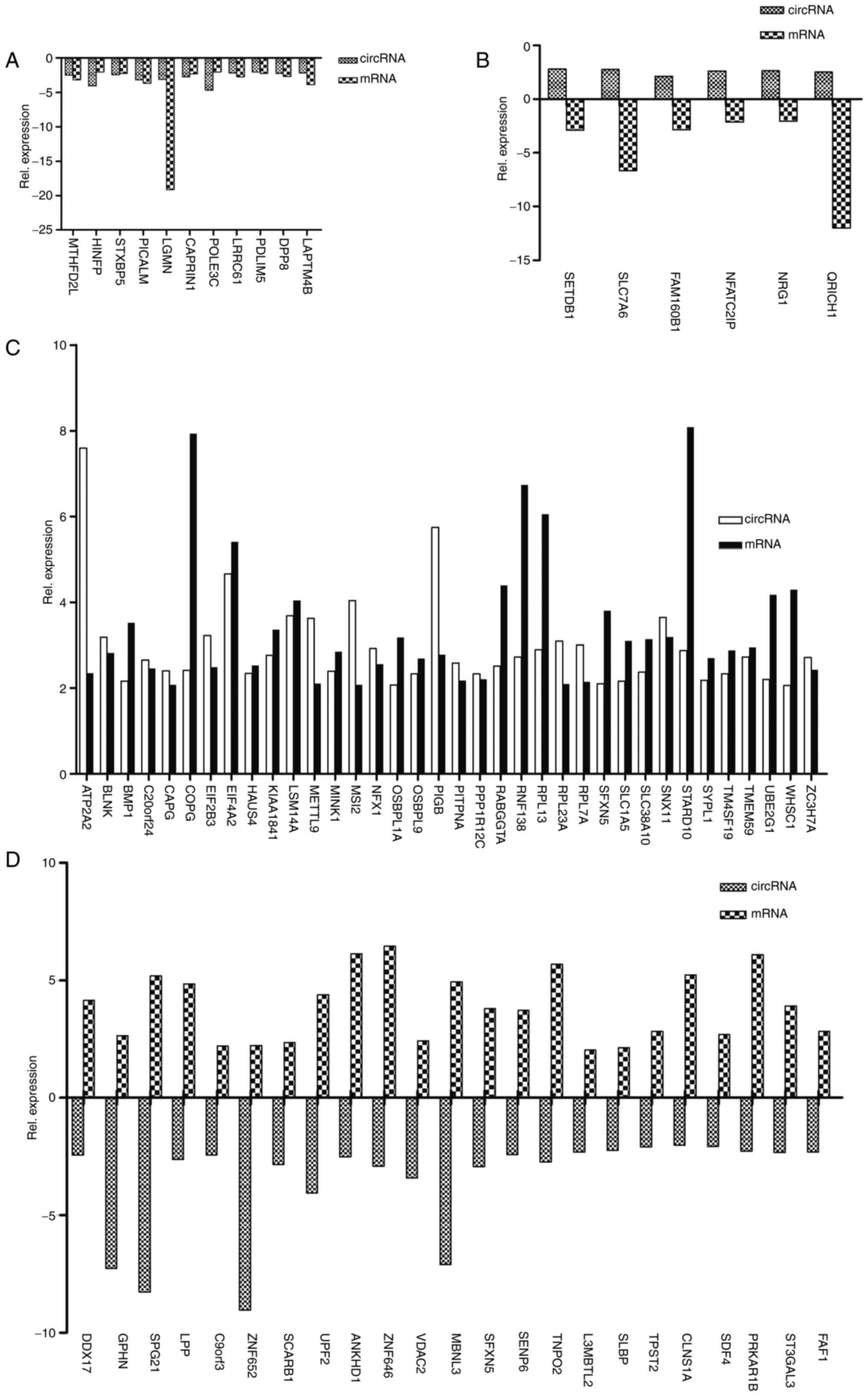
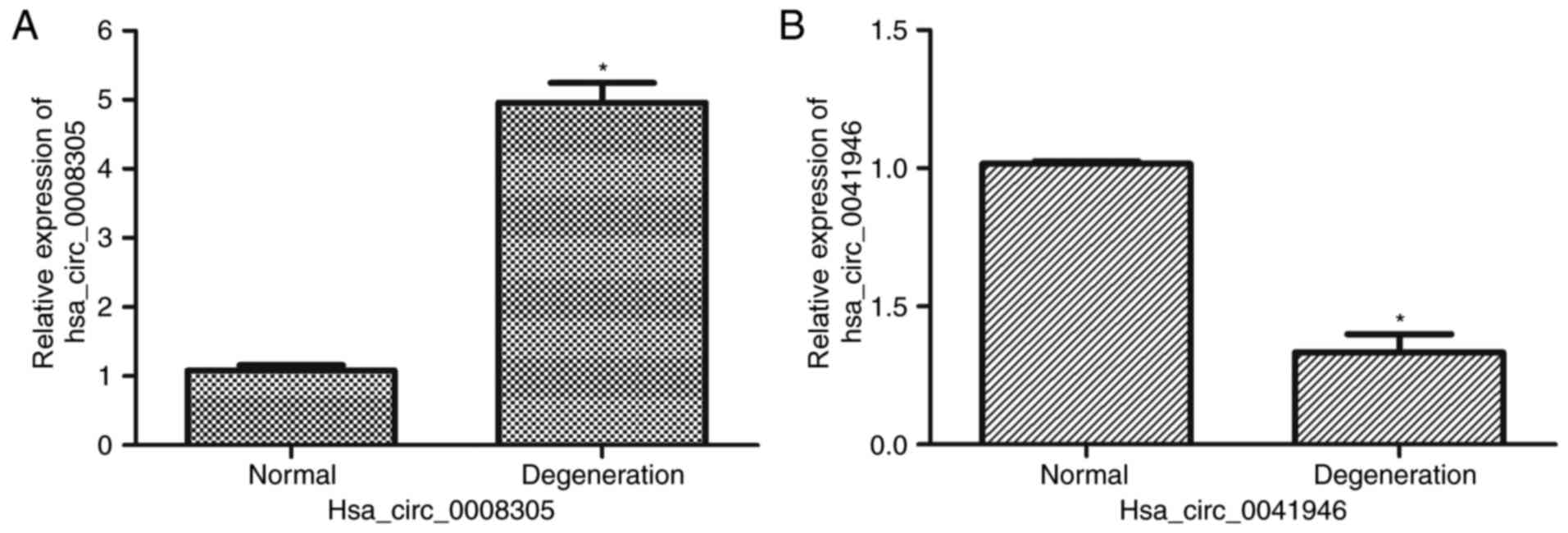
|
1
|
Benjamin RW and Xuejun W: Intervertebral
disc (IVD): Structure, degeneration, repair and regeneration. Mater
Sci Engineer C. 32:61–77. 2012. View Article : Google Scholar
|
|
2
|
Deshpande BR, Katz JN, Solomon DH, Yelin
EH, Hunter DJ, Messier SP, Suter LG and Losina E: Number of persons
with symptomatic knee osteoarthritis in the US: Impact of race and
ethnicity, Age, Sex, and Obesity. Arthritis Care Res (Hoboken).
68:1743–1750. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Song YQ, Karasugi T, Cheung KM, Chiba K,
Ho DW, Miyake A, Kao PY, Sze KL, Yee A, Takahashi A, et al: Lumbar
disc degeneration is linked to a carbohydrate sulfotransferase 3
variant. J Clin Invest. 123:4909–4917. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gruber HE, Hoelscher GL, Ingram JA, Bethea
S, Zinchenko N and Hanley EN Jr: Variations in aggrecan
localization and gene expression patterns characterize increasing
stages of human intervertebral disk degeneration. Exp Mol Pathol.
91:534–539. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gruber HE, Hoelscher GL, Ingram JA and
Hanley EN Jr: Genome-wide analysis of pain-, nerve- and
neurotrophin-related gene expression in the degenerating human
annulus. Mol Pain. 8:632012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sun Z, Wang HQ, Liu ZH, Chang L, Chen YF,
Zhang YZ, Zhang WL, Gao Y, Wan ZY, Che L, et al: Down-regulated CK8
expression in human intervertebral disc degeneration. Int J Med
