Introduction
20-Hydroxyeicosatetraenoic acid (20-HETE) is
primarily produced in the kidney and the liver, and serves a role
in hypertension, metabolic dysfunction and cancers. It has been
reported previously that 20-HETE functions in a tissue-specific
manner, particularly in hemorheology, which may account for its
‘prohypertensive’ function in peripheral arteries like pulmonary
arteries (1) and its
‘antihypertensive’ role in coronary arteries (2). Our previous study developed a
cytochrome P450 family 4 subfamily F member 2 (CYP4F2)
transgenic mouse that overproduced 20-HETE in kidney and liver,
exhibiting a hypertension phenotype (3). Our previous study also demonstrated
that Nedd4-2-mediated ubiquitin-proteasome pathway was induced in
the transgenic mice with high salt intake, which resulted in the
degradation of kidney-specific Na-K-Cl symporter2 (NKCC2), and thus
affected natriuresis and blood pressure (4). Therefore, the present study
hypothesized that production of the multifunctional 20-HETE
metabolite in a tissue-specific pattern may be related to its role
in modulating Nedd4-2.
As an E3 ubiquitin ligase of the Nedd4 family,
Nedd4-2 probably modulates and binds to multiple membrane proteins
to aid in their internalization and turnover (5,6).
Dysfunctions of Nedd4-2 have been implicated in many human
pathologies through the targeted membrane proteins; for example,
inhibition of the interaction between Nedd4-2 and epithelial sodium
channel (ENaC) leads to Liddle's syndrome (7), downregulation of voltage-gated sodium
channels by Nedd4-2 contributes to neuropathic pain (8), altered levels of Nedd4-2 are linked
to multiple tumor types (9,10)
and genetic variation in Nedd4-2 is associated with hypertension
(11). Therefore, the present
study examined post-translational modifications to explore the
potential mechanism of 20-HETE-regulated Nedd4-2 expression in the
CYP4F2 transgenic mouse model.
Neddylation is a post-translational modification
whereby the ubiquitin-like protein, Nedd8, is conjugated to target
proteins. Like the ubiquitin pathway, neddylation is a cascade
pathway of specific E1, E2 and E3 enzymes, which deliver Nedd8 to
substrates in a covalent bond. So far defined, the only E1 that is
involved in the neddylation process is a heterodimer comprising
amyloid β precursor protein-binding protein 1 and ubiquitin-like
modifier-activating enzyme3. There are two E2 enzymes involved in
the neddylation process: Ubiquitin-conjugating enzyme E2 M and
ubiquitin-conjugating enzyme E2 F (12,13).
Certain enzymes have been reported to be of the E3 type in
neddylation, such as regulator of cullins 1/RING-box protein 1
(12–14) and Smurf1 (15). This post-translational modification
was reported to be involved in an extensive pathophysiology process
depending on the specific E3, and the best studied of these E3
ligases belong to the cullin-RING ubiquitin ligase (CRL) family
(16). Neddylation has been
reported to promote the ubiquitination activity of CRLs for the
activation of cullin 3-adaptor-E3 ubiquitin ligase complex and the
prevention of binding to the structural-based inhibitory factor
(17). Neddylation of CRLs may be
reversed through deneddylation, a process that removes Nedd8
through the isopeptidase activity of the COP9 signalosome complex
(CSN) (18). Therefore, both
neddylation and deneddylation, which cycle E3 ligases between
activated and inactivated states, may be essential for the
ubiquitination activities of E3 ligases. Furthermore, an inhibitor
of neddylation E1, MLN4924, has been reported to stop the
neddylation cycle (19) and to
effectively suppress the development of multiple tumor types
(20,21). As aforementioned, it was reported
that HECT-type E3 ubiquitin-protein ligase Smurf1 was activated by
neddylation through a thioester bond at C426, which was revealed to
be correlated with colon cancer progression and poor prognosis
(15).
The present study aimed to elucidate the neddylation
of Nedd4-2 and to determine the tissue-specific influence of
20-HETE on Nedd4-2. Nedd4-2 expressions in the liver and the kidney
were compared between CYP4F2 transgenic mice and wild-type
mice, and the interaction of Nedd4-2 with Nedd8 was examined. The
results may provide the first evidence that 20-HETE regulated
Nedd4-2 through neddylation modification in kidney and liver.
Materials and methods
Antibodies and reagents
Rabbit anti-Nedd8 polyclonal antibody was obtained
from Enzo Life Sciences, Inc. (catalog no. ALX-210-194-R200;
Farmingdale, NY, USA); rabbit anti-Senp8 polyclonal antibody was
obtained from Abcam (catalog no. ab58423; Cambridge, UK); rabbit
anti-Nedd4L polyclonal antibody (catalog no. 13690-1-AP); rabbit
anti-tubulin polyclonal antibody (catalog no. 10094-1-AP) and mouse
anti-GAPDH monoclonal antibody (catalog no. 60004-1-Ig) were
purchased from ProteinTech Group, Inc. [(Chicago, IL, USA). Fetal
bovine serum (FBS) and RPMI-1640 culture medium were purchased from
Biological Industries, USA, Inc. (Cromwell, CN, USA)], and 20-HETE
was purchased from Cayman Chemical Company (catalog no. 90030; Ann
Arbor, MI, USA). MLN4924 was purchased from Active Biochem (A-1139;
Kowloon, Hong Kong).
Transgenic mouse
This study was approved by the Ethics committee of
Shengjing Hospital (Shenyang, China). All animal-related
experiments conformed to the Guide for the Care and Use of
Laboratory Animals published by the US National Institutes of
Health (NIH Publication no. 85-23, revised 1996). The CYP4F2
transgenic mice overexpressing CYP4F2 was a FVB strain (3). Experiments were performed on 12- to
16-week-old male transgenic mice weighing 24–30 g. All mice were
matched in weight and age with littermate wild-type FVB mice as
controls (a gift from Animal Laboratory of China Medical
University). All mice were provided with food and water ad
libitum under a 12-h light-dark cycle and controlled
temperature (23±2°C).
