNAMap, PicTar2, PITA, RNA22, RNAhybrid
and TargetScan, and the three validated databases were miRTarBase,
TarBase and miRecords. For predicting the target genes, those ones
which were repeated in four or more databases were selected for the
next step. The overlapping hits of the selected predicted genes and
all the validated genes were entered into a gene-enrichment pathway
analysis. Function analysis of the potential target genes from the
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway annotations
and Gene Ontology (GO) terms were provided by Database for
Annotation, Visualization and Integrated Discovery 6.7 (DAVID 6.7,
https://david.ncifcrf.gov/) (40). The protein interaction analysis was
showed by protein-protein interaction (PPI) Networks from Search
Tool for the Retrieval of Interacting Genes/Proteins (STRING,
http://www.string-db.org/).
Statistical analysis
Experimental data were analyzed using SPSS 20.0
(SPSS, Inc., Chicago, IL, USA) statistical software using methods
such as the Independent Samples t-test, χ2 test and
Spearman's correlation coefficient analysis, and α=0.05 was
considered as the test threshold. When the two-sided P-values was
<0.05, the result was considered to indicate a statistically
significant difference.
Results
The expression levels of miR-146a-5p
in 4 cell lines
Total RNA extracted from the H460, PC9, 1299 and
A549 cell lines were analyzed for concentration and purity. H460
cells presented the lowest expression level of miR-146a-5p than the
other three cell lines; thus, H460 cells were selected to construct
the CAM xenograft tumor model. The relative expression levels of
miR-146a-5p in the 4 NSCLC cell lines were showed that H460 cells
had the lowest miR-146a-5p expression among the cell lines
tested.
Lentivirus transduction outcomes
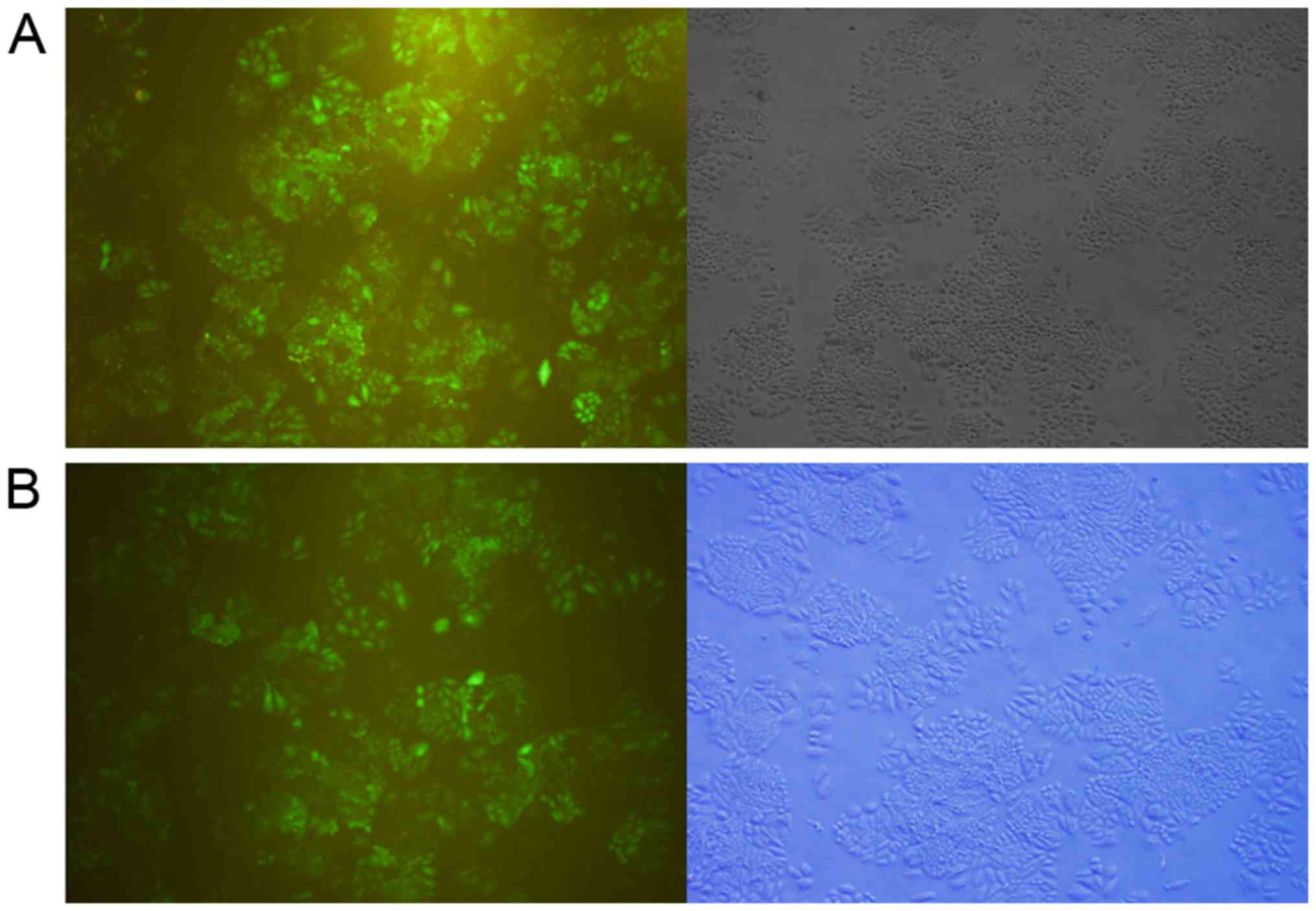
H460 cells were transduced with the optimal ratios
for either LV-hsa-miR-146a or LV-no load (Fig. 1). The miR-146a-5p expression levels
in H460 cells transduced with LV-hsa-miR-146a and LV-no load group
were detected by qRT-PCR. The results showed that the H460 cells
were successfully transduced with lentivirus and that the
miR-146a-5p in LV-hsa-miR-146a groups presented obviously
overexpression than the negative LV-no load group.
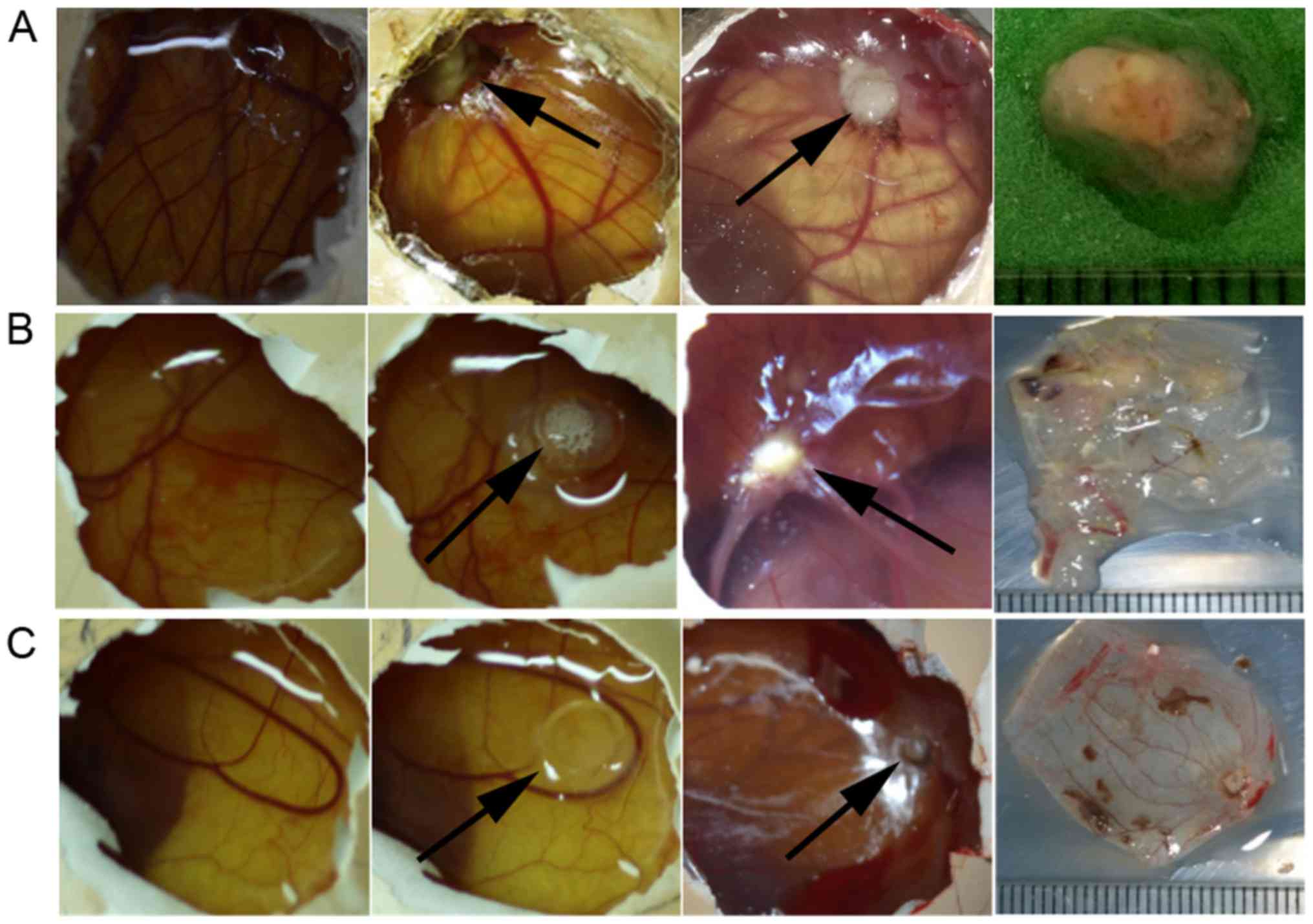
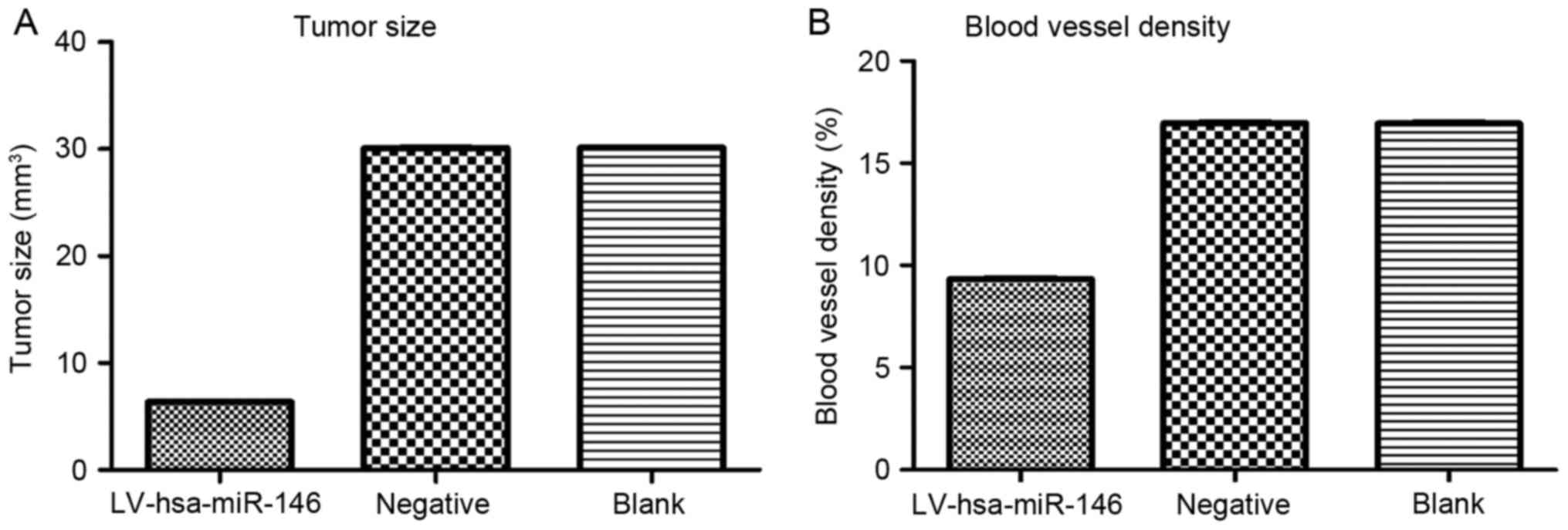
CAM xenograft tumor model
The results of the xenograft tumor sizes from day 1
(24 h after inoculation, or day 8 of chick embryo growth) to day 5
