Introduction
Dental pulp stem cells (DPSCs) are important
mesenchymal stem cells in dental pulp tissue. Due to simple
isolation and culture and certain amplification ability and
multipotentiality DPSCs provide a new cell source for odontogenic
tissue-engineered repair. The cell is usually obtained from
naturally shedding teeth or extracted teeth in pathological
situation, which provide a rich source, extremely promoting the
application of DPSCs in the treatment of dental diseases (1–4). It
has been proved that DPSCs have the ability of odontoblast
differentiation. In the case of dentin or odontoblast damage, DPSCs
can also differentiate into new functional odontoblasts to form
dentin structure, accomplishing tooth restoration (2,5). In
recent years, how to optimize the function of DPSCs differentiating
to odontoblasts and better achieve regeneration of dentin has been
a hot issue in the field of oral medicine (4). The process of DPSCs differentiation
involves many transcription factors associated with stem cell
differentiation. Previous research has indicated that DPSCs serve
the role of injury repair mainly by activating the Wnt signaling
pathway (6). As an important
transcription factor, SRY-box 2 (SOX2) has important significance
in maintaining pluripotency of stem cells and somatic cell
reprogramming (7–10). Simultaneously, it is also
demonstrated in some research that SOX2 can induce neural
differentiation of stem cells. In addition, SOX2, coupled with
other transcription factors, can directly transform somatic cells
without differentiation ability into nerve cells, which provides a
novel technological solution to the cell therapy of nervous system
disease (11,12).
Our previous research indicated that the
proliferation, migration and adhesion of DPSCs could were increased
by SOX2 overexpression in DPSCs, which demonstrated that SOX2 has a
certain effect on the biological characteristics of DPSCs (13). Though SOX2 can induce neural
differentiation of stem cells at a certain degree, it is still not
clear whether SOX2 has a certain effect on odontoblast
differentiation of DPSCs. By referring to the previous research
method (13), the present study
was carried out to further explore the effect of SOX2 on
odontoblast differentiation of DPSCs by regulating the Wnt
signaling pathway.
Materials and methods
Reagents and equipment
Dulbecco's modified Eagle's medium/F12, BME culture
medium and fetal bovine serum (FBS) were purchased from Hyclone;
Thermo Fisher Scientific, Inc. (Waltham, MA, USA). Sodium
β-glycerophosphate, dexamethasone and vitamin C were bought from
Sigma-Aldrich; Merck KGaA (Darmstadt, Germany). Transforming growth
factor (TGF)-β1 was purchased from R&D Systems, Inc.
(Minneapolis, MN, USA). TRIzol was purchased from Invitrogen;
Thermo Fisher Scientific, Inc. RNA reverse transcription kits and
the quantitative polymerase chain reaction (qPCR) machine were
bought from Takara Biotechnology Co., Ltd. (Dalian, China). All
primer sequences were synthesized by Sangon Biotech Co., Ltd.
(Shanghai, China; www.sangon.com/). Mouse anti-human dentin matrix
protein-1 (DMP-1; cat. no. BS7995), anti-dentin sialophosphoprotein
(DSPP; cat. no. BS71212) and anti-Wnt7b monoclonal antibodies (cat.
no. BS71443) were purchased from Bioworld Technology, Inc. (St.
Louis Park, MN, USA). A goat anti-mouse immunoglobulin (Ig)
G-fluorescein isothiocyanate (FITC) polyclonal antibody (cat. no.
BS20509) was bought from Invitrogen; Thermo Fisher Scientific, Inc.
A human cell genomic expression microarray was purchased from
Agilent Technologies, Inc. (Santa Clara, CA, USA).
An IX-70 inverted phase microscope was purchased
from Olympus Corporation (Tokyo, Japan). PCR equipment was
purchased from Eastwin Life Sciences, Inc. (Bejing, China;
http://www.eastwin.com.cn/). CFX96 qPCR
equipment was bought from Bio-Rad Laboratories, Inc. (Hercules, CA,
USA) Company. A FACSCalibur flow cytometer was purchased from BD
Biosciences (Franklin Lakes, NJ, USA).