Cell culture
The mouse M1 kidney cell line and the mouse NCTC1469
liver cell line were purchased from the Type Culture Collection of
the Chinese Academy of Sciences (Shanghai, China) and were cultured
in DMEM and RPMI-1640 medium, respectively, at 37°C in a humidified
5% CO2 atmosphere. Cells were cultured in 6-well plates
until 75% confluence, after which the cells were placed in fetal
bovine serum-free medium (99% DMEM or 1640 with 1% mycillin) for 4
h at 37°C in a humidified 5% CO2 atmosphere. Control
cells were treated with vehicle (0.1% ethanol). Experimental cell
cultures were treated with 20-HETE at different concentrations
(0.1, 0.5, 1, 2.5 or 10 µM for 1 h at 37°C in a humidified 5%
CO2 atmosphere), followed by incubation with 1 µM
20-HETE for 10, 30, 60, 120 or 240 min in a 37°C incubator. The
greatest change of protein expression in Nedd4-2 occurred at 1 µM
at 120 min. Subsequent analyses were performed under these
conditions. The medium was replaced with medium containing 10%
fetal bovine serum and 1% mycillin 30 min prior to harvest. For
MLN4924 treatments, the cells were treated with MLN4924 at
different concentrations (0.1, 1 or 2.5 µM for 24 h at 37°C for 120
min after 1 µM 20-HETE was added). The greatest change in protein
expression of Nedd4-2 was observed with 2.5 µM MLN4924 treatment,
and subsequent experiments were performed under these
conditions.
Immunohistochemical staining
Mice were sacrificed by decapitation. Their kidney
and liver were quickly removed and then fixed in 10% formalin
solution for 48 h at room temperature. The tissues were subjected
to routine processing for paraffin embedding, and sections (4 µm)
were placed onto glass slides coated with triethoxysilane,
air-dried, fixed in a 50% acetone/methanol solution at 4°C for 2 h,
deparaffinized with xylene, rehydrated in an alcohol series and
washed with PBS. Some of the sections were stained with hematoxylin
and eosin for 3 min at room temperature to determine the
histological type. For the remaining sections, antigen retrieval
was performed in 10 mM sodium citrate buffer (pH 6) for 10 min at
100°C, then the sections were subjected to blocking of endogenous
peroxidases by incubation for 30 min in 3%
H2O2 solution at room temperature, and
blocking non-specific binding sites with 1/100 diluted goat serum
for 40 min at room temperature. After blocking, the sections were
incubated with primary antibodies (1:100) at 4°C overnight.
Sections were washed with PBS and incubated with biotin-conjugated
secondary antibodies for 30 min. The sections were washed with PBS
and avidin-biotin complex (1:1,000) was added to the sections for
60 min at room temperature. Following extensive washing with PBS,
immunoreactive products were visualized by reaction with
3,3′-diaminobenzidine, catalyzed by horseradish peroxidase in the
presence of H2O2. Sections were
counterstained with Gill's hematoxylin for 3 min at room
temperature, dehydrated in ascending methanol series, cleared with
xylene and mounted under a coverslip. All Images were observed
using light microscope (Nikon Eclipse CI; Nikon Corporation, Tokyo,
Japan).
Western blot analysis
Total protein was extracted from cells (6 well
plates, 2.5×106 cells in each well) or tissues (weighing
150 µg) and the concentration was determined with the Bradford
method. Proteins (40 µg) were denatured at 95°C for ≥5 min and were
separated by one-dimensional 10% SDS-PAGE and transferred onto
polyvinylidene fluoride (PVDF) membranes (catalog no. ISEQ00010;
EMD Millipore, Billerica, MA, USA). Membranes were subsequently
blocked with skimmed milk powder [5%, diluted in 1% TBS containing
0.5% Tween-20 (TBST)] at room temperature for 2 h. Then the PVDF
membrane was washed with TBST. After that the membrane was
incubated with primary antibodies against Nedd4-2 (1:1,000), Nedd8
(1:1,000), Senp8 (1:1,000), tubulin (1:10,000) or GAPDH (1:10,000)
at 4°C overnight with gentle shaking. Membranes were incubated with
horseradish peroxidase-conjugated goat anti-rabbit IgG (catalog no.
ab97051; 1:5,000) or goat anti-mouse IgG (catalog no. ab97023;
1:5,000) (both from Abcam) for >1 h at room temperature.
Following washing with TBST, protein bands were visualized with an
Enhanced Chemiluminescence kit (catalog no. B18005; Biotool;
Selleck Chemicals, Shanghai, China). Densitometry was performed
using Quantity One software version 4.2 (Bio-Rad Laboratories,
Inc., Hercules, CA, USA) and normalized to GAPDH or tubulin.
Co-immunoprecipitation (co-IP)
Kidney and liver tissue (weighing 800 µg) were
prepared by homogenizing the frozen tissues or cells in four 75
cm2 cell culture flasks in lysis buffer containing
protease inhibitors, and the concentration was determined by the
Bradford method. A total of 500 µg protein was incubated with
Nedd4-2 primary antibody (1:500, catalog no. 13690-1-AP,
ProteinTech Group, Inc. Chicago, IL, USA) rotating at 4°C for 3 h
overnight. Subsequently, protein A/G PLUS-agarose (30 µl; Santa
Cruz Biotechnology, Inc., Dallas, TX USA) was added and incubated
with rotation at 4°C overnight. The protein A/G beads were then
collected by centrifugation (4°C, 2,200 × g, 5 min) and washed
three times with TBST. Immunoprecipitates were incubated with 2X
loading buffer (Beijing TransGen Biotech, Co., Ltd., Beijing,
China) and immediately denatured 95°C for >5 min and analyzed by
western blotting.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA (1 µg) was extracted from tissues (50 µg)
and cells (6 well plates, 2.5×106 cells in each) using
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA), and was reverse transcribed with random primers
using the Reverse Transcription Reagent kit (Takara Biotechnology
Co., Ltd., Dalian, China)), following the manufacturer's protocol.
qPCR was performed using SYBR Premix Ex Taq II (catalog no. RR820;
Takara Biotechnology Co., Ltd.), according to the manufacturer's
protocol, and the Applied Biosystems 7500 Real-Time PCR System
(Thermo Fisher Scientific, Inc.). The qPCR reaction condition was
as follows: 95°C for 5 min; followed by 40 cycles of 95°C for 15
sec and 60°C for 50 sec. As an internal control, GAPDH was measured
under the same conditions in RT-qPCR. Primer sequences were:
Nedd4-2, forward 5′-TGAAGCCCAATGGGTCAGAAATA-3′, reverse
5′-GGACCCTGTTCACAAATCTCCAC-3′; and GAPDH, forward
5′-GAAGGTGAAGGTCGGAGTC-3′, reverse 5′-GAAGATGGTGATGGGATTTC-3′. mRNA
expression levels were quantified using the 2−ΔΔCq and
normalized to the internal reference gene GAPDH (22).