(120 h after inoculation, or day 12 of chick embryo growth) were
showed in Figs. 2 and 3. First, one-way analysis of variance
(ANOVA) was performed, but the results were not statistically
significant. Then, the independent sample t-test was performed by
comparing the sizes with those of the blank control group. The size
of the xenograft tumors in the experimental group (LV-hsa-miR-146a
group, V=6.340±0.066 mm3) were obviously reduced
compared with that in the blank control group (untreated H460 cell
line, V=30.13±0.06 mm3) (t=613.489, P<0.001), and the
tumor size of the negative control group (V=30.09±0.07
mm3) exhibited no statistical significance when compared
with the blank control group (untreated H460 cell line,
V=30.13±0.06 mm3) (t=1.312, P=0.260).
Angiogenesis of xenograft tumors
Based on the duration of the observed incubation
period and final angiogenesis data (Fig. 3), the independent samples t-test
was performed by comparing the results of the blank control group
with the non-significant result of the one-way ANOVA. The
appearance of the xenograft tumors on day 1 was an attached tumor
on the CAM with a few capillary vessels thriving and surrounding
the uneven surface of the xenograft tumor; by day 5, the
angiogenesis conditions of the experimental group (LV-hsa-miR-146a
group, 9.326±0.083) were largely reduced compared with those of the
blank control group (untreated H460 cells, 16.94±0.11) (t=121.207,
P<0.001), while growth situation of vessel showed no significant
difference between the negative control group (LV-no load group,
16.97±0.07) and the blank control group (untreated H460 cells,
16.94±0.11) (t=-0.612, P=0.573). Since tumor growth relies on the
generation of blood vessels in the chick CAM model, the
overexpression of miR-146a-5p in the experimental group inhibited
the angiogenesis of the xenograft tumors and thus restrained tumor
growth.
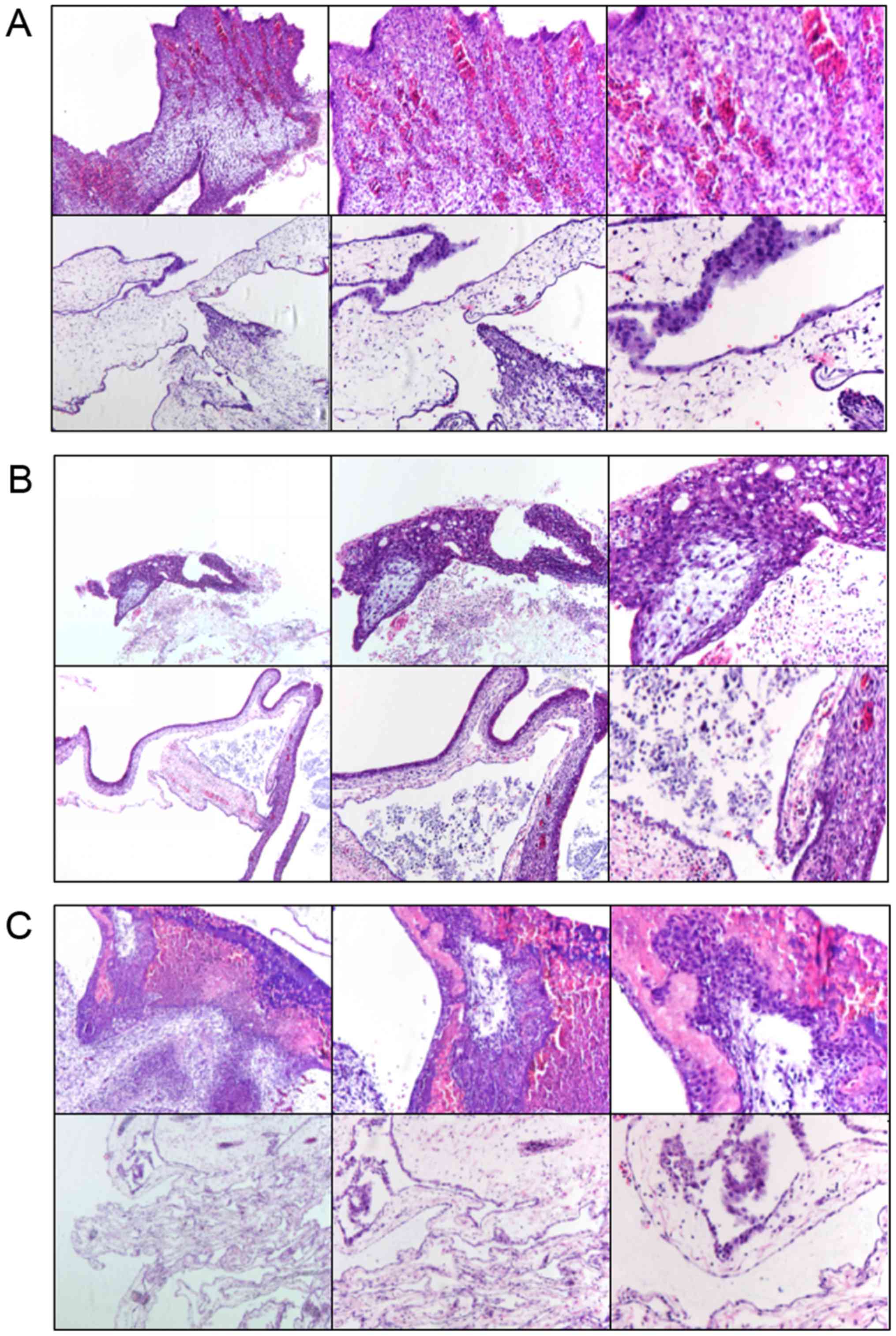
H&E staining
Tumor tissue was removed, paraffin-embedded and
subjected to HE-staining to observe the cell morphology (Fig. 4). Under a light microscope, all
three groups could form tumors on the CAM and showed preservation
of the primitive morphology of cancer, including obvious necrotic
areas and inflammatory cell infiltration. However, the inflammatory
condition of the experimental group (LV-hsa-miR-146a group) was
relatively inconspicuous compared with that of the blank control
group (untreated H460 cells).
