Introduction
Intervertebral disc degeneration (IDD) is a common
chronic, age-related degenerative musculoskeletal disorder with a
deleterious medical and social effect (1). Increasing evidence reveals that IDD
incidence has been growing not only in elderly people but also in
younger adults in the past decade (2). Genetic factors are the main
contributing factor to IDD. Several studies demonstrate that single
nucleotide polymorphisms, coding genes and noncoding RNAs serve
important roles in IDD (3–7).
With the development of whole-genome sequencing
studies, 98% of the human genome is now known to be transcribed
into non-coding RNAs (ncRNAs) (8,9).
NcRNAs can be divided into two subgroups in terms of their genomic
function: Housekeeping and regulatory ncRNAs. Housekeeping ncRNAs
include ribosomal RNA, transfer RNA, small nuclear RNAs and small
nucleolar RNAs (10). Regulatory
ncRNAs are composed of two groups based on the length of their
transcripts. Small ncRNAs >200 nucleotides include microRNAs
(miRNAs), small-interfering RNAs and Piwi-interacting RNAs
(11). The other category is long
non-coding RNAs (lncRNAs) (12,13).
Circular RNAs (circRNAs) are a class of lncRNAs with covalently
linked ends. Although there are not many studies on circRNAs, they
have emerged as critical molecules in post-transcriptional
regulation (14,15). CircRNAs can interact with miRNAs or
proteins functioning as molecular sponges. In addition, CircRNAs
can affect their host genes via cis- or trans-action (16). Recently, a study demonstrated that
circRNAs can interact with polymerase (Pol) II machinery as
positive regulators to regulate Pol II transcription (17–19).
Overall, the interplay between circRNAs and the transcriptional
machinery provides novel insights into strategies for regulating
gene expression.
More recently, Lan et al (20) detected a combination of mRNA,
miRNA, circRNA and lncRNA in the human lumbar disc using microarray
analysis. This is the first discovery of circRNAs in the human
lumbar disc by high-throughput methods. However, the potential
roles of circRNA molecules in IDD remain unclear. To answer this
question, circRNAs were classified by length in the present study
and it was identified that they distributed uniformly in the human
intervertebral disc. Subsequently, a functional enrichment analysis
was performed, and confirmed tissue specific expression of the
circRNAs host genes in IDD. Interactions of 76 pairs of circRNAs
and their host genes were also demonstrated with potential to be
used in the diagnosis and clinical treatment of IDD in the
future.
Materials and methods
Human nucleus pulposus (NP) sample
collection
Human lumbar NP specimens used as normal controls
were collected from 10 patients (4 women and 6 men; mean age, 24.5
years; range, 20–36 years) that had idiopathic scoliosis and were
undergoing deformity correction surgery. The degenerative human
lumbar NP specimens were collected from 15 patients (5 women and 10
men; mean age, 37.2 years; range, 29–62 years) that had IDD and
were undergoing disc excision and spinal fusion surgery. The degree
of IDD was assessed according to the modified Pfirrmann grading
system using preoperative MRI scans (21). The lumbar discs of all patients
with IDD were classified as Grade III–V, and the discs of those
with idiopathic scoliosis were classified as Grade I–II. The
samples were used in RT-qPCR to validate the circRNAs microarray
data. The study protocol was approved by the Clinical Research
Ethics Committee of Huashan Hospital, Fudan University (Shanghai,
China). Written informed consent was obtained from all
participants.
Data source
The gene expression profile of GSE56081 and the
circRNA expression profile of GSE67566 were downloaded from the
NCBI GEO database (www.ncbi.nlm.nih.gov/geo).
Gene ontology (GO) analysis
Differentially expressed circRNAs and their host
genes [which were ascertained using circbase database (http://circbase.org/)] were identified using the GEO
database. Subsequently, the GO enrichment of these genes was
analyzed using STRAP version no. 1.5 (http://www.bumc.bu.edu/cardiovascularproteomics/cpctools/strap/)
with the default parameter settings.
Protein classification and pathways
analysis
Protein classification and pathways analysis was
obtained from the PANTHER Classifications Database (pantherdb.org/).
RNA isolation and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
TRIspin method was used to isolate total RNA of NP
samples (22). Subsequently, RNA
was reversely transcribed using the TransScript One-step gDNA
Removal and cDNA Synthesis SuperMix kit according to the
manufacturer's protocol (Beijing Transgen Biotech, Co., Ltd.,
Beijing, China). Expression of hsa_circ_0008305 and
hsa_circ_0041946 were analyzed on the Bio-Rad IQ5 multicolor
detection system with Power SYBR Green PCR Master Mix (Takara Bio,
Inc., Otsu, Japan). The thermo cycling conditions for the RT-qPCR
step is as follows: initial denaturation at 95°C, one cycle 2 min;
40 cycles of 95°C for 5 sec, 60°C, 30 sec; and final extension. The
primers used were: Hsa_circ_0008305 forwards,
5′-AAATCGCTTTAAACCAACATC-3′ and reverse, 5′-CGGACACACGCCGTCTCTG-3′;
hsa_circ_0041946 forwards, 5′-TGTGATGATGGTGAGGATGGG-3′, and
reverse, 5′-GGCCTTGGAACTCAAGGATGC-3′; human GAPDH forwards,
5′-GGGAAACTGTGGCGTGAT-3′ and reverse, 5′-GAGTGGGTGTCGCTGTTGA-3′.
Expression levels of the two circRNAs were normalized to GAPDH
using the 2−ΔΔCq method (23).
Statistical analysis
RT-qPCR data are expressed as the mean ± standard
error and evaluated with a two-tailed Student's t test. P<0.05
was considered to indicate a statistically significant difference.
All experiments were repeated at least three times.
Results
Landscape of circRNA length in the
human intervertebral disc
The study by Lan et al (20) demonstrated that circRNAs lengths in
human intervertebral disc vary greatly, ranging from 74–433,729 nt.