Statistical analysis
All experimental data were derived from at least
three independent experiments. All expression data were presented
as mean ± standard error of the mean. Statistical analysis of two
samples was performed with Student's t-test analysis. Statistical
analyses among multiple groups were performed using a one-way
analysis of variance followed by Bonferroni's post hoc test.
Analysis was performed using SPSS 17.0 (SPSS, Inc., Chicago, IL,
USA). P<0.05 was considered to indicate a statistically
significant difference.
Results
Comparison of Nedd4-2 expression
between CYP4F2 transgenic mice and wild-type mice
As 20-HETE is produced mainly in the kidney and
liver, the present study aimed to determine the effects of 20-HETE
on Nedd4-2 expression in kidney and liver tissues by comparing the
expression levels of Nedd4-2 between CYP4F2 transgenic mice
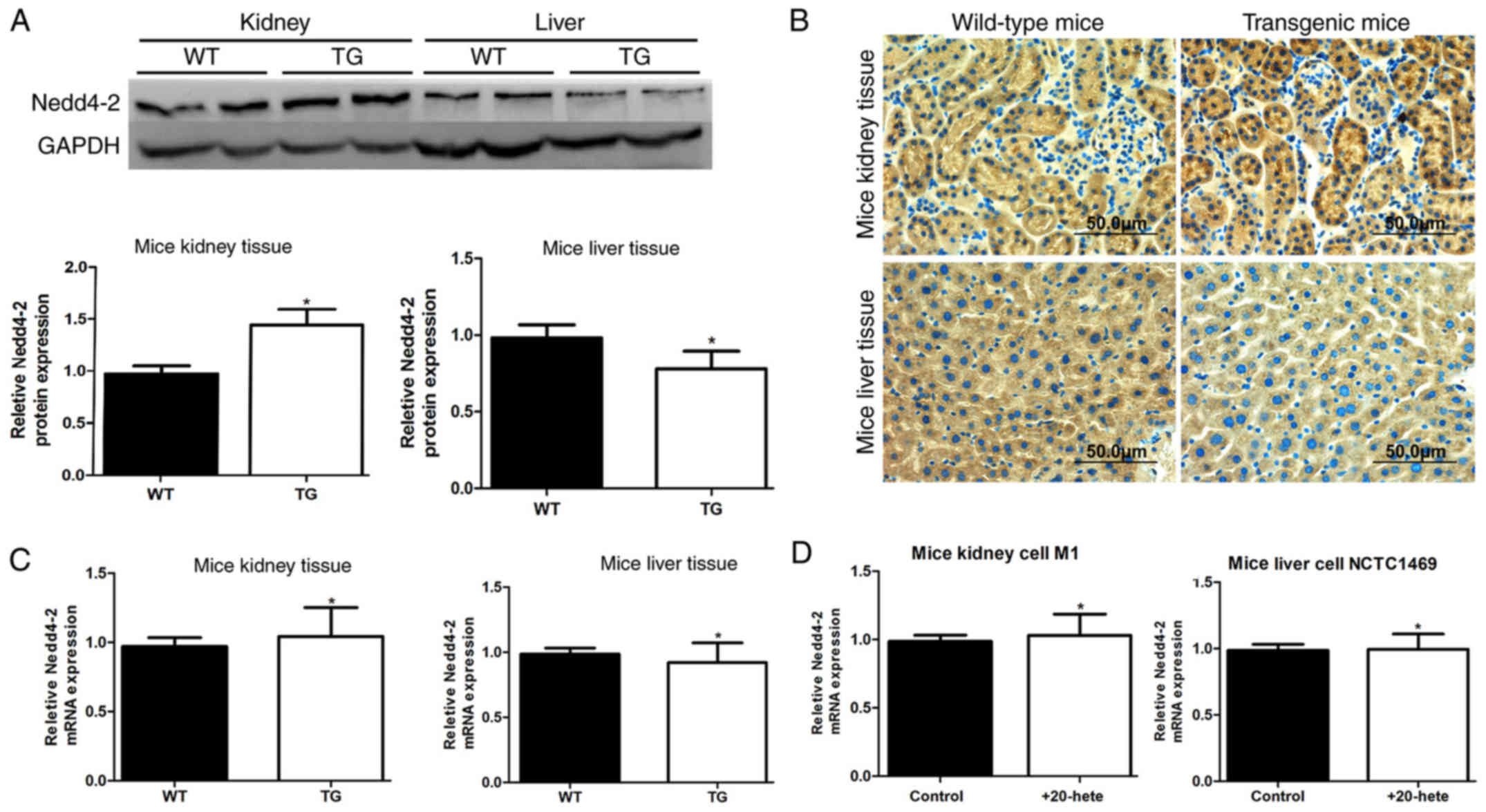
and wild-type control mice. Western blotting results demonstrated
that Nedd4-2 protein is expressed at higher levels in kidney and
lower levels in liver of transgenic mice compared to expression in
the matching tissues of wild-type mice (Fig. 1A). Immunohistochemical analysis
confirmed that Nedd4-2 protein expression was higher in transgenic
kidney tissues and lower in transgenic liver tissues compared with
the wild-type control tissues (Fig.
1B). Conversely, RT-qPCR results identified no significant
differences between the Nedd4-2 mRNA expression levels in kidney
and liver of transgenic mouse compared with wild-type mice
(Fig. 1C). Similarly, no
significant differences were identified between the Nedd4-2 mRNA
expression levels in mouse kidney cell line M1 and mouse liver cell
line NCTC1469 when 20-HETE was added (Figs. 1D and 4B). These results indicated that 20-HETE
was able to regulate Nedd4-2 expression in a tissue-specific
manner, and this regulation was at the post-transcriptional
level.