Potential target genes of miR-146a-5p
based on bioinformatics analysis
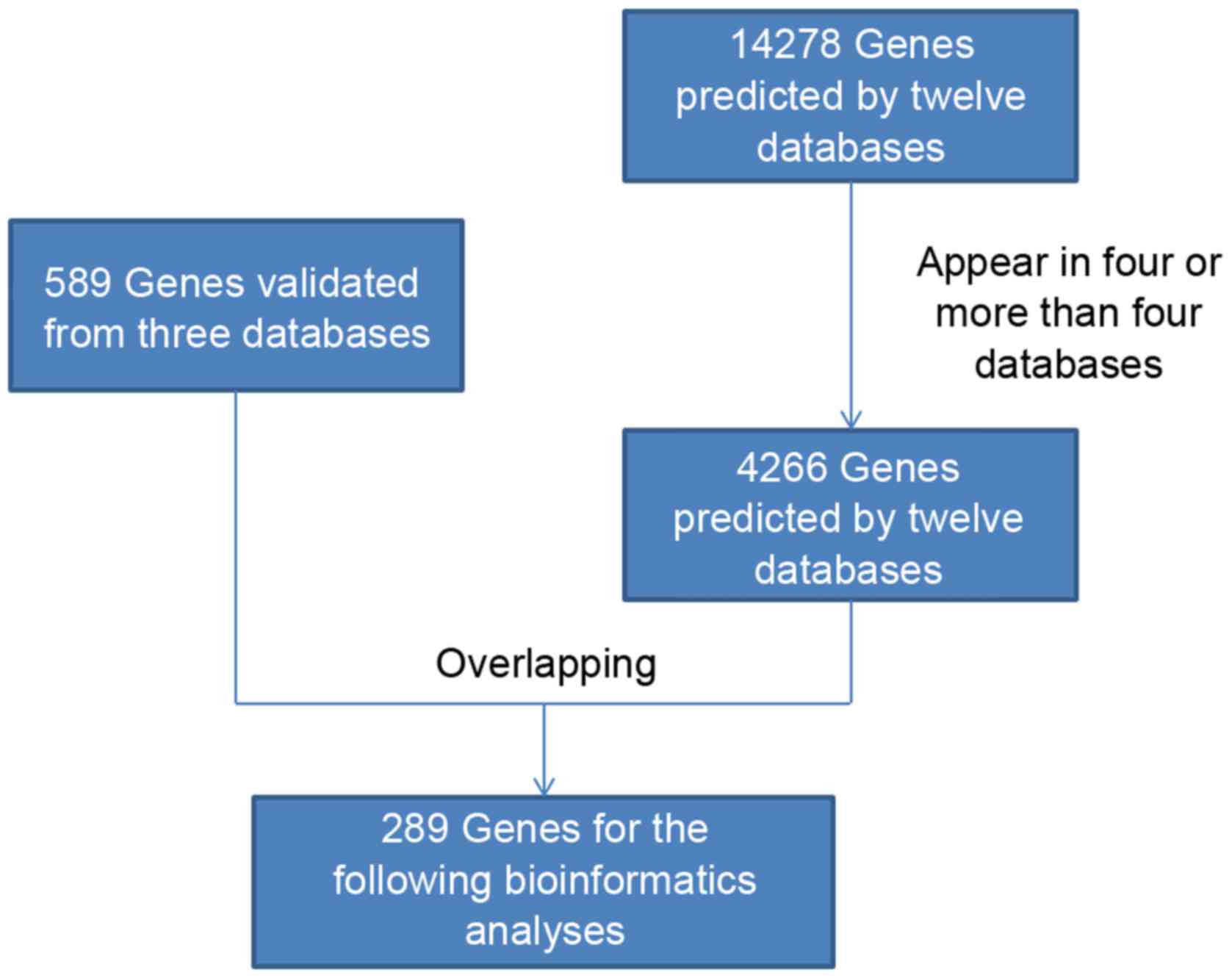
Twelve in silico interaction tools to predict miRNA
targets were used in this study. The databases were miRWalk,
MicroT4, miRanda, miRBridge, miRDB, miRMap, miRNAMap, PicTar2,
PITA, RNA22, RNAhybrid and TargetScan. In total, 14,278 target
mRNAs were listed as predicted genes. Eight genes were predicted by
ten databases in silico (FZD3, KLF7, STRBP, ZBTB2, IQGAP3, UPP2,
JAZF1 and IER5L), which provides strong evidence that these genes
are targets of miR-146a-5p. To improve the credibility of these
target genes, we obtained 4,266 intersecting elements that were
simultaneously predicted by at least four databases. Then, these
elements were overlapped with all 589 validated target genes of
miR-146a-5p using the databases miRTarBase, TarBase and miRecords
to finally obtain 289 target genes (Fig. 5; Table
I).
 | Table I.Potential target genes of
miR-146a-5p. |
Table I.
Potential target genes of
miR-146a-5p.
| IER5L | EIF4G2 | SMAD4 | STC1 | CCDC6 | MTA2 | PHF20L1 | LCOR |
| CCDC117 | THAP5 | ELAVL1 | IRAK1 | NFIX | PRKCE | TCF20 | BTG2 |
| SYNJ1 | AP3S2 | FNBP4 | FBXO3 | BRWD1 | C12orf4 | SRPRB | TRAK2 |
| KCTD15 | ATG9A | CDC73 | TMEM167A | ZNF367 | HIPK1 | ACTBL2 | PTGS2 |
| RAC1 | TIMELESS | RHOBTB3 | BRK1 | ITCH | ATP5G2 | SLC26A2 | EGFR |
| ERBB4 | NR6A1 | MYO6 | NF2 | ROBO1 | PPP1R11 | TSPYL1 | CASK |