However, the distribution density of circRNA length is still not
clear. In the current study, circRNAs from the GEO database were
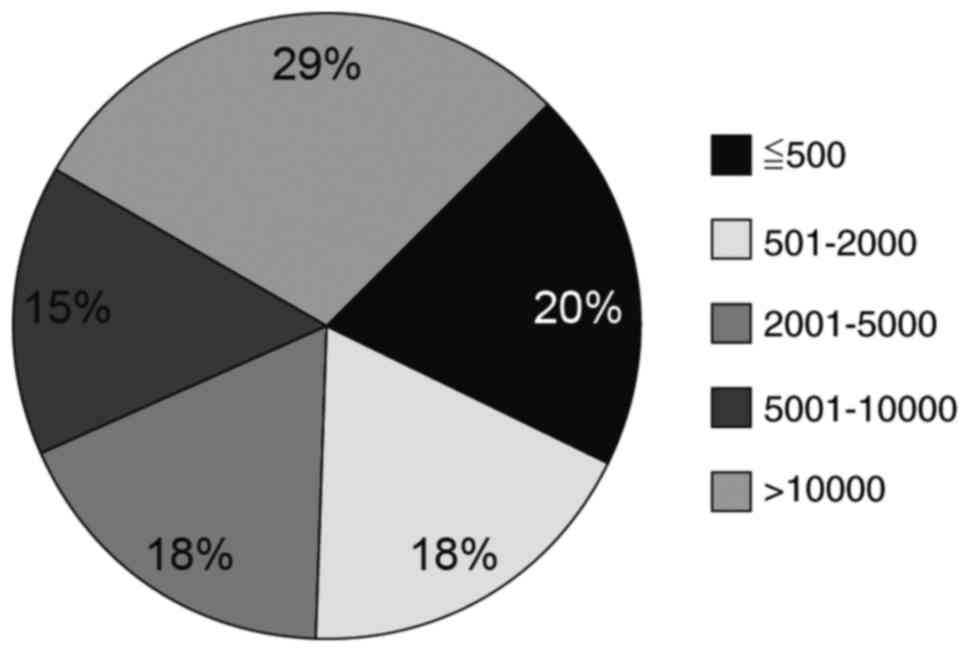
classified according to length and were distributed as follows in
intervertebral discs: CircRNA containing >10,000 nt were at the
highest level (29.34%), whereas circRNA containing 5,001–10,000 nt
were at the lowest (14.79%); other lengths accounted for ~20%
(Fig. 1). The distribution density
of circRNA lengths in the intervertebral disc was very different
from other organs like the heart, in which the majority of circRNAs
are <501 nt (unpublished data). This result suggested that
circRNA expression in humans is tissue-specific.
GO and classification analysis
To understand the potential roles of circRNAs, the
GO categories of the host genes, which expressed the circRNAs were
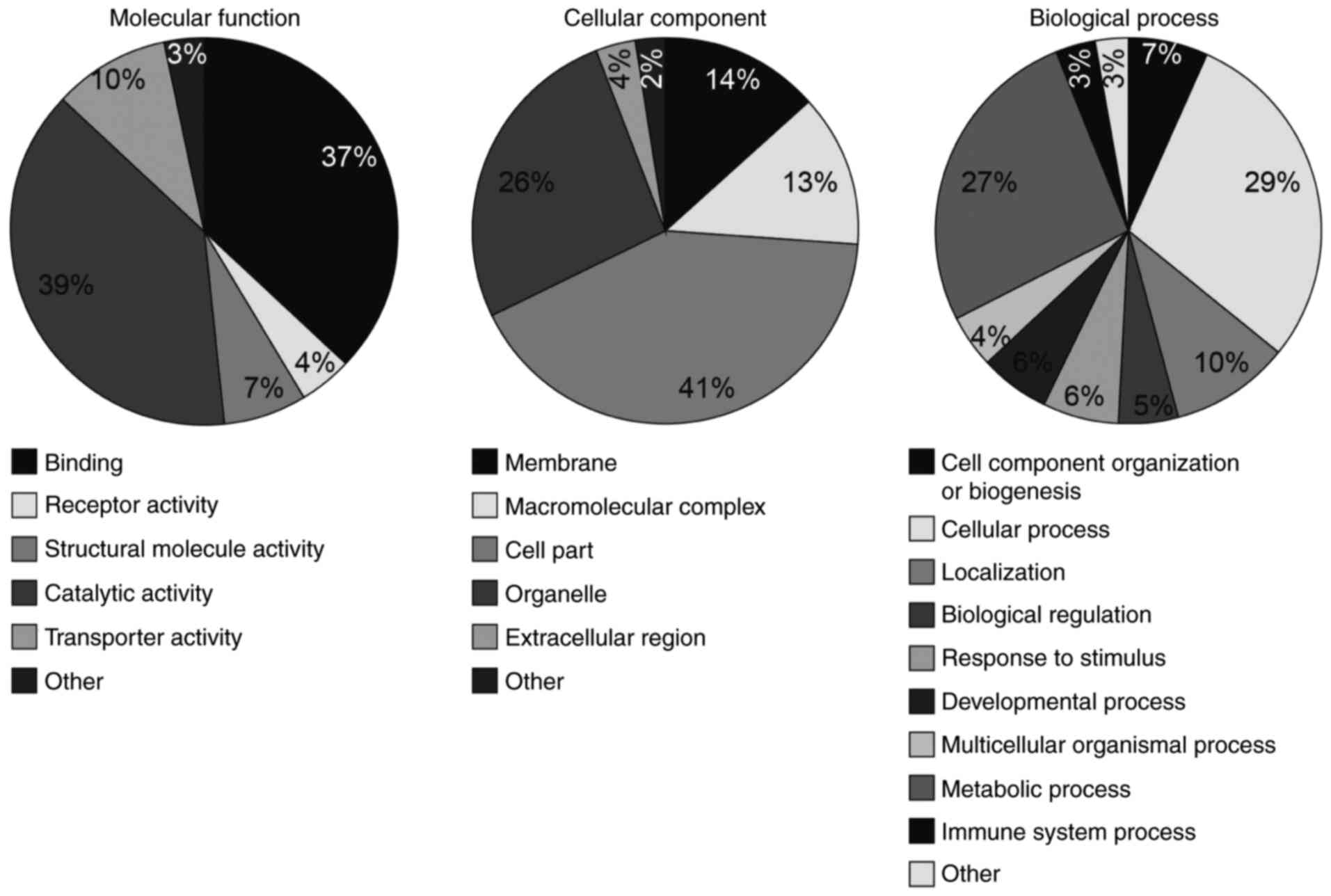
analyzed (Fig. 2). In the
molecular function domain, the circRNA host genes were
predominantly enriched in catalytic activity (39%) and binding
terms (37%), and the remaining terms included receptor, structural
molecule and transporter activity. In the cellular component
domain, the highest host gene term was cell part (41%). Among the
others, host genes were enriched in membrane, macromolecular
complex, organelle and extracellular region terms. In biological
process, host genes were part of the cellular (29%) and metabolic
process terms (27%). In addition, the host genes were from the
cellular component organization or biogenesis, localization,
biological regulation, response to stimulus, developmental process,
multicellular organismal process and immune system process
terms.