Cell lines
Isolation and identification of primary DPSCs from
human subjects was performed as described previously (13). A total of 37 patients ranging from
20–65 years old (16 males, 21 females) were screened, out of the
patients treated in Henan Provincial People's Hospital (Zhengzhou,
China) in December 2015 and the pulp tissue was collected. DPSCs
were isolated and incubated under sterile conditions in research
center of the Henan Provincial People's Hospital. The empty vector
control-infected (DPSCs-vector) and the SOX2-overexpressing
(DPSCs-SOX2) cell lines were generated as described previously
(13), and the stable cell lines
were used in the present study. The present study was approved by
the ethics committee of Henan Provincial People's Hospital
(Zhengzhou, China), and written informed consent was obtained from
all patients.
Odontoblast differentiation of
DPSCs
The method of odontoblast differentiation of DPSCs
performed as described previously (14). DPSCs in the logarithmic growth
phase were collected, and seeded into 24-well plates at a density
of 2×105 cells/well. The cells were cultivated with
odontoblast differentiation medium (10% FBS, 10−3 mm
dexamethasone, 10 mm sodium β-glycerophosphate, 50 mg/l vitamin C
and 5 ng/ml TGF-β1 were added into BME basal medium), and
differentiated for 3 weeks.
Flow cytometry
After the induction and differentiation, flow
cytometry was used to test the expression levels of DSPP and DMP-1
in the DPSC, DPSC-vector and DPSC-SOX2 groups to analyze the effect
of odontoblast differentiation of DPSCs. Then DPSCs in each
experimental group were fixed and penetrated with 4%
paraformaldehyde at room temperature with 20 mins, and mouse
anti-human DSPP and DMP-1 monoclonal antibodies (1:50) in FBS
(Hyclone; GE Healthcare Life Sciences, Logan, UT, USA) were
incubated with cells for 30 min at room temperature. After cleaning
twice with PBS, a goat anti-mouse IgG-FITC polyclonal antibody
(1:400) was incubated with the cells for 30 min at room
temperature. Following cleaning twice with PBS again, the cells
were detected using a FACSCalibur flow cytometer (FACS101; BD
Biosciences).
For analysis of Wnt7b expression, a mouse anti-human
Wnt7b monoclonal antibody was diluted to 1:50, and a goat
anti-mouse IgG-FITC polyclonal antibody was diluted to 1:400. The
assay was performed at room temperature for 1 h. After cleaning
with PBS, these antibodies were detected using a FACSCalibur flow
cytometer.
Microarray analysis
A microarray analysis comparing the gene expression
profiles of the DPSCs-vector and the DPSCs-SOX2 cell lines was
performed as previously described (13), and the results are publicly
available in the Gene Expression Omnibus database (accession no.
GSE73548).
Reverse transcription (RT)-qPCR
TRIzol was used to extract RNA from cells. cDNA was
generated using an RNA reverse transcription kit. RT-qPCR was used
to confirm the expression of genes in the Wnt signaling pathway
that were identified as significantly altered by the previous
microarray analysis but were not described or confirmed previously
(15). Primer sequences for
RT-qPCR were: Wnt1 forward, 5′-CGATGGTGGGGTATTGTGAAC-3′ and
reverse, 5′-CCGGATTTTGGCGTATCAGAC-3′; Wnt7b forward,
5′-GAAGCAGGGCTACTACAACCA-3′ and reverse,
5′-CGGCCTCATTGTTATGCAGGT-3′; Wnt8a forward,
5′-GAACTGCCCTGAAAATGCTCT-3′ and reverse,
5′-TCGAAGTCACCCATGCTACAG-3′; Wnt11 forward,
5′-GACCTCAAGACCCGATACCTG-3′ and reverse;
5′-TAGACGAGTTCCGAGTCCTTC-3′; and β-actin forward,
5′-CCCAGAGCAAGAGAGG-3′ and reverse, 5′-GTCCAGACGCAGGATG-3′. With
the fluorophore SYBR Advantage qPCR Premix (cat. nos. 638321 and
639676; Takara Bio, Inc., Otsu, Japan), used so any alterations
were clearly observable. The 2−∆∆Cq method was used for
quantification (15). As the mRNA
expression of Wnt7b was revealed to be the most significantly
upregulated, the protein expression level of Wnt7b was subsequently
assessed by flow cytometry. The thermocycling conditions were:
Predenaturing at 95°C, 5 min; denaturing at 94°C, 30 sec, annealing
at 64°C, 30 sec, extension (72°C, 30 sec) with 32 cycles and
terminal extension (72°C, 5 min).