| NUMB | SERTAD2 | MED13 | WASF2 | CARD8 | MKRN2 | FBXL3 | C16orf72 |
| CARD10 | FAM8A1 | TMEM214 | PLEKHG5 | EPB41L4A | NSD1 | DDHD1 | TSPAN14 |
| VANGL1 | NACC1 | ARL8A | SESN3 | SAMD9L | TMEM136 | CD80 | CD40LG |
| CDKN3 | EIF4EBP2 | CFH | MR1 | HSPA1A | IFIT3 | PMAIP1 | POU3F1 |
| PSMD3 | RBL1 | SFRP1 | SLC2A3 | STAT1 | SF1 | LSM4 | METTL7A |
| RUFY2 | STARD7 | RFX7 | LIMD2 | MOB1B | ZNF257 | EDARADD | WDR36 |
| PTAR1 | CCDC83 | EFNA5 | FANCF | IRF5 | LBR | MID1 | TLL1 |
| TRAF6 | BCL7B | HYOU1 | ARPP19 | ZHX1 | RAB18 | AVL9 | PDS5A |
| TNRC6A | BABAM1 | TULP4 | GOPC | ATP13A3 | EDEM3 | TMPRSS5 | SLC38A1 |
| STK40 | RHPN2 | TMEM67 | SESTD1 | EPSTI1 | NACC2 | HORMAD2 | OLFML2A |
| SKA2 | TMPPE | BRCA1 | BRCA2 | CD86 | GART | GPM6B | SMAD2 |
| MKLN1 | MVD | PPP2R4 | CCL5 | SDCBP | SLC1A5 | UMPS | CDS2 |
| CD84 | AKAP8 | CCR9 | AAK1 | ZNF629 | ZNF117 | ZDHHC13 | GIMAP4 |
| PBLD | C1orf21 | UTP15 | USP48 | ST6GAL2 | RAB2B | ZNF493 | ZNF260 |
| TRIM22 | IGF2BP1 | ERRFI1 | MFSD6 | RAB20 | APMAP | GATAD2B | RAPH1 |
| PARD6B | BACH1 | CALU | COPA | GNAI2 | HOXB8 | IREB2 | NF1 |
| NFE2L1 | RORA | SRPK1 | RAD54L | SQSTM1 | CPNE3 | DEDD | ZBTB22 |
| SLK | TXNIP | SERBP1 | RBM26 | PAPD5 | GRPEL1 | ISG20L2 | RASSF5 |
| TMEM101 | ITPRIPL2 | UBN2 | ATXN1L | C16orf52 | AKT2 | LY75 | POLR2E |
| TLR4 | TPD52 | ZNF264 | NUPL1 | BTN2A2 | COPS8 | SNRNP27 | CARHSP1 |
| STMN3 | AEN | SIKE1 | ATOH8 | SYT12 | VWCE | PLIN2 | HLA-C |
| LAMC2 | STIM1 | ULK1 | MSC | RAPGEF5 | MDFIC | PPHLN1 | CYBRD1 |