To further understand the function of circRNAs in
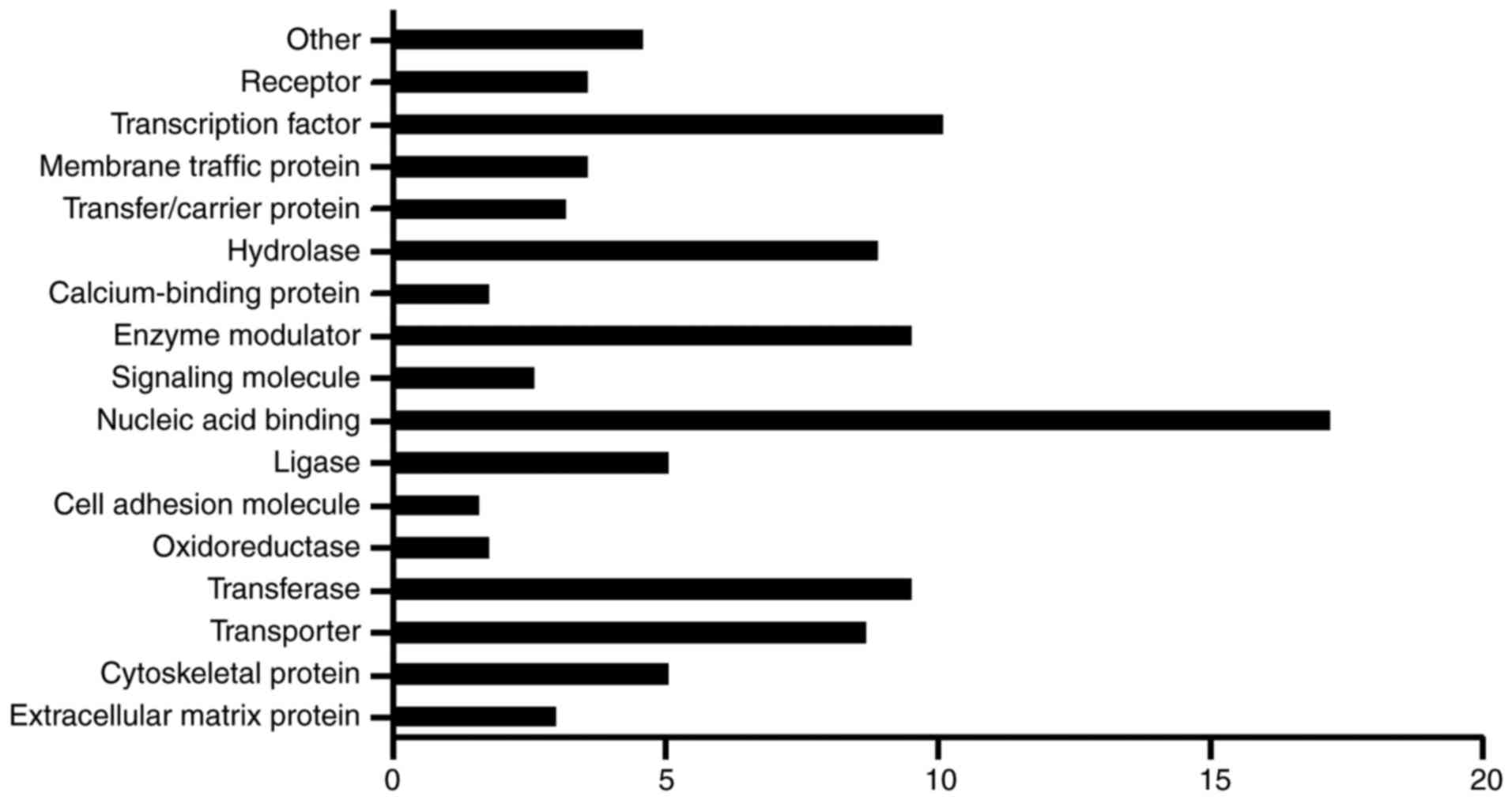
IDD, the host genes were classified on the basis of GO analysis.
The genes were divided into 16 protein function classes and were
ranked according to the percentage of proteins in the class
(Fig. 3). Nucleic acid binding
genes were ranked as the first (17.2%), and another class that
included >10% of host genes was transcription factors (10.1%).
In addition, transferases (9.5%), enzyme modulators (9.5%),
hydrolases (8.9%) and transporters (8.7%) may also serve
significant roles. These results are similar to those of the GO
analysis.