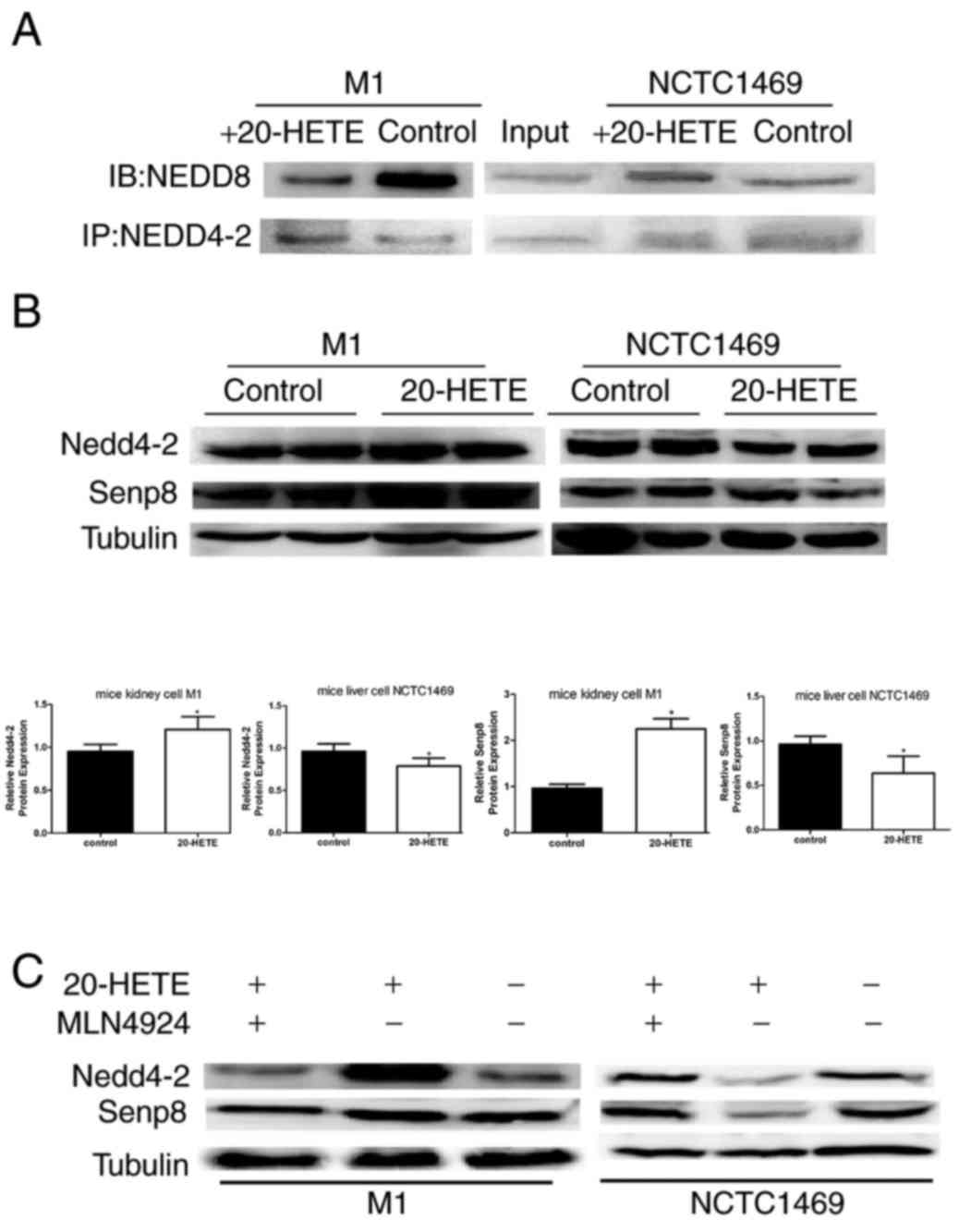 | Figure 4.20-HETE affects the neddylaton of
Nedd4-2 in cell culture through Senp8. (A) In M1 mouse kidney
cells, Nedd4-2 was less modified by Nedd8 following addition of
20-HETE to the culture medium, whereas the opposite was observed in
NCTC1469 mouse liver cells. (B) Western blotting in cell cultures
demonstrated that 20-HETE treatment promoted the expression of
Senp8 in M1 cells, but inhibited its expression in NCTC1469, which
may result in a notable change in Nedd4-2 expression. All
experiments were performed three times independently; n=6 (the
tissues in these 3 independent experiments were from 6 mice
including 3 TG mice and 3 WT mice); *P<0.05 vs. control;
*P<0.05 vs. control. (C) Recovery experiments: In M1 cells
treated with 20-HETE alone, the expression level of Nedd4-2 and
Senp8 were upregulated, whereas when both MLN4924 and 20-HETE were
added, the expression of Nedd4-2 and Senp8 were recovered, compared
with untreated control; the opposite results were observed in
NCTC1469 cells. 20-HETE, 20-hydroxyeicosatetraenoic acid; Nedd4-2,
neural precursor cell expressed developmentally downregulated
4-like, E3 ubiquitin-protein ligase; Senp8, sentrin-specific
protease 8. |
Nedd4-2 is neddylated in kidney and
liver tissues
As neddylation is an important post-translational
modification of E3 ligase, whether or not Nedd4-2 is modified by
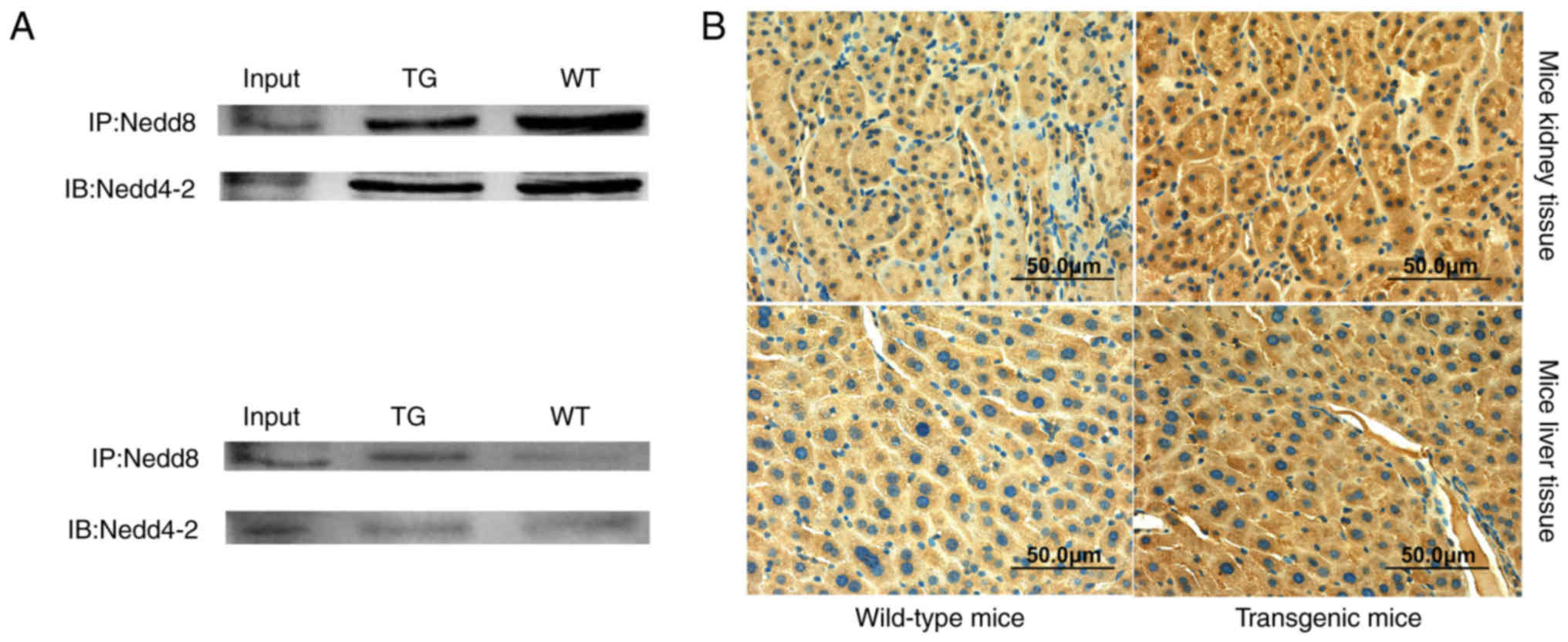
Nedd8 was examined. Co-IP was used to determine the interaction
between Nedd8 and Nedd4-2 in kidney and liver tissues, which
demonstrated that Nedd4-2 was modified by Nedd8 in both kidney and