| DGCR6L | SLFN11 | PRR15 | PA2G4 | KLF9 | CAPN2 | JUN | TNPO1 |
| LNPEP | RPL11 | CCL8 | SOX4 | WASL | LATS1 | DDX21 | ZBTB33 |
| G3BP1 | SUPT16H | MDN1 | STX12 | IFIT5 | CLIC4 | KLHL20 | NDC1 |
| KIAA1432 | SHCBP1 | KLHL15 | CHMP4B | MTPN | DYNLL2 | CPNE8 | IFIT1 |
| LTB | FXR1 | KIAA0040 | ATP11B | PACS2 | UHRF1 | HM13 | MRPL10 |
| ALG10B | ADD1 | CCNA2 | ELK4 | MYLK | SERPINB9 | NAPG | PAPOLA |
| LMTK2 | CBX6 | SLC22A15 | TXLNG | KLHL42 | ACBD3 | DBF4B | KBTBD6 |
| USP54 | CNOT6L | ZNF410 | P2RX5 | RAG1 | CXCL12 | RER1 | ETNK1 |
| TET3 |
|
|
|
|
|
|
|
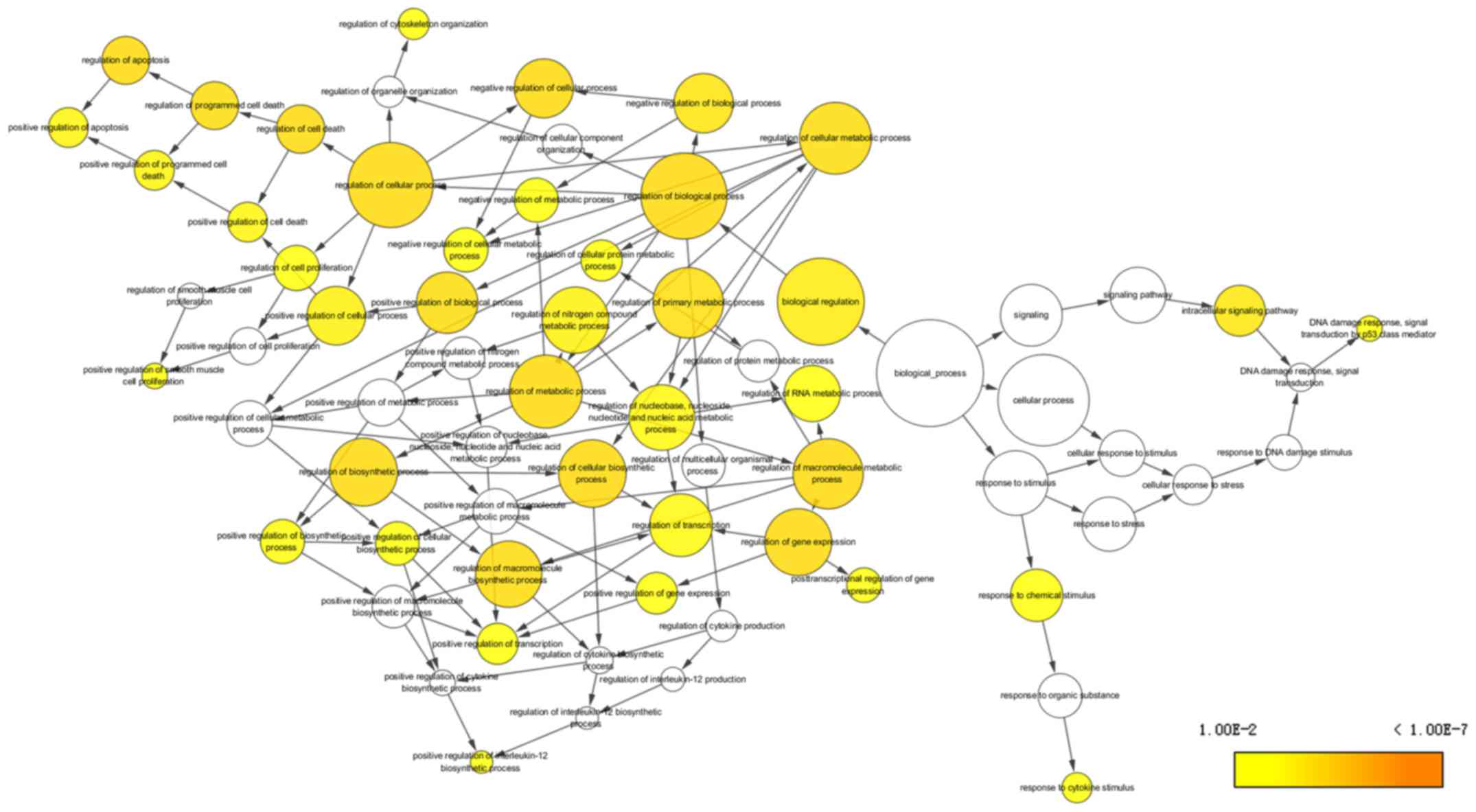
The top 10 most significant KEGG pathway annotations
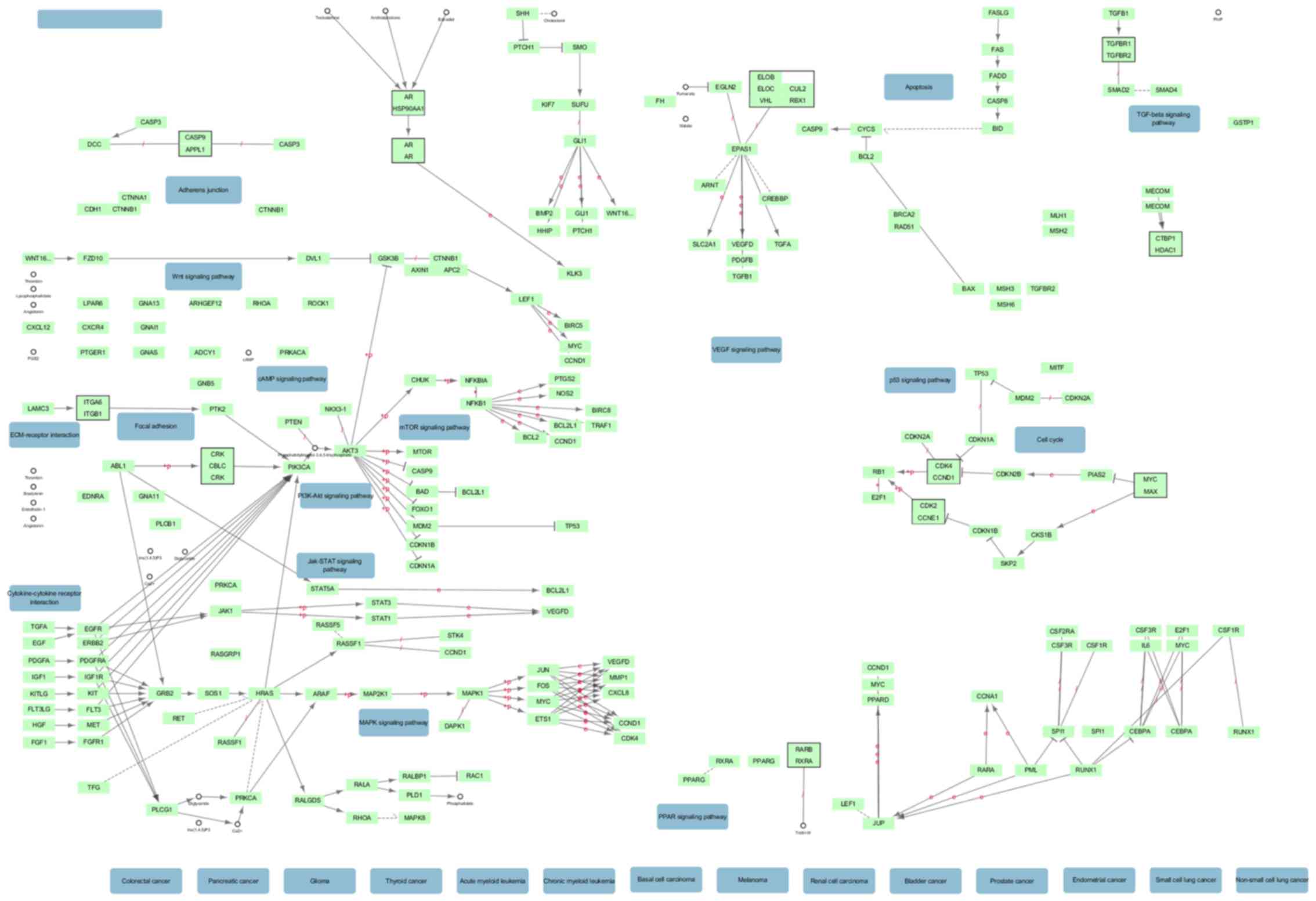
were showed in Table II. The most
significant KEGG pathway was involved in cancer. Moreover, the top
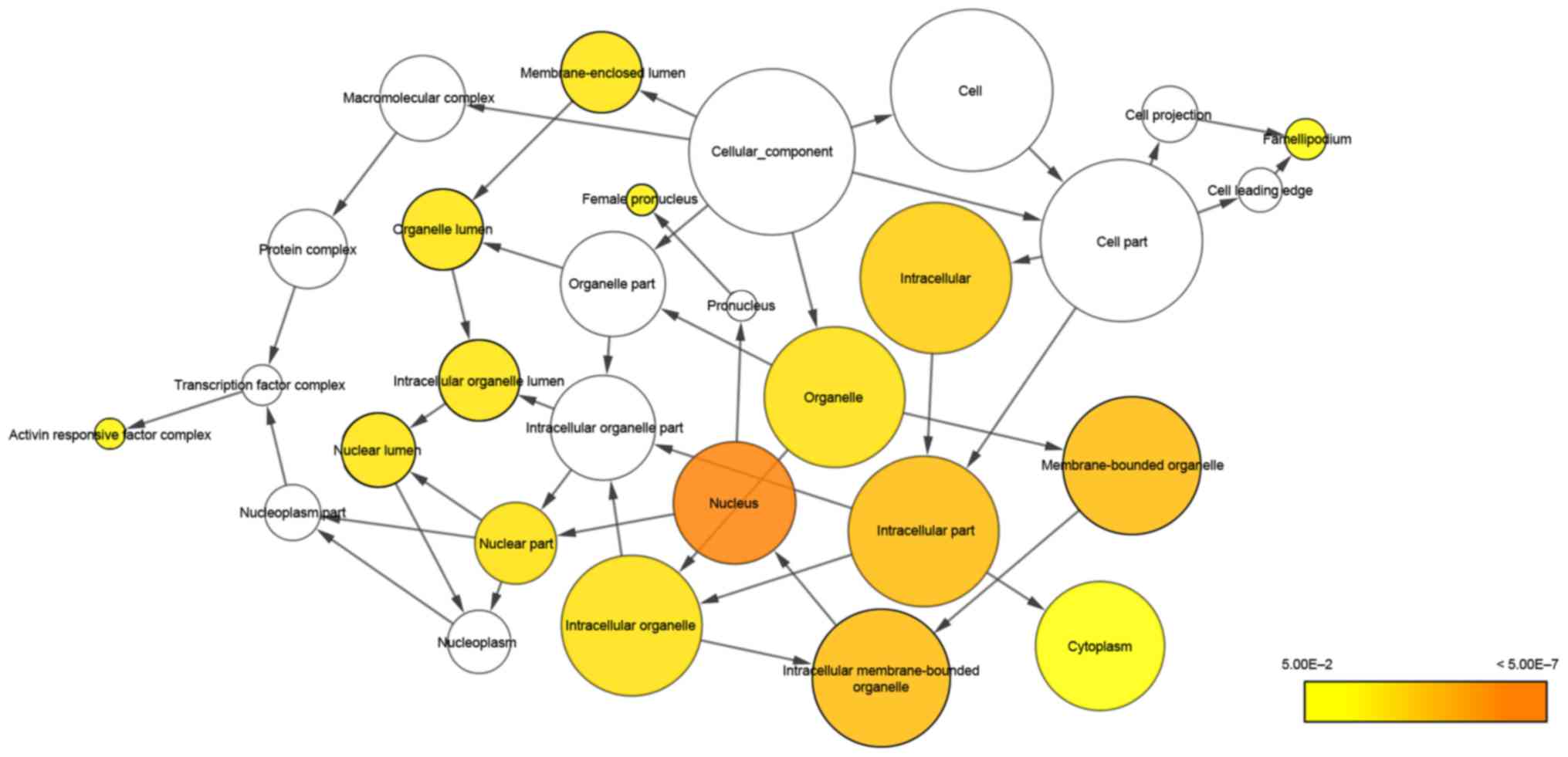
10 GO terms of biological process (BP), cellular component (CC) and
molecular function (MF) are listed in Table III, and the first terms of each
analysis were cellular process, cell part and binding,
respectively. The network of BP and CC are shown in Figs. 6 and 7. The most significant KEGG pathway
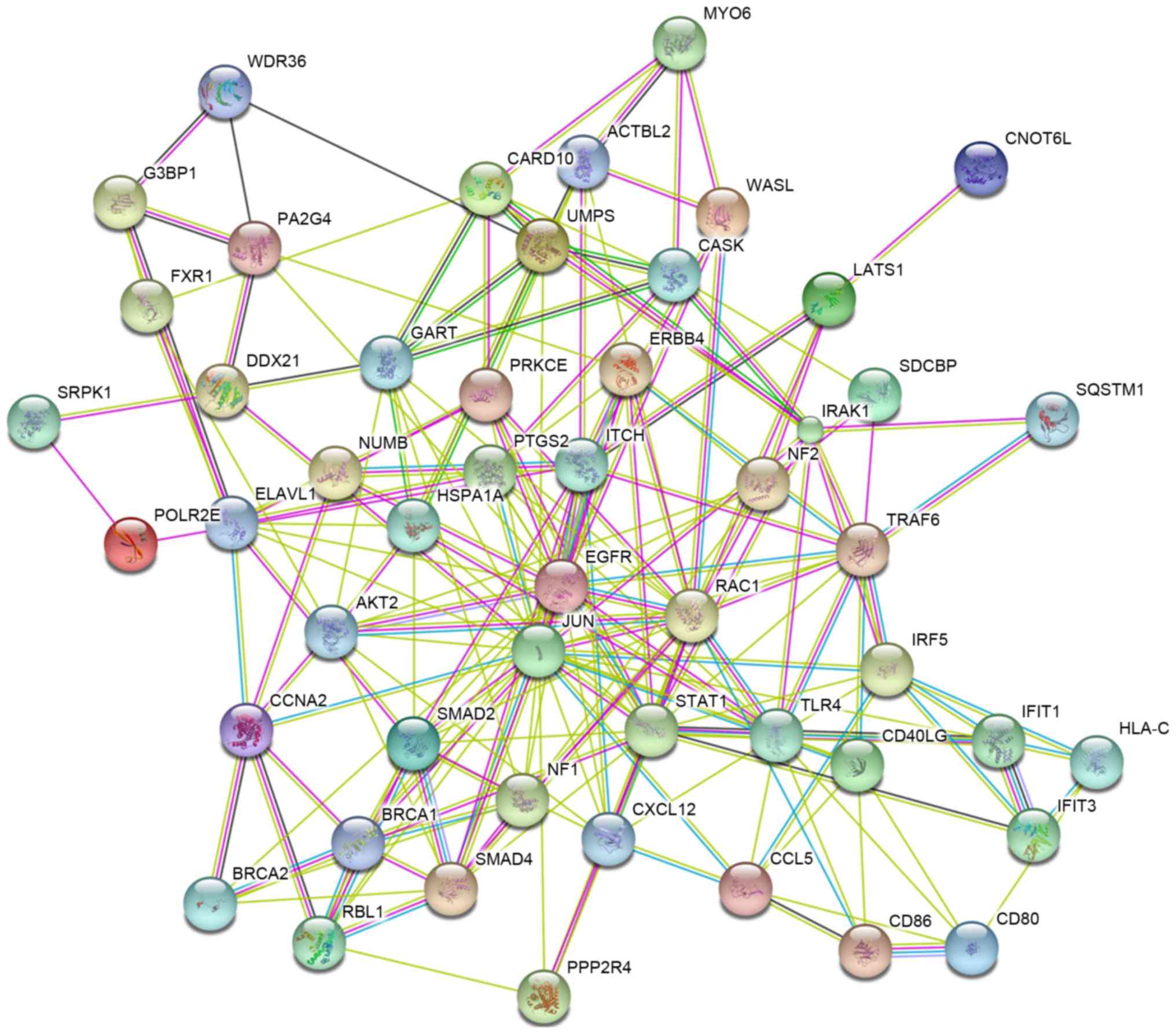
(hsa05200, pathways in cancer) is shown in Fig. 8. The PPI network included 50 hub
genes (Fig. 9). JUN, EGFR and RAC1
were the most relevant protein among the selected possible targets
of miR-146a-5p.
 | Table II.KEGG pathway enrichment analysis of
miR-146a-5p. |
Table II.