Pathway analysis
To understand which signaling pathways the circRNAs
are potentially involved in, the signaling pathways of host genes
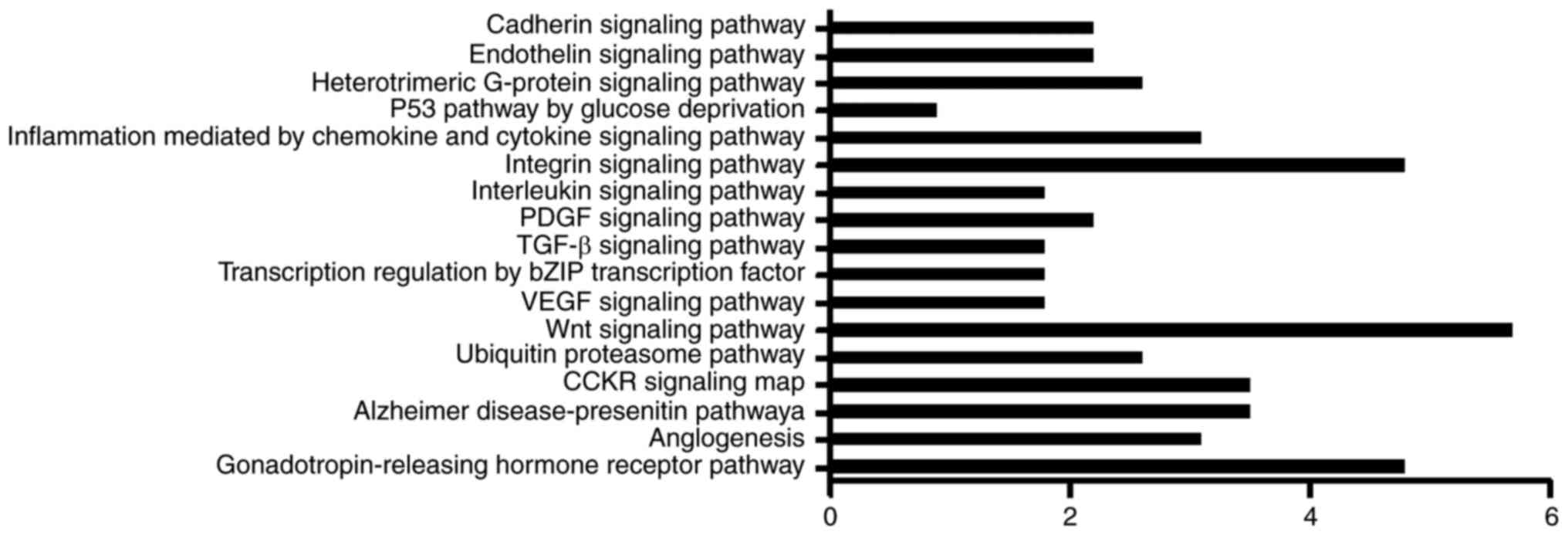
were analyzed. A total of 17 signaling pathways were obtained from
the PANTHER Classifications Database (Fig. 4). The distribution of host genes in
pathways was discrete, with the highest level reaching only 5.7%,
which was the Wnt signaling pathway. The gonadotropin-releasing
hormone receptor and integrin signaling pathways also contained a
high level of host genes. In addition, the p53 pathway was
associated with the lowest number of genes of the pathways
investigated.
Interactions between circRNAs and
their host genes
A recent study demonstrated that circRNAs could
affect their host genes: Frequently there is a decrease in
expression of linear mRNAs when the majority of transcribed exons
are circularized, leading to a negative correlation between linear
and circRNAs (24). Therefore, the
co-expression of circRNAs and their host genes were analyzed. A
total of 76 pairs of circRNAs and their host genes that were both
differentially expressed in IDD were identified from the GEO
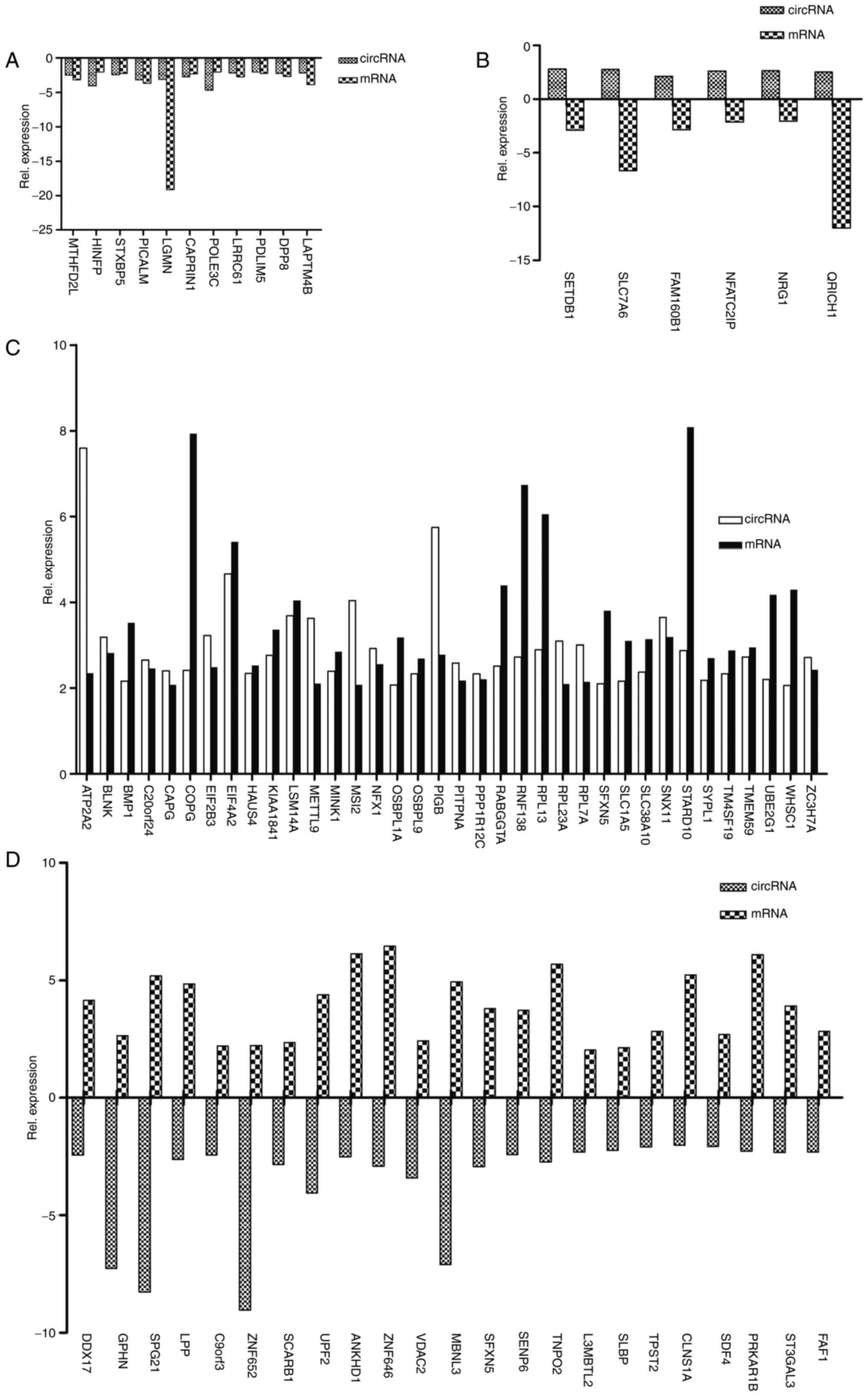
databases (Fig. 5; Table I). They were divided into four
classes of co-expression: Both circRNAs and their host genes
downregulated, circRNAs upregulated and host genes downregulated,
both circRNAs and their host genes upregulated, and circRNAs
downregulated and host genes upregulated were detected. In the
group with circRNAs and their host genes downregulated, there were
11 pairs of RNAs, and the majority were downregulated <5-fold
(Fig. 5A). However, the Legumain
mRNA was downregulated >15-fold (Fig. 5A). In the group with circRNAs
upregulated and host genes downregulated, there were six pairs of
RNAs, and all the RNAs, except glutamine-rich protein 1
(11.97-fold) and solute carrier family 7 member 6 (6.67-fold), were
downregulated <3-fold (Fig.