liver tissue (Fig. 2A). However,
Nedd8 binding to Nedd4-2 was weaker in kidney and stronger in liver
of transgenic mice compared with wild-type controls, which was
opposite to protein expression in kidney and liver. In addition,
Nedd8 protein expression was detected in kidney and liver tissues
by immunohistochemistry (Fig. 2B).
Nedd8 was expressed higher in both liver and kidney tissues of
transgenic mice compared with expression in wild-type control
tissues. These data suggested that 20-HETE may reduce the
neddylation of Nedd4-2 in kidney, which may lead to increased
expression of Nedd4-2, and that 20-HETE may increase neddylation of
Nedd4-2 in liver, resulting in decreased expression of Nedd4-2 in
mice.
Expression of Senp8 in CYP4F2
transgenic mice and wild-type mice
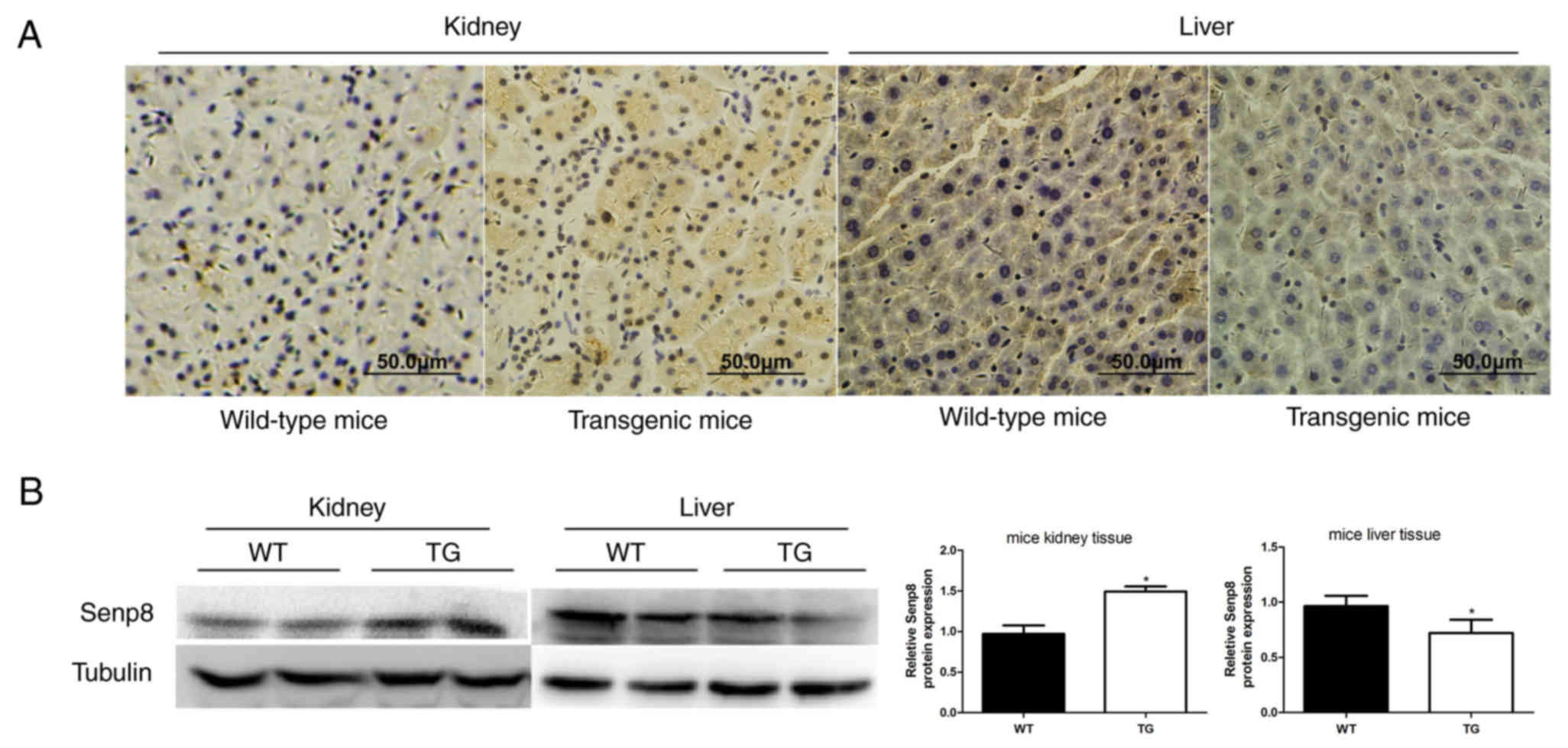
To investigate why 20-HETE regulated Nedd4-2
expression contrary to its neddylation status, expression of the
deneddylation enzyme Senp8 was examined in CYP4F2 transgenic
and in wild-type mice. Western blotting demonstrated that Senp8
expression was higher in the kidney of transgenic mice compared
with wild-type control tissues, and Senp8 expression was lower in
transgenic liver tissues compared with wild-type (Fig. 3A). Similar Senp8 protein expression
results were obtained from western blotting (Fig. 3B). These data implied that 20-HETE
may affect Senp8 expression, which may increase the dissociation of
Nedd8 from Nedd4-2 in kidney and decrease the dissociation of Nedd8
from Nedd4-2 in liver, resulting in the downregulation of Nedd4-2
neddylation in kidney and the upregulation of Nedd4-2 neddylation
in liver.