Statistical analysis
SPSS 17.0 (SPSS, Inc., Chicago, IL, USA) was used to
analyze clinical data. Data are presented as mean ± standard error.
A student's t-test was used for comparison between groups, and
two-way analysis of variance was used for multiple comparisons.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Odontoblast differentiation of
DPSCs
In our previous study, isolated DPSCs were
demonstrated to exhibit a mesenchymal phenotype, to possess
osteogenic and adipogenic differentiation potential, and to have
increased proliferation, migration and adherence function following
SOX2 overexpression (13). In the
present study, the odontoblast differentiation ability among the
normal DPSCs, DPSCs-vector and DPSCs-SOX2 groups was examined. The
results revealed that there was no significant difference among
normal DPSCs, DPSCs-vector and DPSCs-SOX2 groups in morphology, and
that the cells in all groups grew by adherence in long spindle
shapes (Fig. 1). After the
induction of odontoblast differentiation, the cells in three groups
could express the odontoblast special markers DSPP and DMP-1, but
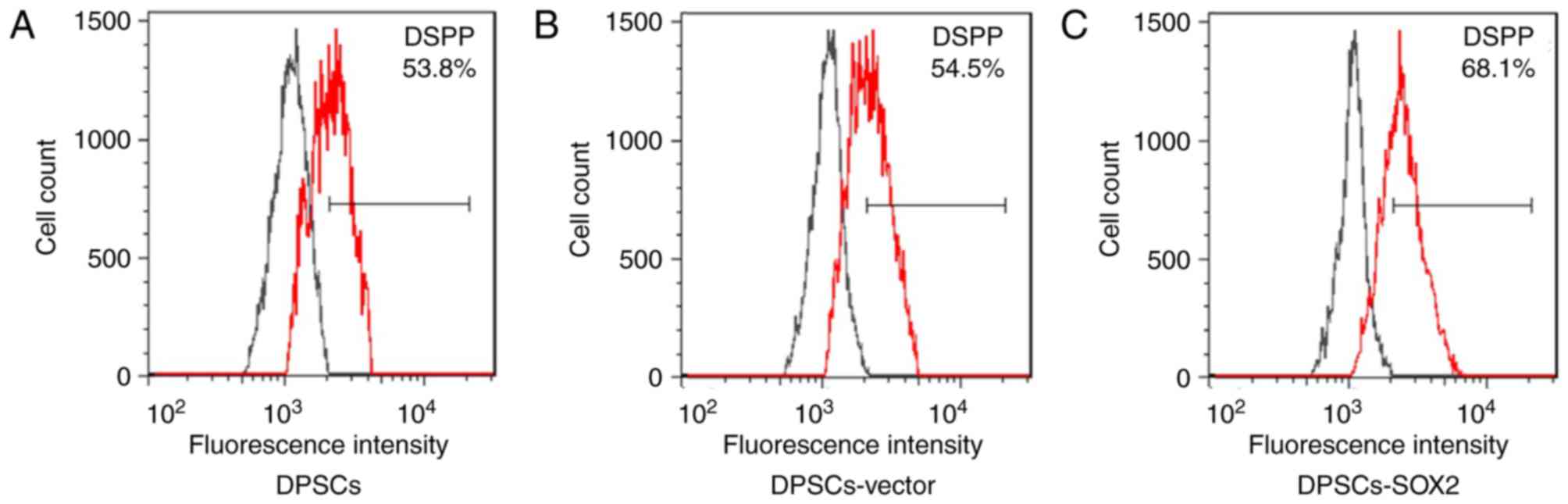
there was some difference in expression degree. After DPSCs-SOX2
was induced to odontoblast differentiation, the expression
efficiency of DSPP in DPSCs-SOX2 group was 68.1%, which was higher
compared with the normal DPSC group (53.8%) and in the DPSCs-vector
group (54.5%; P<0.05; Fig. 2).