5B). In addition, there were 36 pairs of RNAs in the group with
both circRNAs and their host genes upregulated. The majority were
upregulated <5-fold, but two circRNAs (circATP2A2 and circPIGB)
and five mRNAs (coatomer protein complex subunit γ, eukaryotic
translation initiation factor 4A2, ring finger protein 138,
ribosomal protein L13 and StAR related lipid transfer domain
containing 10) were upregulated by >5-fold (Fig. 5C). Additionally, 23 pairs of RNAs
were in the group with circRNAs downregulated and host genes
upregulated. All pairs of RNAs except for SPG21maspardin
(13.46-fold), muscleblind-like splicing regulator 3 (12.02-fold)
and zinc finger protein 652 (11.26-fold) were differentially
expressed <10-fold (Fig. 5D).
These results suggested that these 76 circRNAs may serve vital
roles in regulating mRNAs in IDD (Table I).
 | Table I.A total of 76 differentially expressed
circRNAs and their host genes in intervertebral disc
degeneration. |
Table I.
A total of 76 differentially expressed
circRNAs and their host genes in intervertebral disc
degeneration.
| circRNA | circRNA
expression | Host gene | Gene expression |
| hsa_circ_0074269 | −2.51 | ANKHD1 | 6.12 |
| hsa_circ_0028173 | 7.59 | ATP2A2 |
2.34 |
|
hsa_circ_0000253 |
3.19 | BLNK |
2.81 |
|
hsa_circ_0002075 |
2.17 | BMP1 |
3.51 |
|
hsa_circ_0060194 |
2.65 | C20orf24 |
2.45 |
|
hsa_circ_0002191 | −2.44 | C9orf3 |
2.20 |
|
hsa_circ_0055412 |
2.40 | CAPG |
2.07 |
|
hsa_circ_0000288 | −2.76 | CAPRIN1 | −2.29 |
|
hsa_circ_0023696 | −2.02 | CLNS1A |
5.23 |
|
hsa_circ_0067260 |
2.41 | COPG |
7.93 |
|
hsa_circ_0063331 | −2.44 | DDX17 |
4.13 |
|
hsa_circ_0007527 | −2.24 | DPP8 | −2.70 |
|
hsa_circ_0012185 |
3.22 | EIF2B3 |
2.48 |
|
hsa_circ_0068462 |
4.66 | EIF4A2 |
5.40 |
|
hsa_circ_0004619 | −2.30 | FAF1 |
2.81 |
|
hsa_circ_0020080 |
2.13 | FAM160B1 | −2.87 |
|
hsa_circ_0032253 | −7.26 | GPHN |
2.65 |
|
hsa_circ_0031258 |
2.34 | HAUS4 |
2.52 |
|
hsa_circ_0006932 | −4.08 | HINFP | −2.03 |
|
hsa_circ_0001013 |
2.76 | KIAA1841 |
3.35 |
|
hsa_circ_0001234 | −2.32 | L3MBTL2 |
2.02 |
|
hsa_circ_0085013 | −2.19 | LAPTM4B | −3.90 |
|
hsa_circ_0033010 | −3.13 | LGMN | −19.14 |
|
hsa_circ_0003759 | −2.63 | LPP |
4.84 |
|
hsa_circ_0082892 | −2.20 | LRRC61 | −2.72 |
|
hsa_circ_0006446 |
3.69 | LSM14A |
4.05 |
|
hsa_circ_0091570 | −7.10 | MBNL3 |
4.92 |
|
hsa_circ_0002696 |
3.62 | METTL9 |
2.11 |
|
hsa_circ_0092278 |
2.39 | MINK1 |
2.84 |
|
hsa_circ_0000788 |