Effects of 20-HETE on neddylation of
Nedd4-2
To verify the aforementioned ex vivo results,
which indicated that differential Nedd4-2 expression may be linked
to neddylation status in kidney and liver of transgenic mice, M1
and NCTC1469 cells were treated with 20-HETE. Co-IP analysis
demonstrated that 20-HETE treatment resulted in less binding of
Nedd8 to Nedd4-2 in M1 kidney cells and increased binding in
NCTC1469 liver cells compared with the respective untreated
controls (Fig. 4A). Furthermore,
western blotting demonstrated that Nedd4-2 expression was higher in
20-HETE-treated M1 cells and lower in 20-HETE-treated NCTC1469
cells compared with untreated controls (Fig. 4B). To determine the reason for this
inverse effect of 20-HETE on neddylation of Nedd4-2, Senp8 protein
expression was determined in the same tissues. The results showed
that Senp8 expression was in line with Nedd4-2 expression (Fig. 4B). In addition, the effects of
20-HETE on Nedd4-2 were eliminated by co-culturing cell with the
neddylation inhibitor MLN4924, which demonstrated that the
20-HETE-induced increases of Nedd4-2 and Senp8 protein expression
levels in M1 cells were blocked by MLN4924, and the 20-HETE-induced
decreases of Nedd4-2 and Senp8 expression were restored by MLN4924
in NCTC1469 cells (Fig. 4C).
Therefore, these results suggested that 20-HETE may positively
regulate Nedd4-2 expression through the neddylation pathway in
kidney and negatively regulate Nedd4-2 expression in liver. This
regulation of 20-HETE performed partly through its effect on the
neddylation process.
Disccusion
A previous study suggested that the most well-known
function of neddylation is to enhance E3 ligase activity (23). The ubiquitination activity of E3
ligases depends in part on neddylation. The CRL family was the
first identified and well-studied substrate of neddylation
(14,24). Covalent binding of Nedd8 to a
conserved lysine in the C-terminal domain stimulates CRL
ubiquitination activity and prevents them from binding to the
structural-based inhibitor cullin-associated Nedd8-dissociated
protein 1 (25). However, the
function of Nedd8 covalent conjugation is not only to activate, but
also to confer an intrinsic instability (26). Chung et al demonstrated that
neddylated cullin proteins are recycled and the conjugated Nedd8 is
removed by CSN isopeptidase; otherwise, they are efficiently
degraded (27). Thus, the
neddylation/deneddylation cycle provides a way to maintain normal
cellular levels of activated E3 ubiquitin ligase and to prevent
excessive ubiquitin ligase activity (28). Nedd4-2 acts as an E3 ubiquitin
ligase of the HECT-type ubiquitin ligase family and has potential
Nedd8 covalent conjugation (15).
Neddylation of Nedd4-2 has not been reported to
date. In regard to the regulation of Nedd4-2 function,
serum/glucocorticoid-regulated kinase 1 has been reported to
phosphorylate serine residues 222 and 328 of Nedd4-2, which
promoted its binding to 14-3-3 proteins, and consequently inhibited
the interaction with its substrates, such as NKCC2 and ENaC
(29). It also has been reported
that Nedd4-2 is controlled by auto-ubiquitination; however, it may
weakly bind to the PY motif in its HECT domain through its WW
domain, which suggested that this intramolecular interaction may
inhibit its auto-ubiquitination (30). Using co-IP, the present study
identified an interaction of Nedd8 with Nedd4-2 in kidney and liver
tissues of mice, as well as in M1 and NCTC1469 cell lines. To the
best of our knowledge, this is the first preliminary report to
demonstrate that Nedd4-2 was modified by Nedd8. Consistent with
this, neddylation of Smurf1, such as with HECT-like ligase, has
been reported (15). In addition,
the data from comparisons between CYP4F2 transgenic mice and
wild-type mice demonstrated that 20-HETE reduced the neddylation of
Nedd4-2 in kidney, leading to probably increased expression of
Nedd4-2, and that 20-HETE increased the neddylation of Nedd4-2 in
liver, which probably resulted in decreased expression of Nedd4-2
in mice. To verify 20-HETE positively regulated Nedd4-2 in kidney
and negatively regulated Nedd4-2 in liver, expression of the
deneddylation enzyme Senp8 was examined between transgenic mice and
wide-type mice. Senp8, also known as DCN1, was reported more
efficient to hyper-neddylated cullins (cullins with more than one
combine site of nedd8) and non-cullins compared to mono-neddylated
ones (cullins with only one combine site of nedd8) (17,23)
The results revealed that Senp8 expression was higher in kidney and
lower in liver of transgenic mice compared with wild-type mice,
which was similar to the expression of Nedd4-2. Notably, cell
culture experiments indicated that the effects of 20-HETE on
Nedd4-2 and Senp8 expression were recovered by treatment with the
inhibitor MLN4924, which indicated that 20-HETE regulation of
Nedd4-2 and Senp8 was at least partly dependent on neddylation
modification. Therefore, it was inferred that increased levels of
Senp8 dissociated more Nedd8 from Nedd4-2 in kidney to interfere
with self-ubiquitination and degradation, and that the opposite
occurred in the liver.
Notably, 20-HETE may function in a tissue-specific
manner in the kidney and liver: The present study demonstrated that
20-HETE increased the expression of Nedd4-2 and Senp8 in kidney and
decreased their expression levels in the liver. In addition,
20-HETE decreased the neddylation of Nedd4-2 in kidney and
increased Nedd4-2 neddylation in the liver. Similarly, previous
studies have demonstrated tissue-specific expression in the
vascular system. On the one hand, 20-HETE was reported to be a
vasoconstrictor in renal and cerebral arteries through activation
of the protein kinase C or mitogen-activated protein kinase pathway
(31). In addition, 20-HETE may
also directly affect the L-type Ca2+ channels on the
cell membrane to increase the concentration of intracellular
Ca2+, thus triggering vasoconstriction (32). In addition to the vascular system,
Nowicki et al have reported that 20-HETE inhibited a
cAMP-dependent pathway in the kidney of rats that consumed a
potassium-deficient diet (33). By
contrast, results from our previous study demonstrated that 20-HETE
activated the cAMP pathway in the liver of CYP4F2 transgenic
mice (34). In blood pressure
regulation, 20-HETE exhibited both prohypertensive and
antihypertensive actions through vasoconstriction and natriuresis.