KEGG pathway enrichment analysis of
miR-146a-5p.
| KEGG term | Count (%) | P-value | Genes |
|---|
| hsa05200:Pathways
in cancer | 13 (4.6) | 0.003466 | EGFR, PTGS2, SMAD4,
BRCA2, SMAD2, STAT1, CCDC6, RASSF5, JUN, RAC1, LAMC2, TRAF6,
AKT2 |
| hsa04620:Toll-like
receptor signaling pathway | 11 (3.9) | 2.22E-06 | IRAK1, CD86, IRF5,
CD80, JUN, RAC1, TLR4, TRAF6, STAT1, CCL5, AKT2 |
| hsa04062:Chemokine
signaling pathway | 9
(3.2) | 0.006907 | CCR9, GNAI2, RAC1,
CCL8, WASL, STAT1, CCL5, CXCL12, AKT2 |
|
hsa04144:Endocytosis | 8
(2.8) | 0.02061 | EGFR, PARD6B,
ERBB4, CHMP4B, HLA-C, HSPA1A, ITCH, TRAF6 |
| hsa05212:Pancreatic
cancer | 7
(2.5) | 7.00E-04 | EGFR, RAC1, SMAD4,
BRCA2, SMAD2, STAT1, AKT2 |
| hsa04510:Focal
adhesion | 7
(2.5) | 0.082719 | EGFR, JUN, RAC1,
LAMC2, CAPN2, MYLK, AKT2 |
| hsa05416:Viral
myocarditis | 6
(2.1) | 0.004174 | EIF4G2, CD86, CD80,
CD40LG, RAC1, HLA-C |
| hsa04520:Adherens
junction | 6
(2.1) | 0.005904 | EGFR, WASF2, RAC1,
SMAD4, SMAD2, WASL |
| hsa05210:Colorectal
cancer | 6
(2.1) | 0.008501 | EGFR, JUN, RAC1,
SMAD4, SMAD2, AKT2 |
| hsa04310:Wnt
signaling pathway | 6
(2.1) | 0.077664 | SFRP1, VANGL1, JUN,
RAC1, SMAD4, SMAD2 |
| hsa04672:Intestinal
immune network for IgA production | 5
(1.8) | 0.006149 | CCR9, CD86, CD80,
CD40LG, CXCL12 |
| hsa04666:Fcγ
R-mediated phagocytosis | 5
(1.8) | 0.055033 | WASF2, RAC1, WASL,
PRKCE, AKT2 |
| hsa05330:Allograft
rejection | 4
(1.4) | 0.016657 | CD86, CD80, CD40LG,
HLA-C |
| hsa05320:Autoimmune
thyroid disease | 4
(1.4) | 0.041376 | CD86, CD80, CD40LG,
HLA-C |
| hsa04621:NOD-like
receptor signaling pathway | 4
(1.4) | 0.066868 | CARD8, CCL8, TRAF6,
CCL5 |
| hsa05120:Epithelial
cell signaling in Helicobacter pylori infection | 4
(1.4) | 0.083179 | EGFR, JUN, RAC1,
CCL5 |
 | Table III.Top 10 enrichment GO functional
annotations for related targets of miR-146a-5p. |
Table III.
Top 10 enrichment GO functional
annotations for related targets of miR-146a-5p.
| GO ID | GO term | Count (%) | P-value | Gene symbol |
|---|
| Biological
process |
|
|
|
|
|
GO:0009987 | cellular
process | 189 (67.0) | 8.64E-04 | GRPEL1, DBF4B,
PTGS2, SLC22A15, CASK, TLR4, PMAIP1, RORA |
|
GO:0065007 | biological
regulation | 142 (50.4) | 0.002032365 | PTGS2, CASK, TLR4,
PMAIP1, RORA, CXCL12, CBX6, EIF4EBP2 |
|
GO:0050789 | regulation of
biological process | 140 (49.6) | 3.29E-04 | PTGS2, CASK, TLR4,
PMAIP1, RORA, CXCL12, CBX6, EIF4EBP2 |
|
GO:0050794 | regulation of
cellular process | 136 (48.2) | 2.54E-04 | PTGS2, CASK, TLR4,
PMAIP1, RORA, CXCL12, CBX6, EIF4EBP2 |
|
GO:0008152 | metabolic
process | 134 (47.5) | 0.075818672 | GRPEL1, PTGS2,
CASK, RORA, CBX6, ACBD3, USP54, EIF4EBP2 |
|
GO:0044238 | primary metabolic
process | 125 (44.3) | 0.038487984 | GRPEL1, PTGS2,
CASK, RORA, CBX6, ACBD3, USP54, EIF4EBP2 |
|
GO:0044237 | cellular metabolic
process | 121 (42.9) | 0.032393623 | GRPEL1, PTGS2,
CASK, RORA, CBX6, USP54, EIF4EBP2, TRAK2 |
|
GO:0043170 | macromolecule
metabolic process | 111 (39.4) | 0.005972279 | GRPEL1, CASK, RORA,
CBX6, USP54, EIF4EBP2, TRAK2, MDFIC, AAK1 |
|
GO:0044260 | cellular
macromolecule metabolic process | 105 (37.2) | 0.002504917 | GRPEL1, CASK, RORA,
CBX6, USP54, EIF4EBP2, TRAK2, MDFIC |
|
GO:0019222 | regulation of
metabolic process | 95 (33.7) | 7.86E-08 | BACH1, ZBTB33,
DEDD, NR6A1, CASK, TLR4, RORA, LATS1 |
| Cellular
component |
|
|
|
|
|
GO:0044464 | cell part | 240 (85.1) | 0.00492047 | HM13, DBF4B, PTGS2,
TMPPE, RORA, TPD52, ACBD3, TRAK2 |
|
GO:0005623 | cell | 240 (85.1) | 0.004971296 | HM13, DBF4B, PTGS2,
TMPPE, RORA, TPD52, ACBD3, TRAK2 |
|
GO:0005622 | intracellular | 202 (71.6) | 4.68E-06 | HM13, DBF4B, PTGS2,
RORA, TPD52, ACBD3, TRAK2, RAPGEF5 |
|
GO:0044424 | intracellular
part | 200 (70.9) | 5.32E-07 | HM13, DBF4B, PTGS2,
RORA, TPD52, ACBD3, TRAK2, RAPGEF5 |
|
GO:0043229 | intracellular
organelle | 163 (57.8) | 0.001498556 | GRPEL1, HM13,
DBF4B, PTGS2, CHMP4B, CASK, PMAIP1, RORA |
|
GO:0043226 | organelle | 163 (57.8) | 0.001618783 | GRPEL1, HM13,
DBF4B, PTGS2, CHMP4B, CASK, PMAIP1, RORA |
|
GO:0043231 | intracellular
membrane-bounded organelle | 153 (54.3) | 1.46E-04 | GRPEL1, DBF4B,
HM13, PTGS2, CHMP4B, CASK, PMAIP1, RORA |
|
GO:0043227 | membrane-bounded
organelle | 153 (54.3) | 1.54E-04 | GRPEL1, DBF4B,
HM13, PTGS2, CHMP4B, CASK, PMAIP1, RORA |
|
GO:0005737 | cytoplasm | 136 (48.2) | 0.003037975 | GRPEL1, HM13,
PTGS2, CHMP4B, CASK, TLR4, PMAIP1, TPD52 |
|
GO:0005634 | nucleus | 117 (41.5) | 3.70E-07 | DBF4B, PTGS2, CASK,
RORA, CBX6, TRAK2, MDFIC, LSM4 |
| Molecular
function |
|
|
|
|
|
GO:0005488 | binding | 223 (79.1) | 8.09E-06 | HM13, DBF4B, PTGS2,
TMPPE, RORA, TPD52, ACBD3, USP54 |
|
GO:0005515 | protein
binding | 169 (59.9) | 1.90E-07 | GRPEL1, HM13,
PTGS2, CASK, TLR4, PMAIP1, RORA, TPD52 |
|
GO:0003676 | nucleic acid
binding | 72
(25.5) | 0.002025204 | BACH1, ZBTB33,
DBF4B, DEDD, SYNJ1, NR6A1, RORA, MKRN2 |
|
GO:0003677 | DNA binding | 52
(18.4) | 0.009158415 | BACH1, ZBTB33,
DEDD, NR6A1, RORA, SLK, ATOH8, LBR |
|
GO:0000166 | nucleotide
binding | 46
(16.3) | 0.054477028 | ACTBL2, GRPEL1,
ERBB4, GNAI2, MVD, IGF2BP1, CASK, HSPA1A |
|
GO:0030528 | transcription
regulator activity | 42
(14.9) | 4.31E-04 | BACH1, NR6A1,
ZNF367, SOX4, NFIX, RORA, TCF20, ELK4 |
|
GO:0017076 | purine nucleotide
binding | 42
(14.9) | 0.027572564 | ACTBL2, GRPEL1,
ERBB4, GNAI2, MVD, CASK, HSPA1A, LATS1 |
|
GO:0032555 | purine
ribonucleotide binding | 41
(14.5) | 0.022470925 | ACTBL2, ERBB4,
GNAI2, MVD, CASK, HSPA1A, LATS1, STK40 |
|
GO:0032553 | ribonucleotide
binding | 41
(14.5) | 0.022470925 | ACTBL2, ERBB4,
GNAI2, MVD, CASK, HSPA1A, LATS1, STK40 |
|
GO:0030554 | adenyl nucleotide
binding | 35
(12.4) | 0.0392542 | ACTBL2, GRPEL1,
ERBB4, MVD, CASK, HSPA1A, LATS1, STK40 |
Discussion
Lung cancer has been consistently regarded as the
most aggressive carcinoma worldwide and accounts for a large
percentage of cancer-related deaths (41). Although there is substantial
amelioration in the application of chemical and molecular-targeted
treatments, the prognosis of patients with lung carcinoma is still
embarrassing (42). Since the
consensus behaviors of cancer cells-invasion and metastasis-are the
major hurdles for the clinical treatment of NSCLC, more attention
should be focused on the verification of small molecules that can
suppress the invasion, metastasis and angiogenesis of NSCLC.