4.04 | MSI2 |
2.07 |
|
hsa_circ_0069977 | −2.46 | MTHFD2L | −3.21 |
|
hsa_circ_0038885 |
2.60 | NFATC2IP | −2.11 |
|
hsa_circ_0086645 |
2.92 | NFX1 |
2.55 |
|
hsa_circ_0007279 |
2.62 | NRG1 | −2.06 |
|
hsa_circ_0047288 |
2.07 | OSBPL1A |
3.17 |
|
hsa_circ_0002039 |
2.32 | OSBPL9 |
2.68 |
|
hsa_circ_0005770 | −2.08 | PDLIM5 | −2.22 |
|
hsa_circ_0002513 | −3.17 | PICALM | −3.64 |
|
hsa_circ_0035381 |
5.74 | PIGB |
2.77 |
|
hsa_circ_0041252 |
2.58 | PITPNA |
2.17 |
|
hsa_circ_0013912 | −4.66 | POLR3C | −2.04 |
|
hsa_circ_0000958 |
2.33 | PPP1R12C |
2.20 |
|
hsa_circ_0079040 | −2.28 | PRKAR1B |
6.09 |
|
hsa_circ_0092288 |
2.51 | QRICH1 | −11.97 |
|
hsa_circ_0007750 |
2.51 | RABGGTA |
4.39 |
|
hsa_circ_0047378 |
2.72 | RNF138 |
6.73 |
|
hsa_circ_0092337 |
2.89 | RPL13 |
6.05 |
|
hsa_circ_0092360 |
3.09 | RPL23A |
2.09 |
|
hsa_circ_0092328 |
3.00 | RPL7A |
2.14 |
|
hsa_circ_0029340 | −2.84 | SCARB1 |
2.34 |
|
hsa_circ_0009244 | −2.07 | SDF4 |
2.69 |
|
hsa_circ_0001614 | −2.42 | SENP6 |
3.71 |
|
hsa_circ_0006352 |
2.81 | SETDB1 | −2.90 |
|
hsa_circ_0055201 | −2.93 | SFXN5 |
3.79 |
|
hsa_circ_0004699 |
2.10 | SFXN5 |
3.79 |
|
hsa_circ_0068850 | −2.25 | SLBP |
2.13 |
|
hsa_circ_0002311 |
2.17 | SLC1A5 |
3.09 |
|
hsa_circ_0046123 |
2.37 | SLC38A10 |
3.14 |
|
hsa_circ_0005623 |
2.75 | SLC7A6 | −6.67 |
|
hsa_circ_0002069 |
3.65 | SNX11 |
3.19 |
|
hsa_circ_0035875 | −8.27 | SPG21 |
5.18 |
|
hsa_circ_0012107 | −2.34 | ST3GAL3 |
3.89 |
|
hsa_circ_0002383 |
2.87 | STARD10 |
8.08 |
|
hsa_circ_0078150 | −2.43 | STXBP5 | −2.22 |
|
hsa_circ_0081873 |
2.18 | SYPL1 |
2.70 |
|
hsa_circ_0068641 |
2.33 | TM4SF19 |
2.87 |
|
hsa_circ_0012634 |
2.72 | TMEM59 |
2.95 |
|
hsa_circ_0000894 | −2.73 | TNPO2 |
5.68 |
|
hsa_circ_0062682 | −2.09 | TPST2 |
2.82 |
|
hsa_circ_0041555 |
2.20 | UBE2G1 |
4.17 |
|
hsa_circ_0017713 | −4.05 | UPF2 |
4.39 |
|
hsa_circ_0018909 | −3.41 | VDAC2 |
2.42 |
|
hsa_circ_0068895 |
2.05 | WHSC1 |
4.29 |
|
hsa_circ_0005394 |
2.71 | ZC3H7A |
2.42 |
|
hsa_circ_0000691 | −2.91 | ZNF646 |
6.45 |
|
hsa_circ_0044413 | −9.04 | ZNF652 |
2.22 |
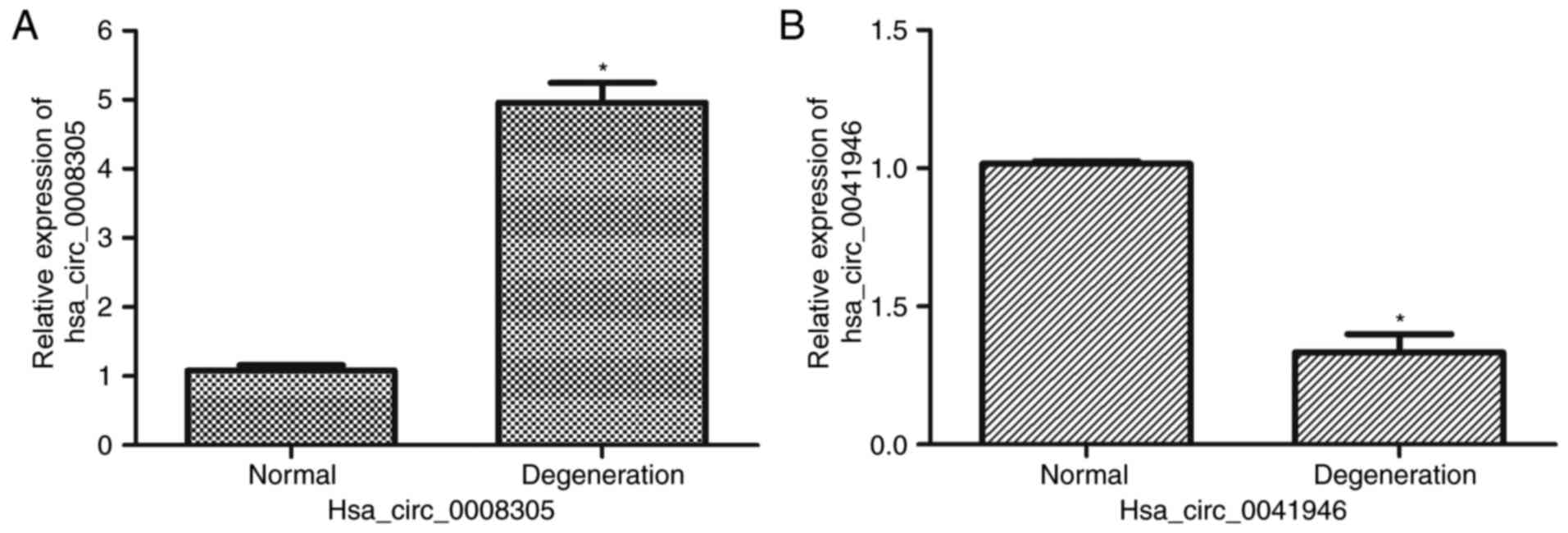
RT-qPCR validation
To validate the circRNAs microarray, one upregulated
circRNA (hsa_circ_0008305) and one downregualted circRNA
(hsa_circ_0041946) was selected randomly for further evaluation.
RT-qPCR demonstrated that circ_0008305 was significantly
upregulated (P<0.05) in degenerative disc samples compared with
normal samples (Fig. 6A).