This dual role was also evident in or CYP4F2 transgenic
mice: 20-HETE had a prohypertensive role in the transgenic mice fed
a normal salt diet, whereas 20-HETE exhibited an antihypertensive
role by promoting natriuresis in transgenic mice fed a high salt
diet (4). This demonstrated that
20-HETE serves different roles in various tissues. The role of
20-HETE is extensive and complicated. These data may provide an
insight into the function of 20-HETE on neddylation modification of
Nedd4-2. 20-HETE not only participated in blood pressure regulation
but was also involved in tumor formation and progression (35), kidney diseases (36) and plasma glucose regulation
(34). The present study
demonstrated that 20-HETE regulates the expression of Nedd4-2 and
Senp8 positively in kidney and negatively in liver. One possible
explanation for the differential regulation is that 20-HETE may
activate different pathways in different tissues, as 20-HETE has
high lipid solubility and is able to freely pass through the cell
membrane and exert multiple functions (37). Further examinations of
20-HETE-induced pathways in kidney and liver are required. Another
possible explanation is that 20-HETE may have its own receptor that
may be distributed differently in various tissues. Recently,
several studies have sought to identify a 20-HETE receptor
(38,39), which may help to explain the
regulation of 20-HETE in kidney and liver.
In summary, the present study demonstrated that
20-HETE upregulated Senp8 expression, which enhanced the
deneddylation of Nedd4-2 and interfered with Nedd4-2 degradation,
and subsequently resulted in an increased expression level of
Nedd4-2 in kidney, and that the opposite effects occured in the
liver. The mechanisms by which 20-HETE positively regulated Nedd4-2
in kidney and negatively regulated Nedd4-2 in the liver remain to
be fully understood, but the regulation of 20-HETE between
neddylation and deneddylation of Nedd4-2 may be involved.
Acknowledgements
This study was supported by The National Natural
Science Foundation of China (grant nos. 81270343 and 31571198) and
The Outstanding Scientific Fund of Shengjing Hospital (grant no.
201601).
Glossary
Abbreviations
Abbreviations:
|
20-HETE
|
20-hydroxyeicosatetraenoic acid
|
|
Nedd4-2
|
neural precursor cell expressed
developmentally downregulated 4-like, E3 ubiquitin-protein
ligase
|
|
CYP4F2
|
cytochrome P450 family 4 subfamily F
member 2
|
References
|
1
|
Medhora M, Chen Y, Gruenloh S, Harland D,
Bodiga S, Zielonka J, Gebremedhin D, Gao Y, Falck JR, Anjaiah S and
Jacobs ER: 20-HETE increases superoxide production and activates
NAPDH oxidase in pulmonary artery endothelial cells. Am J Physiol
Lung Cell Mol Physiol. 294:L902–L911. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Pratt PF, Falck JR, Reddy KM, Kurian JB
and Campbell WB: 20-HETE relaxes bovine coronary arteries through
the release of prostacyclin. Hypertension. 31:237–241. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Liu X, Zhao Y, Wang L, Yang X, Zheng Z,
Zhang Y, Chen F and Liu H: Overexpression of cytochrome P450 4F2 in
mice increases 20-hydroxyeicosatetraenoic acid production and
arterial blood pressure. Kidney Int. 75:1288–1296. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wu J, Liu X, Lai G, Yang X, Wang L and
Zhao Y: Synergistical effect of 20-HETE and high salt on NKCC2
protein and blood pressure via ubiquitin-proteasome pathway. Hum
Genet. 132:179–187. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Harvey KF, Dinudom A, Cook DI and Kumar S:
The Nedd4-like protein KIAA0439 is a potential regulator of the
epithelial sodium channel. J Biol Chem. 276:8597–8601. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rougier JS, van Bemmelen MX, Bruce MC,
Jespersen T, Gavillet B, Apothéloz F, Cordonier S, Staub O, Rotin D
and Abriel H: Molecular determinants of voltage-gated sodium
channel regulation by the Nedd4/Nedd4-like proteins. Am J Physiol
Cell Physiol. 288:C692–C701. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kamynina E, Debonneville C, Bens M,
Vandewalle A and Staub O: A novel mouse Nedd4 protein suppresses
the activity of the epithelial Na+ channel. FASEB J. 15:204–214.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Laedermann CJ, Cachemaille M, Kirschmann
G, Pertin M, Gosselin RD, Chang I, Albesa M, Towne C, Schneider BL,
Kellenberger S, et al: Dysregulation of voltage-gated sodium
channels by ubiquitin ligase NEDD4-2 in neuropathic pain. J Clin
Invest. 123:3002–3013. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hellwinkel OJ, Asong LE, Rogmann JP,
Sültmann H, Wagner C, Schlomm T and Eichelberg C: Transcription
alterations of members of the ubiquitin-proteasome network in
prostate carcinoma. Prostate Cancer Prostatic Dis. 14:38–45. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kito Y, Bai J, Goto N, Okubo H, Adachi Y,
Nagayama T and Takeuchi T: Pathobiological properties of the
ubiquitin ligase Nedd4L in melanoma. Int J Exp Pathol. 95:24–28.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wen H, Lin R, Jiao Y, Wang F, Wang S, Lu
D, Qian J, Jin L and Wang X: Two polymorphisms in NEDD4L gene and
essential hypertension in Chinese Hans-a population-based
case-control study. Clin Exp Hypertens. 30:87–94. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Whitby FG, Xia G, Pickart CM and Hill CP:
Crystal structure of the human ubiquitin-like protein NEDD8 and
interactions with ubiquitin pathway enzymes. J Biol Chem.