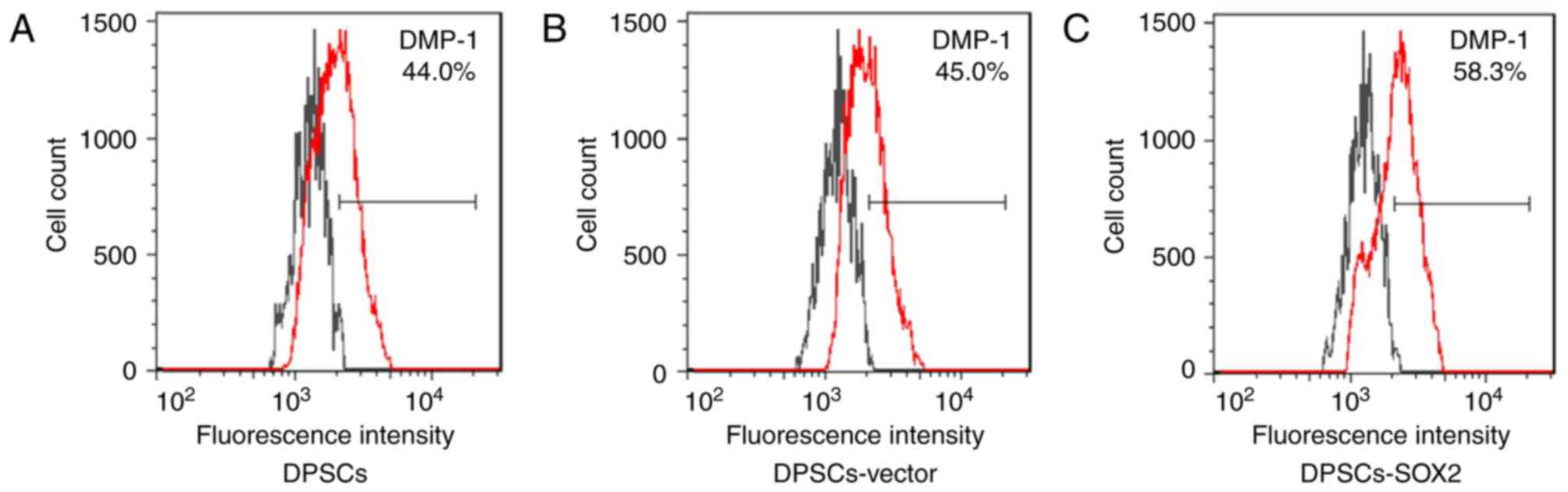
The expression efficiency of DMP-1 in the DPSCs-SOX2 group was
58.3%, which was higher than the DPSC group (44.0%) and the
DPSC-vector group (45.0%; Fig.
3).
Analysis on mechanism of SOX2
affecting osteogenic differentiation of DPSCs
The change of signaling pathways in DPSCs after SOX2
overexpression was preliminarily evaluated by genomic expression
microarray, and the effect of SOX2 on odontoblast differentiation
of DPSCs was analyzed. The results from a previously published
microarray analysis comparing the expression profiles of the
DPSCs-vector and DPSCs-SOX2 cells indicated that several genes
involved in the Wnt pathway were significantly upregulated
following SOX2 overexpression (16). The Wnt signaling pathway serves an
important role in regulating function in tooth development, and
also has a significant effect on dentinal formation (17). Therefore, the Wnt signalling
pathway may have a role in the SOX2-mediated induction of
odontoblast differentiation.