Furthermore, hsa_circ_0041946 expression was reduced (P<0.05) in
degenerative samples compared with control samples (Fig. 6B). This result suggested the
circRNAs microarray data was credible.
Discussion
IDD is one of most common chronic and age-related
degenerative musculoskeletal disorders, and one of the key factors
leading to IDD is genetics (1–7). One
class of recently discovered ncRNAs is circRNAs, which have been
reported to be involved in IDD (20). In the present study, circRNAs were
classified according to lengths, and it was demonstrated that
different circRNA lengths were homogeneously distributed in the
human intervertebral disc. Using functional enrichment analysis,
the tissue-specific expression of circRNAs host genes in IDD was
confirmed. By performing co-expression analysis, 76 pairs of
differentially expressed circRNAs and their host genes were
identified. Finally, RT-qPCR was performed to confirm
hsa_circ_0008305 upregulation and hsa_circ_0041946 downregulation
in IDD compared with normal lumbar NP specimens.
It has been reported that circRNAs have
spatio-temporal features. Xia et al (25) identified >300,000 circRNAs in
humans and mice, and 10.4% of human circRNAs and 34.3% of mouse
circRNAs were tissue-specific. Werfel et al (26) identified 16,427 circRNAs in the
human heart; however, only 2,202 of these were homologous to mouse
circRNAs. The present analysis identified that circRNAs in the
intervertebral disc are very different from heart circRNAs and that
their lengths in the intervertebral disc are distributed uniformly.
The results of the present study indicate that circRNAs may serve a
key role in tissue development or disease occurrence.
In the process of IDD, extracellular matrix (ECM)
metabolism becomes dysfunctional and the expression of growth
factors and cytokines leads to inflammation (27). Subsequently, the ECM is degraded by
catabolic factors (28). The GO
data demonstrated that the circRNAs that interact with catalytic
activity and binding genes are differentially expressed in IDD. The
protein classification analysis illustrated that circRNAs that
associated with genes that encode nucleic acid binding proteins,
transferases, enzyme modulators and hydrolases maybe involved in
IDD. All these results suggest that circRNAs may participate in ECM
metabolism. However, the underlying molecular mechanisms need to be
explored further.
Several signaling pathways have been reported to be
involved in IDD, including the Wnt/β-catenin, the P53/P21, the
interleukin, and the tumor growth factor-β pathways (26,29–33).
These signaling pathways have important roles in regulating IDD.
Notably, many host genes of the differentially expressed circRNAs
in IDD are part of these pathways. For example, the circRNAs that
bind mRNAs with roles in the Wnt signaling pathways were expressed
in high levels. This result suggests that not only the mRNAs, but
circRNAs in these signaling pathways serve key roles in IDD.
An important function of circRNAs is thought to be
the regulation of mRNA expression. circRNAs may counteract
miRNA-mediated repression or suppression of mRNA expression by
competing with linear splicing (17,34).
In total, 76 pairs of circRNAs and their host genes that could
interact with each other were identified in IDD samples. Fas
ligand, which mediates the apoptosis, was expressed in NP cells in
IDD processes (35). Capping actin
protein gelsolin like, a Fas apoptosis pathway gene, and its
circRNA (hsa_circ_0055412) were both upregulated in IDD. However,
the mRNA of another Fas pathway gene, Fas associated factor 1, was
upregulated in IDD but its circRNA (hsa_circ_0004619) was
downregulated. Growth factors serve important roles in IDD
(36). Neuregulin 1 (NRG1)
is a member of the epidermal growth factor receptor pathway that
mediates cell-cell signaling and serves a critical role in growth
and development (37). NRG1
mRNA was downregulated in IDD, but its circRNA (hsa_circ_0007279)
was upregulated. Bone morphogenetic protein 1 (BMP 1) is
another growth factor (38).
BMP1 mRNA and circRNA (hsa_circ_0002075) were upregulated in
IDD. These differentially expressed circRNAs may serve key roles in
IDD and have the potential to be used in the diagnosis and clinical
treatment of IDD in the future.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant no. 81601914 and
81472036).
References
|
1
|
Benjamin RW and Xuejun W: Intervertebral
disc (IVD): Structure, degeneration, repair and regeneration. Mater
Sci Engineer C. 32:61–77. 2012. View Article : Google Scholar
|
|
2
|
Deshpande BR, Katz JN, Solomon DH, Yelin
EH, Hunter DJ, Messier SP, Suter LG and Losina E: Number of persons
with symptomatic knee osteoarthritis in the US: Impact of race and
ethnicity, Age, Sex, and Obesity. Arthritis Care Res (Hoboken).
68:1743–1750. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Song YQ, Karasugi T, Cheung KM, Chiba K,
Ho DW, Miyake A, Kao PY, Sze KL, Yee A, Takahashi A, et al: Lumbar
disc degeneration is linked to a carbohydrate sulfotransferase 3
variant. J Clin Invest. 123:4909–4917. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gruber HE, Hoelscher GL, Ingram JA, Bethea
S, Zinchenko N and Hanley EN Jr: Variations in aggrecan
localization and gene expression patterns characterize increasing
stages of human intervertebral disk degeneration. Exp Mol Pathol.
91:534–539. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gruber HE, Hoelscher GL, Ingram JA and
Hanley EN Jr: Genome-wide analysis of pain-, nerve- and
neurotrophin-related gene expression in the degenerating human
annulus. Mol Pain. 8:632012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sun Z, Wang HQ, Liu ZH, Chang L, Chen YF,
Zhang YZ, Zhang WL, Gao Y, Wan ZY, Che L, et al: Down-regulated CK8
expression in human intervertebral disc degeneration. Int J Med
Sci. 10:948–956. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sun Z, Guo YS, Yan SJ, Wan ZY, Gao B, Wang
L, Liu ZH, Gao Y, Samartzis D, Lan LF, et al: CK8 phosphorylation
induced by compressive loads underlies the downregulation of CK8 in
human disc degeneration by activating protein kinase C. Lab Invest.
93:1323–1330. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Brosnan CA and Voinnet O: The long and the
short of noncoding RNAs. Curr Opin Cell Biol. 21:416–425. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Batista PJ and Chang HY: Long noncoding
RNAs: Cellular address codes in development and disease. Cell.