273:34983–34991. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Huang DT, Hunt HW, Zhuang M, Ohi MD,
Holton JM and Schulman BA: Basis for a ubiquitin-like protein
thioester switch toggling E1-E2 affinity. Nature. 445:394–398.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Pan ZQ, Kentsis A, Dias DC, Yamoah K and
Wu K: Nedd8 on cullin: Building an expressway to protein
destruction. Oncogene. 23:1985–1997. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Xie P, Zhang M, He S, Lu K, Chen Y, Xing
G, Lu Y, Liu P, Li Y, Wang S, et al: The covalent modifier Nedd8 is
critical for the activation of Smurf1 ubiquitin ligase in
tumorigenesis. Nat Commun. 5:37332014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Boh BK, Smith PG and Hagen T:
Neddylation-induced conformational control regulates cullin RING
ligase activity in vivo. J Mol Biol. 409:136–145. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Goldenberg SJ, Cascio TC, Shumway SD,
Garbutt KC, Liu J, Xiong Y and Zheng N: Structure of the
Cand1-Cul1-Roc1 complex reveals regulatory mechanisms for the
assembly of the multisubunit cullin-dependent ubiquitin ligases.
Cell. 119:517–528. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kapelari B, Bech-Otschir D, Hegerl R,
Schade R, Dumdey R and Dubiel W: Electron microscopy and
subunit-subunit interaction studies reveal a first architecture of
COP9 signalosome. J Mol Biol. 300:1169–1178. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wei W, Guo H, Liu X, Zhang H, Qian L, Luo
K, Markham RB and Yu XF: A first-in-class NAE inhibitor, MLN4924,
blocks lentiviral infection in myeloid cells by disrupting
neddylation-dependent Vpx-mediated SAMHD1 degradation. J Virol.
88:745–751. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sarantopoulos J, Shapiro GI, Cohen RB,
Clark JW, Kauh JS, Weiss GJ, Cleary JM, Mahalingam D, Pickard MD,
Faessel HM, et al: Phase I study of the investigational
NEDD8-activating enzyme inhibitor pevonedistat (TAK-924/MLN4924) in
patients with advanced solid tumors. Clin Cancer Res. 22:847–857.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shah JJ, Jakubowiak AJ, O'Connor OA,
Orlowski RZ, Harvey RD, Smith MR, Lebovic D, Diefenbach C, Kelly K,
Hua Z, et al: Phase I study of the novel investigational
NEDD8-activating enzyme inhibitor Pevonedistat (MLN4924) in
patients with relapsed/refractory multiple myeloma or lymphoma.
Clin Cancer Res. 22:34–43. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bornstein G and Grossman C:
COP9-Signalosome deneddylase activity is enhanced by simultaneous
neddylation: Insights into the regulation of an enzymatic protein
complex. Cell Div. 10:52015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wu K, Chen A and Pan ZQ: Conjugation of
Nedd8 to CUL1 enhances the ability of the ROC1-CUL1 complex to
promote ubiquitin polymerization. J Biol Chem. 275:32317–32324.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kurz T, Ozlü N, Rudolf F, Luke B, Hofmann
K, Hyman AA, Bowerman B and Peter M: The conserved protein
DCN-1/Dcn1p is required for cullin neddylation in C. Elegans and S.
Cerevisiae. Nature. 435:1257–1261. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kurz T, Chou YC, Willems AR,
Meyer-Schaller N, Hecht ML, Tyers M, Peter M and Sicheri F: Dcn1
functions as a scaffold-type E3 ligase for cullin neddylation. Mol
Cell. 29:23–35. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chung D and Dellaire G: The Role of the
COP9 signalosome and neddylation in DNA damage signaling and
repair. Biomolecules. 5:2388–2416. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wu JT, Lin HC, Hu YC and Chien CT:
Neddylation and deneddylation regulate Cul1 and Cul3 protein
accumulation. Nat Cell Biol. 7:1014–1020. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chandran S, Li H, Dong W, Adams C,
Alexandrova L, Chien A, Hallows KR and Bhalla V: Neural precursor
cell-expressed developmentally down-regulated protein 4-2 (Nedd4-2)
regulation by 14-3-3 protein binding at canonical serum and
glucocorticoid kinase 1 (SGK1) phosphorylation sites. J Biol Chem.
286:37830–37840. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bruce MC, Kanelis V, Fouladkou F,
Debonneville A, Staub O and Rotin D: Regulation of Nedd4-2
self-ubiquitination and stability by a PY motif located within its
HECT-domain. Biochem J. 415:155–163. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Singh H and Schwartzman ML: Renal vascular
cytochrome P450-derived eicosanoids in androgen-induced
hypertension. Pharmacol Rep. 60:29–37. 2008.PubMed/NCBI
|
|
32
|
Mulligan SJ and MacVicar BA: Calcium
transients in astrocyte endfeet cause cerebrovascular
constrictions. Nature. 431:195–199. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Nowicki S, Chen SL, Aizman O, Cheng XJ, Li
D, Nowicki C, Nairn A, Greengard P and Aperia A:
20-Hydroxyeicosa-tetraenoic acid (20 HETE) activates protein kinase
C. Role in regulation of rat renal Na+,K+-ATPase. J Clin Invest.
99:1224–1230. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lai G, Wu J, Liu X and Zhao Y: 20-HETE
induces hyperglycemia through the cAMP/PKA-PhK-GP pathway. Mol
Endocrinol. 26:1907–1916. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yu W, Chen L, Yang YQ, Falck JR, Guo AM,
Li Y and Yang J: Cytochrome P450 ω-hydroxylase promotes
angiogenesis and metastasis by upregulation of VEGF and MMP-9 in
non-small cell lung cancer. Cancer Chemother Pharmacol. 68:619–629.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Park F, Sweeney WE, Jia G, Roman RJ and
Avner ED: 20-HETE mediates proliferation of renal epithelial cells
in polycystic kidney disease. J Am Soc Nephrol. 19:1929–1939. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Escalante B, Erlij D, Falck JR and McGiff
JC: Cytochrome P450-dependent arachidonate metabolites affect renal
transport in the rabbit. J Cardiovasc Pharmacol. 22 Suppl
2:S106–S108. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ward NC, Chen K, Li C, Croft KD and Keaney
JF Jr: Chronic activation of AMP-activated protein kinase prevents
20-hydroxyeicosatetraenoic acid-induced endothelial dysfunction.
Clin Exp Pharmacol Physiol. 38:328–333. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Sodhi K, Wu CC, Cheng J, Gotlinger K,
Inoue K, Goli M, Falck JR, Abraham NG and Schwartzman ML:
CYP4A2-induced hypertension is 20-hydroxyeicosatetraenoic acid- and
angiotensin II-dependent. Hypertension. 56:871–878. 2010.
View Article : Google Scholar : PubMed/NCBI
|