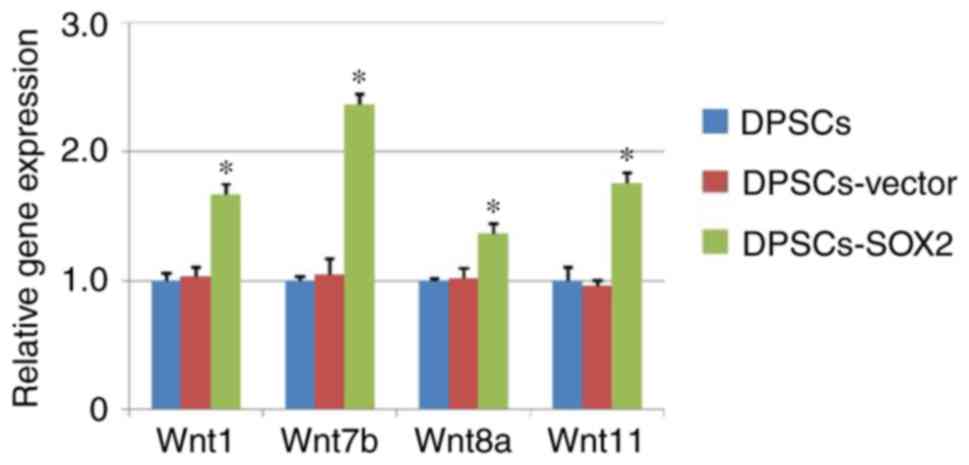
According to analysis results of genome expression
microarray, promoter genes (Wnt1, Wnt7b, Wnt8a and Wnt11) in the
Wnt signaling pathway were evaluated by RT-qPCR and flow cytometry.
The results of RT-qPCR demonstrated that Wnt1, Wnt7b, Wnt8a and
Wnt11 in the DPSCs-SOX2 group all were upregulated compared with
the DPSC and DPSCs-vector groups. Among these promoter genes, the
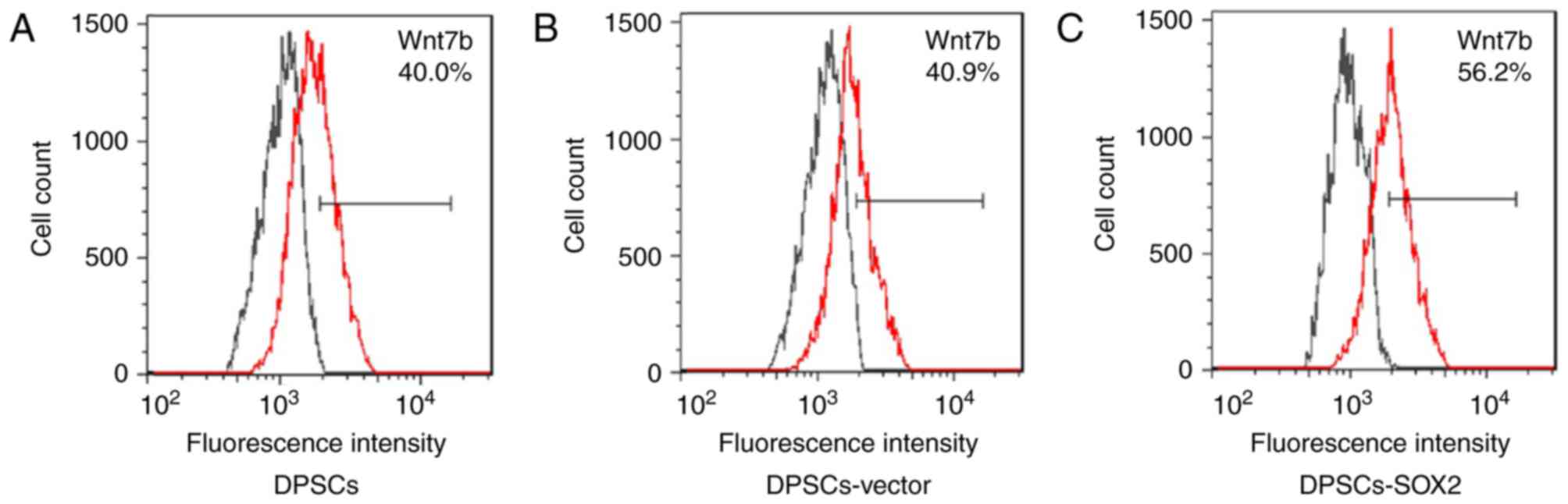
upregulation of Wnt7b was highest (Fig. 4). Therefore, the expression level
of Wnt7b at the protein level in each experimental group was
further analyzed by flow cytometry. The results demonstrated that
the expression Wnt7b protein in each experimental group was
consistent with the result of RT-qPCR. The expression efficacy of
Wnt7b in the DPSCs-SOX2 group (56.2%) was highest compared with the
DPSC (40.0%) and DPSC-vector (40.9%) groups (Fig. 5).