Recently, evidence demonstrated that miRNAs can be
used as diagnostic and prognostic biomarkers of leukemia, lung
cancer and colon cancer (43).
miRNAs may also be new therapeutic agents for antitumor therapies
in humans (44). One study found
that miRNAs can regulate the signal transduction of the EGFR
signaling pathway in a wide variety of tumor cells (45), including lung cancer (46). Identifying the molecules within the
EGFR signaling pathway that are targeted by miRNAs may be a
potential therapeutic approach for treating lung cancer.
Previously, we found that low expression of miR-146a-5p in NSCLC
cells inhibited cell proliferation and metastasis as well as
induced apoptosis through the EGFR signaling pathway by using
functional experiments (30);
these results were concomitant with another study conducted by Li
et al (31). In contrast to
other studies on miR-146a-5p expression in NSCLC, this study
verified the reliability of our previous experimental study in
vivo to identify differences of biological growth properties
and molecular variations among the CAM xenograft tumors comprising
blank control, negative control or experimental cells.
Our previous study identified EGFR as a downstream
regulatory target of miR-146a at both the mRNA and protein level
(30). The high expression of
miR-146a inhibited the proliferation of NSCLC cells by
downregulating the expression of EGFR. EGFR is a type of
transmembrane glycoprotein receptor that functions as a tyrosine
kinase (TK) (47). Its
conformational changes can cause receptor polymerization and induce
the activation of the intracellular TK subregion to activate
multiple signaling pathways, including the PLC-γ/PKCPI-3K/AKT,
RAS-RAF-MEK-MAPK and STAT/NF-κB A pathways (47,48).
In different cells or at various differentiation stages, the EGFR
configuration changes to activate different signaling pathways, and
the cells react to the activation or inhibition of a series of
downstream molecules in the signaling pathway (49). This different signaling is in
accordance with many other malignant tumors, such as gliomas and
prostate cancer (50,51). Moreover, EGFR mutations were found
in cancerous and adjacent tissues of 10–40% of the lung cancer
patients, among which 30% were Asian female non-smokers diagnosed
with lung adenocarcinoma (52).
miR-146a-5p was showed to be an important regulatory factor in
tumor formation mediated by EGFR, which provided a new theoretical
basis for the treatment of patients with lung cancer. Monoclonal
antibody D2-40, a specific biomarker of lymphatic epithelial cells,
can show via immunohistochemical staining the profiles of small
lymphatic vessels stretching from the alveolar space to the small
blood vessels in the lung lobules. As a result, this technique can
be applied to the identification of lymphatic vessel tumor emboli
(53,54).
In this study, we conducted a target mRNA prediction
of miR-146a-5p using in silico methods. When combining the results
of twelve prediction-based databases, we obtained theoretical
target genes of miR-146a-5p. However, this artificial prediction
could have limitations in this single computational algorithm, and
this process still needs further experimental verification. Based
on the predicted genes, KEGG pathways and GO enrichment analysis
were conducted. Pathways involved in cancer and toll-like receptor
signaling were the most two significant pathway groups of the
target genes. In addition, the protein localization of the mRNAs
was enriched in the cell membrane and functioned in cellular
processes. For molecular function, the predicted genes were
associated with binding. Then, we could predict that miR-146a-5p
may participate in tumor-related intercellular protein or nucleic
acid binding signaling behavior. We plan to conduct further
experimental research on miR-146a-5p regarding the development of
tumorigenesis. For the PPI network, network nodes represent
proteins, and edges represent protein-protein associations. We
obtained 50 hub genes to input into the PPI and found that JUN,
EGFR and RAC1 were the most relevant protein names among the
selected possible targets of miR-146a-5p.
miR-146a-5p was overexpressed in an NSCLC cell line
and could inhibit the tumorigenesis and angiogenesis in a CAM
xenograft tumor model. The in silico analysis revealed that its
target genes are found in pathways related to cancer. miR-146a-5p
is a potential tumor suppressor gene in NSCLC. The carcinogenic
mechanism and its prognostic value in lung cancer needs further
research.
Acknowledgements
The study was supported by the funds of the National
Natural Science Foundation of China (NSFC81360327, NSFC81560469),
the Natural Science Foundation of Guangxi, China
(2015GXNSFCA139009), Innovation Project of Guangxi Graduate
Education (YCSZ2015106) and the Guangxi Zhuang Autonomous Region
University Student Innovative Plan (No. 201610598003). The funders
had no role in the study design, the data collection and analysis,
the decision to publish, or the preparation of the manuscript.
Wen-Ting Huang and Wei-Luan Cen contributed equally as co-first
authors, and Xiao-Hua Hu and Gang Chen contributed equally as
co-corresponding authors of this study.
References
|
1
|
Global Burden of Disease Cancer
Collaboration, ; Fitzmaurice C, Dicker D, Pain A, Hamavid H,
Moradi-Lakeh M, MacIntyre MF, Allen C, Hansen G, Woodbrook R, et
al: The global burden of cancer 2013. JAMA Oncol. 1:505–527. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Dietel M, Bubendorf L, Dingemans AM, Dooms
C, Elmberger G, García RC, Kerr KM, Lim E, López-Ríos F, Thunnissen
E, et al: Diagnostic procedures for non-small-cell lung cancer
(NSCLC): Recommendations of the European Expert Group. Thorax.
71:177–184. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Travis WD, Brambilla E, Müller-Hermelink
HK and Harris CC: World Health Classification of Tumours. Pathology
and Genetics of Tumours of the Lung, Pleura, Thymus and Heart. IARC
Press/Oxford University Press/Oxford University Press.
(distributor). Lyon: 2004
|
|
4
|
Ul Hussain M: Micro-RNAs (miRNAs): Genomic
organisation, biogenesis and mode of action. Cell Tissue Res.
349:405–413. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang B, Pan X, Cobb GP and Anderson TA:
microRNAs as oncogenes and tumor suppressors. Dev Biol. 302:1–12.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Baehrecke EH: miRNAs: Micro managers of
programmed cell death. Curr Biol. 13:R473–R475. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Michael MZ, O' Connor SM, van Holst
Pellekaan NG, Young GP and James RJ: Reduced accumulation of
specific mircoRNAs in colorectal neoplasia. Mol Cancer Res.
1:882–891. 2003.PubMed/NCBI
|
|
10
|
Croce CM: Causes and consequences of
microRNA dysregulation in cancer. Nat Rev Genet. 10:704–714. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Weber JA, Baxter DH, Zhang S, Huang DY,
Huang KH, Lee MJ, Galas DJ and Wang K: The microRNA spectrum in 12
body fluids. Clin Chem. 56:1733–1741. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bishop JA, Benjamin H, Cholakh H, Chajut
A, Clark DP and Westra WH: Accurate classification of non-small
cell lung carcinoma using a novel microRNA-based approach. Clin
Cancer Res. 16:610–619. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yanaihara N, Caplen N, Bowman E, Seike M,
Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, et
al: Unique microRNA molecular profiles in lung cancer diagnosis and
prognosis. Cancer Cell. 9:189–198. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yu SL, Chen HY, Chang GC, Chen CY, Chen
HW, Singh S, Cheng CL, Yu CJ, Lee YC, Chen HS, et al: MicroRNA
signature predicts survival and relapse in lung cancer. Cancer
Cell. 13:48–57. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Raponi M, Dossey L, Jatkoe T, Wu X, Chen
G, Fan H and Beer DG: MicroRNA classifiers for predicting prognosis
of squamous cell lung cancer. Cancer Res. 69:5776–5783. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Patnaik SK, Kannisto E, Knudsen S and
Yendamuri S: Evaluation of microRNA expression profiles that may
predict recurrence of localized stage I non-small cell lung cancer
after surgical resection. Cancer Res. 70:36–45. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jazdzewski K, Boguslawska J, Jendrzejewski
J, Liyanarachchi S, Pachucki J, Wardyn KA, Nauman A and de la
Chapelle A: Thyroid hormone receptor beta (THRB) is a major target
gene for microRNAs deregulated in papillary thyroid carcinoma
(PTC). J Clin Endocrinol Metab. 96:E546–E553. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sun M, Fang S, Li W, Li C, Wang L, Wang F
and Wang Y: Associations of miR-146a and miR-146b expression and
clinical characteristics in papillary thyroid carcinoma. Cancer
Biomark. 15:33–40. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zheng T, Chou J, Zhang F, Liu Y, Ni H, Li
X, Zheng L, Tang T, Jin L and Xi T: CXCR4 3′UTR functions as a
ceRNA in promoting metastasis, proliferation and survival of MCF-7
cells by regulating miR-146a activity. Eur J Cell Biol. 94:458–469.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xu B, Wang N, Wang X, Tong N, Shao N, Tao
J, Li P, Niu X, Feng N, Zhang L, et al: miR-146a suppresses tumor
growth and progression by targeting EGFR pathway and in a
p-ERK-dependent manner in castration-resistant prostate cancer.