152:1298–1307. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jarroux J, Morillon A and Pinskaya M:
History, discovery, and classification of lncRNAs. Adv Exp Med
Biol. 1008:1–46. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Maxmen A: RNA: The genome's rising stars.
Nature. 496:127–129. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Brown CJ, Ballabio A, Rupert JL,
Lafreniere RG, Grompe M, Tonlorenzi R and Willard HF: A gene from
the region of the human X inactivation centre is expressed
exclusively from the inactive X chromosome. Nature. 349:38–44.
1991. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bartolomei MS, Zemel S and Tilghman SM:
Parental imprinting of the mouse H19 gene. Nature. 351:153–155.
1991. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jeck WR, Sorrentino JA, Wang K, Slevin MK,
Burd CE, Liu J, Marzluff WF and Sharpless NE: Circular RNAs are
abundant, conserved, and associated with ALU repeats. RNA.
19:141–157. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Greene J, Baird AM, Brady L, Lim M, Gray
SG, McDermott R and Finn SP: Circular RNAs: Biogenesis, Function
and role in human diseases. Front Mol Biosci. 4:382017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ashwal-Fluss R, Meyer M, Pamudurti NR,
Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N and
Kadener S: circRNA biogenesis competes with pre-mRNA splicing. Mol
Cell. 56:55–66. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tay Y, Rinn J and Pandolfi PP: The
multilayered complexity of ceRNA crosstalk and competition. Nature.
505:344–352. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lan PH, Liu ZH, Pei YJ, Wu ZG, Yu Y, Yang
YF, Liu X, Che L, Ma CJ, Xie YK, et al: Landscape of RNAs in human
lumbar disc degeneration. Oncotarget. 7:63166–63176. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Pfirrmann CW, Metzdorf A, Zanetti M,
Hodler J and Boos N: Magnetic resonance classification of lumbar
intervertebral disc degeneration. Spine (Phila Pa 1976).
26:1873–1878. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Reno C, Marchuk L, Sciore P, Frank CB and
Hart DA: Rapid isolation of total RNA from small samples of
hypocellular, dense connective tissues. Biotechniques.
22:1082–1086. 1997.PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Huang S, Yang B, Chen BJ, Bliim N,
Ueberham U, Arendt T and Janitz M: The emerging role of circular
RNAs in transcriptome regulation. Genomics. Jun 26–2017.(Epub ahead
of print). View Article : Google Scholar
|
|
25
|
Xia S, Feng J, Lei L, Hu J, Xia L, Wang J,
Xiang Y, Liu L, Zhong S, Han L and He C: Comprehensive
characterization of tissue-specific circular RNAs in the human and
mouse genomes. Brief Bioinform: Agu. 20–2016.(Epub ahead of print).
View Article : Google Scholar
|
|
26
|
Werfel S, Nothjunge S, Schwarzmayr T,
Strom TM, Meitinger T and Engelhardt S: Characterization of
circular RNAs in human, mouse and rat hearts. J Mol Cell Cardiol.
98:103–107. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Le Maitre CL, Pockert A, Buttle DJ,
Freemont AJ and Hoyland JA: Matrix synthesis and degradation in
human intervertebral disc degeneration. Biochem Soc Trans.
35:652–655. 2017. View Article : Google Scholar
|
|
28
|
Wang J, Markova D, Anderson DG, Zheng Z,
Shapiro IM and Risbud MV: TNF-α and IL-1β promote a
disintegrin-like and metalloprotease with thrombospondin type I
motif-5-mediated aggrecan degradation through syndecan-4 in
intervertebral disc. J Biol Chem. 286:39738–39749. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Jia H, Ma J, Lv J, Ma X, Xu W, Yang Y,
Tian A, Wang Y, Sun L, Xu L, et al: Oestrogen and parathyroid
hormone alleviate lumbar intervertebral disc degeneration in
ovariectomized rats and enhance Wnt/β-catenin pathway activity. Sci
Rep. 6:275212016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Xie H, Jing Y, Xia J, Wang X, You C and
Yan J: Aquaporin 3 protects against lumbar intervertebral disc
degeneration via the Wnt/β-catenin pathway. Int J Mol Med.
37:859–864. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhou N, Lin X, Dong W, Huang W, Jiang W,
Lin L, Qiu Q, Zhang X, Shen J, Song Z, et al: SIRT1 alleviates
senescence of degenerative human intervertebral disc cartilage
endo-plate cells via the p53/p21 pathway. Sci Rep. 6:226282016.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang Z, Wang G, Zhu X, Geng D and Yang H:
Interleukin-2 is upregulated in patients with a prolapsed lumbar
intervertebral disc and modulates cell proliferation, apoptosis and
extracellular matrix metabolism of human nucleus pulposus cells.
Exp Ther Med. 10:2437–2443. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hiyama A, Sakai D, Tanaka M, Arai F,
Nakajima D, Ab K and Mochida J: The relationship between the
Wnt/β-catenin and TGF-β/BMP signals in the intervertebral disc
cell. J Cell Physiol. 226:1139–1148. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Cortes-López M and Miura P: Emerging
functions of circular RNAs. Yale J Biol Med. 89:527–537.
2016.PubMed/NCBI
|
|
35
|
Kaneyama S, Nishida K, Takada T, Suzuki T,
Shimomura T, Maeno K, Kurosaka M and Doita M: Fas ligand expression
on human nucleus pulposus cells decreases with disc degeneration
processes. J Orthop Sci. 13:130–135. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Masuda K, Oegema TR Jr and An HS: Growth
factors and treatment of intervertebral disc degeneration. Spine
(Phila Pa 1976). 29:2757–2769. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Garratt AN, Britsch S and Birchmeier C:
Neuregulin, a factor with many functions in the life of a schwann
cell. Bioessays. 22:987–996. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Sedlmeier G and Sleeman JP: Extracellular
regulation of BMP signaling: Welcome to the matrix. Biochem Soc
Trans. 45:173–181. 2017. View Article : Google Scholar : PubMed/NCBI
|