Discussion
Previous studies have demonstrated that DPSCs has an
ability of odontoblast differentiation. DPSCs serve an important
role in dentin regeneration and injury repair, especially in the
case of tooth injury (1–4). However, as a pluripotent regulatory
gene and a neurogenic differentiation factor, it is still unclear
about the effect of SOX2 on odontoblast differentiation of DPSCs.
This study was carried out to investigate this issue. On the basis
of differentiation experiments and mechanism analysis, the
experimental results in this research primarily demonstrated that
SOX2 overexpression in DPSCs could upregulate the expression of
DSPP and DMP-1, and promote odontoblast differentiation of DPSCs.
Simultaneously, it was demonstrated by microarray analysis that the
Wnt signaling pathway was more active in the DPSCs-SOX2 group
compared with the DPSC group; many key genes were upregulated,
which was also revealed by RT-qPCR and flow cytometry. The
activation of the Wnt signaling pathway might be an important
mechanism of SOX2 promoting the odontoblast differentiation of
DPSCs.
The Wnt signaling pathway has an important effect on
tooth development, and is an important signal in the process of
dentin formation (18,19). By using techniques like microarray,
the present results indicated that Wnt1, Wnt7b, Wnt8a and Wnt11 all
were upregulated following SOX2 overexpression. These factors are
activators in the Wnt signaling pathway, which could effectively
activate downstream genes in Wnt signaling (20,21).
Previous research has demonstrated that overexpression of Wnt
signaling molecules in DPSCs is beneficial for their odontoblast
differentiation (22). Therefore,
the Wnt signaling pathway may be a vital approach for SOX2
affecting odontoblast differentiation of DPSCs. However, it is
still unclear whether the upregulation of Wnt signaling molecules
is influenced by SOX2 directly or indirectly. It is necessary to
further analyze the issue. If the issue is solved, it will be more
conducive to the development of novel technical approaches in order
to effectively regulate and control odontoblast differentiation of
DPSCs.
In conclusion, the results of present study
demonstrated that SOX2 overexpression in DPSCs promoted the
regulation of odontoblast differentiation of DPSCs and provided a
theoretical basis for the application of DPSCs in dental pulp
injury repair. However, this study mainly assessed the underlying
mechanisms in vitro; further in vivo studies are
required to investigate the odontoblast differentiation and injury
repair ability of DPSCs for directly transplanting DPSCs-SOX2 to
damaged teeth.
Acknowledgements
The authors would like to thank all those who have
helped during the writing of this manuscript, and gratefully
acknowledge the help of Professor Zhao Lei in his patience,
encouragement, and professional instructions during manuscript
writing.
References
|
1
|
Chang CC, Chang KC, Tsai SJ, Chang HH and
Lin CP: Neurogenic differentiation of dental pulp stem cells to
neuron-like cells in dopaminergic and motor neuronal inductive
media. J Formos Med Assoc. 113:956–965. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gronthos S, Mankani M, Brahim J, Robey PG
and Shi S: Postnatal human dental pulp stem cells (DPSCs) in vitro
and in vivo. Proc Natl Acad Sci USA. 97:pp. 13625–13630. 2000;
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Paino F, La Noce M, Tirino V, Naddeo P,
Desiderio V, Pirozzi G, De Rosa A, Laino L, Altucci L and Papaccio
G: Histone deacetylase inhibition with valproic acid downregulates
osteocalcin gene expression in human dental pulp stem cells and
osteoblasts: Evidence for HDAC2 involvement. Stem Cells.