Sci. 10:948–956. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sun Z, Guo YS, Yan SJ, Wan ZY, Gao B, Wang
L, Liu ZH, Gao Y, Samartzis D, Lan LF, et al: CK8 phosphorylation
induced by compressive loads underlies the downregulation of CK8 in
human disc degeneration by activating protein kinase C. Lab Invest.
93:1323–1330. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Brosnan CA and Voinnet O: The long and the
short of noncoding RNAs. Curr Opin Cell Biol. 21:416–425. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Batista PJ and Chang HY: Long noncoding
RNAs: Cellular address codes in development and disease. Cell.
152:1298–1307. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jarroux J, Morillon A and Pinskaya M:
History, discovery, and classification of lncRNAs. Adv Exp Med
Biol. 1008:1–46. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Maxmen A: RNA: The genome's rising stars.
Nature. 496:127–129. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Brown CJ, Ballabio A, Rupert JL,
Lafreniere RG, Grompe M, Tonlorenzi R and Willard HF: A gene from
the region of the human X inactivation centre is expressed
exclusively from the inactive X chromosome. Nature. 349:38–44.
1991. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bartolomei MS, Zemel S and Tilghman SM:
Parental imprinting of the mouse H19 gene. Nature. 351:153–155.
1991. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jeck WR, Sorrentino JA, Wang K, Slevin MK,
Burd CE, Liu J, Marzluff WF and Sharpless NE: Circular RNAs are
abundant, conserved, and associated with ALU repeats. RNA.