Prostate. 72:1171–1178. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sun Q, Zhao X, Liu X, Wang Y, Huang J,
Jiang B, Chen Q and Yu J: miR-146a functions as a tumor suppressor
in prostate cancer by targeting Rac1. Prostate. 74:1613–1621. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Xu B, Huang Y, Niu X, Tao T, Jiang L, Tong
N, Chen S, Liu N, Zhu W and Chen M: hsa-miR-146a-5p modulates
androgen-independent prostate cancer cells apoptosis by targeting
ROCK1. Prostate. 75:1896–1903. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li Y, Vandenboom TG II, Wang Z, Kong D,
Ali S, Philip PA and Sarkar FH: miR-146a suppresses invasion of
pancreatic cancer cells. Cancer Res. 70:1486–1495. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yao Q, Cao Z, Tu C, Zhao Y, Liu H and
Zhang S: MicroRNA-146a acts as a metastasis suppressor in gastric
cancer by targeting WASF2. Cancer Lett. 335:219–224. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kogo R, Mimori K, Tanaka F, Komune S and
Mori M: Clinical significance of miR-146a in gastric cancer cases.
Clin Cancer Res. 17:4277–484. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hou Z, Yin H, Chen C, Dai X, Li X, Liu B
and Fang X: microRNA-146a targets the L1 cell adhesion molecule and
suppresses the metastatic potential of gastric cancer. Mol Med Rep.
6:501–506. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sha M, Ye J, Zhang LX, Luan ZY and Chen
YB: Celastrol induces apoptosis of gastric cancer cells by miR-146a
inhibition of NF-κB activity. Cancer Cell Int. 13:502013.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhou L, Zhao X, Han Y, Lu Y, Shang Y, Liu
C, Li T, Jin Z, Fan D and Wu K: Regulation of UHRF1 by miR-146a/b
modulates gastric cancer invasion and metastasis. FASEB J.
27:4929–4939. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang RJ, Zheng YH, Wang P and Zhang JZ:
Serum miR-125a-5p, miR-145 and miR-146a as diagnostic biomarkers in
non-small cell lung cancer. Int J Clin Exp Pathol. 8:765–771.
2015.PubMed/NCBI
|
|
30
|
Chen G, Umelo IA, Lv S, Teugels E, Fostier
K, Kronenberger P, Dewaele A, Sadones J, Geers C and De Grève J:
miR-146a inhibits cell growth, cell migration and induces apoptosis
in non-small cell lung cancer cells. PLoS One. 8:e603172013.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li YL, Wang J, Zhang CY, Shen YQ, Wang HM,
Ding L, Gu YC, Lou JT, Zhao XT, Ma ZL and Jin YX: MiR-146a-5p
inhibits cell proliferation and cell cycle progression in NSCLC
cell lines by targeting CCND1 and CCND2. Oncotarget. 7:59287–59298.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Mira E, Lacalle RA, Gómez-Moutón C,
Leonardo E and Mañes S: Quantitative determination of tumor cell
intravasation in a real-time polymerase chain reaction-based assay.
Clin Exp Metastasis. 19:313–318. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Busch C, Krochmann J and Drews U: The
chick embryo as an experimental system for melanoma cell invasion.
PLoS One. 8:e539702013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kunzi-Rapp K, Genze F, Küfer R, Reich E,
Hautmann RE and Gschwend JE: Chorioallantoic membrane assay:
Vascularized 3-dimensional cell culture system for human prostate
cancer cells as an animal substitute model. J Urol. 166:1502–1507.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liu M, Scanlon CS, Banerjee R, Russo N,
Inglehart RC, Willis AL, Weiss SJ and D'Silva NJ: The histone
methyltransferase EZH2 mediates tumor progression on the chick
chorioallantoic membrane assay, a novel model of head and neck
squamous cell carcinoma. Transl Oncol. 6:273–281. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ribatti D, Vacca A, Roncali L and Dammacco
F: The chick embryo chorioallantoic membrane as a model for in vivo
research on angiogenesis. Int J Dev Biol. 40:1189–1197.
1996.PubMed/NCBI
|
|
37
|
Lokman NA, Elder AS, Ricciardelli C and
Oehler MK: Chick chorioallantoic membrane (CAM) assay as an in vivo
model to study the effect of newly identified molecules on ovarian
cancer invasion and metastasis. Int J Mol Sci. 13:9959–9970. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lester RD, Jo M, Montel V, Takimoto S and
Gonias SL: uPAR induces epithelial-mesenchymal transition in
hypoxic breast cancer cells. J Cell Biol. 178:425–436. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Chen PY and Long QC: Effects of
cyclooxygenase 2 inhibitors on biological traits of nasopharyngeal
carcinoma cells. Acta Pharmacol Sin. 25:943–949. 2004.PubMed/NCBI
|
|
40
|
da W Huang, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009.PubMed/NCBI
|
|
41
|
McGuire S: World Cancer Report 2014.
Geneva, Switzerland: World Health Organization, International
Agency for Research on Cancer, WHO Press, 2015. Adv Nutr.
7:418–419. 2016.doi: 10.3945/an.116.012211.
|
|
42
|
Beasley MB, Dembitzer FR and Flores RM:
Surgical pathology of early stage non-small cell lung carcinoma.
Ann Transl Med. 4:2382016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Di Leva G and Croce CM: Roles of small
RNAs in tumor formation. Trends Mol Med. 16:257–267. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Weidhaas JB, Babar I, Nallur SM, Trang P,
Roush S, Boehm M, Gillespie E and Slack FJ: MicroRNAs as potential
agents to alter resistance to cytotoxic anticancer therapy. Cancer
Res. 67:11111–11116. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Webster RJ, Giles KM, Price KJ, Zhang PM,
Mattick JS and Leedman PJ: Regulation of epidermal growth factor
receptor signaling in human cancer cells by microRNA-7. J Biol
Chem. 284:5731–5741. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Yamaguchi G, Takanashi M, Tanaka M, Fujita
K, Ohira T, Kuroda M and Ikeda N: Isolation of miRNAs that target
EGFR mRNA in human lung cancer. Biochem Biophys Res Commun.
420:411–426. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Jost M, Kari C and Rodeck U: The EGF
receptor - an essential regulator of multiple epidermal functions.
Eur J Dermatol. 10:505–510. 2000.PubMed/NCBI
|
|
48
|
Ullrich A, Coussens L, Hayflick JS, Dull
TJ, Gray A, Tam AW, Lee J, Yarden Y, Libermann TA, Schlessinger J,
et al: Human epidermal growth factor receptor cDNA sequence and
aberrant expression of the amplified gene in A431 epidermoid
carcinoma cells. Nature. 309:418–425. 1984. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Quadros MR, Peruzzi F, Kari C and Rodeck
U: Complex regulation of signal transducers and activators of
transcription 3 activation in normal and malignant keratinocytes.
Cancer Res. 64:3934–3939. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Rajan P, Elliott DJ, Robson CN and Leung
HY: Alternative splicing and biological heterogeneity in prostate
cancer. Nat Rev Urol. 6:454–460. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Nicholas MK, Lukas RV, Jafri NF, Faoro L
and Salgia R: Epidermal growth factor receptor - mediated signal
transduction in the development and therapy of gliomas. Clin Cancer
Res. 12:7261–7270. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Shigematsu H and Gazdar AF: Somatic
mutations of epidermal growth factor receptor signaling pathway in
lung cancers. Int J Cancer. 118:257–262. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
van den Eynden GG, Van der Auwera I, Van
Laere SJ, Colpaert CG, van Dam P, Dirix LY, Vermeulen PB and Van
Marck EA: Distinguishing blood and lymph vessel invasion in breast
cancer: A prospective immunohistochemical study. Br J Cancer.
94:1643–1649. 2006.PubMed/NCBI
|
|
54
|
Kambouchner M and Bernaudin JF:
Intralobular pulmonary lymphatic distribution in normal human lung
using D2-40 antipodoplanin immunostaining. J Histochem Cytochem.
57:643–648. 2009. View Article : Google Scholar : PubMed/NCBI
|