32:279–289. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tatullo M, Marrelli M, Shakesheff KM and
White LJ: Dental pulp stem cells: Function, isolation and
applications in regenerative medicine. J Tissue Eng Regen Med.
9:1205–1216. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zheng Y, Wang XY, Wang YM, Liu XY, Zhang
CM, Hou BX and Wang SL: Dentin regeneration using deciduous pulp
stem/progenitor cells. J Dent Res. 91:676–682. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bakopoulou A, Leyhausen G, Volk J,
Papachristou E, Koidis P and Geurtsen W: Wnt/β-catenin signaling
regulates Dental Pulp Stem Cells' responses to pulp injury by
resinous monomers. Dent Mater. 31:542–555. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cai J, Li W, Su H, Qin D, Yang J, Zhu F,
Xu J, He W, Guo X, Labuda K, et al: Generation of human induced
pluripotent stem cells from umbilical cord matrix and amniotic
membrane mesenchymal cells. J Biol Chem. 285:11227–11234. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Esteban MA, Wang T, Qin B, Yang J, Qin D,
Cai J, Li W, Weng Z, Chen J, Ni S, et al: Vitamin C enhances the
generation of mouse and human induced pluripotent stem cells. Cell
Stem Cell. 6:71–79. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Takahashi K, Tanabe K, Ohnuki M, Narita M,
Ichisaka T, Tomoda K and Yamanaka S: Induction of pluripotent stem
cells from adult human fibroblasts by defined factors. Cell.
131:861–872. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Takahashi K and Yamanaka S: Induction of
pluripotent stem cells from mouse embryonic and adult fibroblast
cultures by defined factors. Cell. 126:663–676. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Feng R and Wen J: Overview of the roles of
Sox2 in stem cell and development. Biol Chem. 396:883–891. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Thier M, Wörsdörfer P, Lakes YB, Gorris R,
Herms S, Opitz T, Seiferling D, Quandel T, Hoffmann P, Nöthen MM,
et al: Direct conversion of fibroblasts into stably expandable
neural stem cells. Cell Stem Cell. 10:473–479. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liu P, Cai J, Dong D, Chen Y, Liu X, Wang
Y and Zhou Y: Effects of SOX2 on proliferation, migration and
adhesion of human dental pulp stem cells. PLoS One.
10:e01413462015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yang KC, Kitamura Y, Wu CC, Chang HH, Ling
TY and Kuo TF: Tooth germ-like construct transplantation for
whole-tooth regeneration: An in vivo study in the miniature pig.
Artif Organs. 40:E39–E50. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lin X, Dong R, Diao S, Yu G, Wang L, Li J
and Fan Z: SFRP2 enhanced the adipogenic and neuronal
differentiation potentials of stem cells from apical papilla. Cell
Biol Int. 41:534–543. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu F and Millar SE: Wnt/beta-catenin
signaling in oral tissue development and disease. J Dent Res.
89:318–330. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu B, Chen S, Cheng D, Jing W and Helms
JA: Primary cilia integrate hedgehog and Wnt signaling during tooth
development. J Dent Res. 93:475–482. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yuan G, Yang G, Zheng Y, Zhu X, Chen Z,
Zhang Z and Chen Y: The non-canonical BMP and Wnt/β-catenin
signaling pathways orchestrate early tooth development.
Development. 142:128–139. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Maye P, Zheng J, Li L and Wu D: Multiple
mechanisms for Wnt11-mediated repression of the canonical Wnt
signaling pathway. J Biol Chem. 279:24659–24665. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Katoh M: Regulation of WNT signaling
molecules by retinoic acid during neuronal differentiation in NT2
cells: Threshold model of WNT action (Review). Int J Mol Med.
10:683–687. 2002.PubMed/NCBI
|
|
22
|
Koizumi Y, Kawashima N, Yamamoto M,
Takimoto K, Zhou M, Suzuki N, Saito M, Harada H and Suda H: Wnt11
expression in rat dental pulp and promotional effects of Wnt
signaling on odontoblast differentiation. Congenit Anom (Kyoto).
53:101–108. 2013. View Article : Google Scholar : PubMed/NCBI
|