19:141–157. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Greene J, Baird AM, Brady L, Lim M, Gray
SG, McDermott R and Finn SP: Circular RNAs: Biogenesis, Function
and role in human diseases. Front Mol Biosci. 4:382017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ashwal-Fluss R, Meyer M, Pamudurti NR,
Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N and
Kadener S: circRNA biogenesis competes with pre-mRNA splicing. Mol
Cell. 56:55–66. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tay Y, Rinn J and Pandolfi PP: The
multilayered complexity of ceRNA crosstalk and competition. Nature.
505:344–352. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lan PH, Liu ZH, Pei YJ, Wu ZG, Yu Y, Yang
YF, Liu X, Che L, Ma CJ, Xie YK, et al: Landscape of RNAs in human
lumbar disc degeneration. Oncotarget. 7:63166–63176. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Pfirrmann CW, Metzdorf A, Zanetti M,
Hodler J and Boos N: Magnetic resonance classification of lumbar
intervertebral disc degeneration. Spine (Phila Pa 1976).
26:1873–1878. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Reno C, Marchuk L, Sciore P, Frank CB and
Hart DA: Rapid isolation of total RNA from small samples of
hypocellular, dense connective tissues. Biotechniques.
22:1082–1086. 1997.PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Huang S, Yang B, Chen BJ, Bliim N,
Ueberham U, Arendt T and Janitz M: The emerging role of circular
RNAs in transcriptome regulation. Genomics. Jun 26–2017.(Epub ahead
of print). View Article : Google Scholar
|
|
25
|
Xia S, Feng J, Lei L, Hu J, Xia L, Wang J,
Xiang Y, Liu L, Zhong S, Han L and He C: Comprehensive
characterization of tissue-specific circular RNAs in the human and
mouse genomes. Brief Bioinform: Agu. 20–2016.(Epub ahead of print).
View Article : Google Scholar
|
|
26
|
Werfel S, Nothjunge S, Schwarzmayr T,
Strom TM, Meitinger T and Engelhardt S: Characterization of
circular RNAs in human, mouse and rat hearts. J Mol Cell Cardiol.
98:103–107. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Le Maitre CL, Pockert A, Buttle DJ,
Freemont AJ and Hoyland JA: Matrix synthesis and degradation in
human intervertebral disc degeneration. Biochem Soc Trans.
35:652–655. 2017. View Article : Google Scholar
|
|
28
|
Wang J, Markova D, Anderson DG, Zheng Z,
Shapiro IM and Risbud MV: TNF-α and IL-1β promote a
disintegrin-like and metalloprotease with thrombospondin type I
motif-5-mediated aggrecan degradation through syndecan-4 in
intervertebral disc. J Biol Chem. 286:39738–39749. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Jia H, Ma J, Lv J, Ma X, Xu W, Yang Y,
Tian A, Wang Y, Sun L, Xu L, et al: Oestrogen and parathyroid
hormone alleviate lumbar intervertebral disc degeneration in
ovariectomized rats and enhance Wnt/β-catenin pathway activity. Sci
Rep. 6:275212016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Xie H, Jing Y, Xia J, Wang X, You C and
Yan J: Aquaporin 3 protects against lumbar intervertebral disc
degeneration via the Wnt/β-catenin pathway. Int J Mol Med.
37:859–864. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhou N, Lin X, Dong W, Huang W, Jiang W,
Lin L, Qiu Q, Zhang X, Shen J, Song Z, et al: SIRT1 alleviates
senescence of degenerative human intervertebral disc cartilage
endo-plate cells via the p53/p21 pathway. Sci Rep. 6:226282016.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang Z, Wang G, Zhu X, Geng D and Yang H:
Interleukin-2 is upregulated in patients with a prolapsed lumbar
intervertebral disc and modulates cell proliferation, apoptosis and
extracellular matrix metabolism of human nucleus pulposus cells.
Exp Ther Med. 10:2437–2443. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hiyama A, Sakai D, Tanaka M, Arai F,
Nakajima D, Ab K and Mochida J: The relationship between the
Wnt/β-catenin and TGF-β/BMP signals in the intervertebral disc
cell. J Cell Physiol. 226:1139–1148. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Cortes-López M and Miura P: Emerging
functions of circular RNAs. Yale J Biol Med. 89:527–537.
2016.PubMed/NCBI
|
|
35
|
Kaneyama S, Nishida K, Takada T, Suzuki T,
Shimomura T, Maeno K, Kurosaka M and Doita M: Fas ligand expression
on human nucleus pulposus cells decreases with disc degeneration
processes. J Orthop Sci. 13:130–135. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Masuda K, Oegema TR Jr and An HS: Growth
factors and treatment of intervertebral disc degeneration. Spine
(Phila Pa 1976). 29:2757–2769. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Garratt AN, Britsch S and Birchmeier C:
Neuregulin, a factor with many functions in the life of a schwann
cell. Bioessays. 22:987–996. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Sedlmeier G and Sleeman JP: Extracellular
regulation of BMP signaling: Welcome to the matrix. Biochem Soc
Trans. 45:173–181. 2017. View Article : Google Scholar : PubMed/NCBI
|