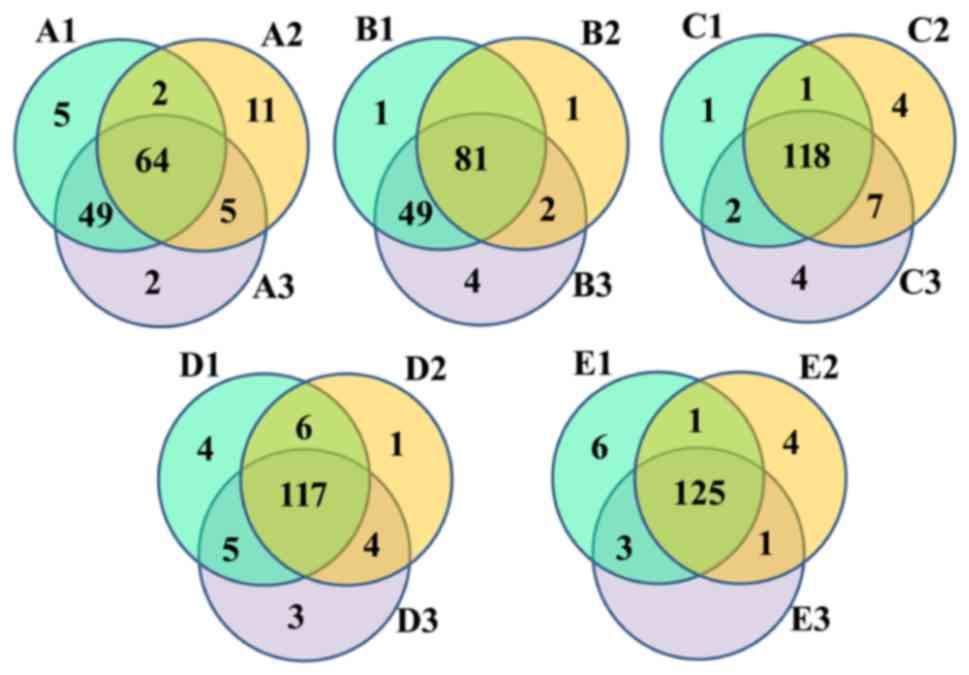
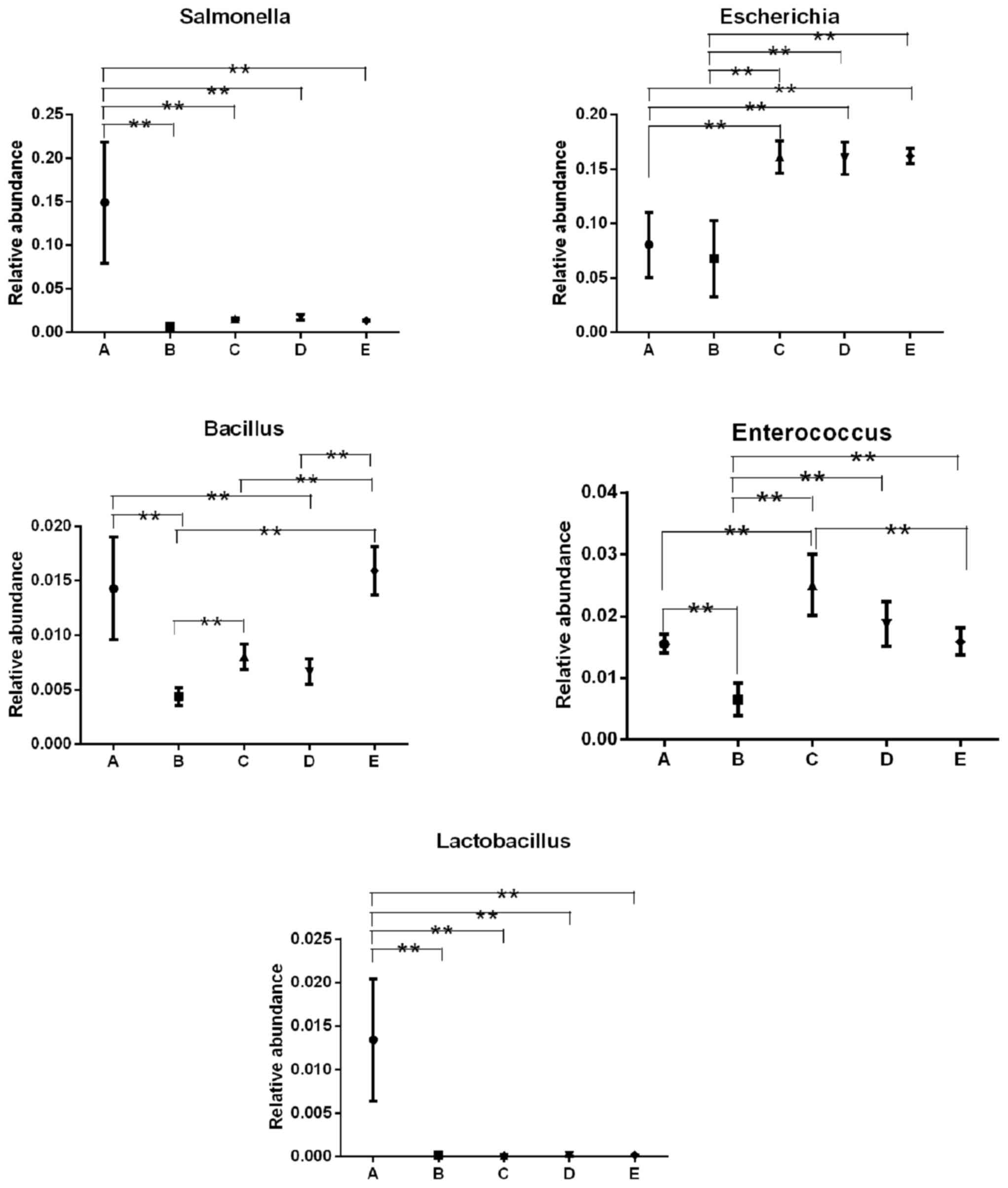
|
1
|
Shokralla S, Spall JL, Gibson JF and
Hajibabaei M: Next-generation sequencing technologies for
environmental DNA research. Mol Ecol. 21:1794–1805. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yue-Xin M, Holmstrom C and Webb J:
Application of denaturing gradient gel electrophoresis (DGGE) in
microbial ecology. Acta Ecologica Sinica. 23:1561–1569. 2003.
|
|
3
|
Yu X, Wu X, Qiu L, Wang D, Gan M, Chen X,
Wei H and Xu F: Analysis of the intestinal microbial community
structure of healthy and long-living elderly residents in Gaotian
village of Liuyang City. Appl Microbiol Biot. 99:9085–9095. 2015.
View Article : Google Scholar
|
|
4
|
Sivan A, Corrales L, Hubert N, Williams
JB, Aquino-Michaels K, Earley ZM, Benyamin FW, Lei YM, Jabri B,
Alegre ML, et al: Commensal Bifidobacterium promotes
antitumor immunity and facilitates anti-PD-L1 efficacy. Science.
350:1084–1089. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhernakova A, Kurilshikov A, Bonder MJ,
Tigchelaar EF, Schirmer M, Vatanen T, Mujagic Z, Vila AV, Falony G,
Vieira-Silva S, et al: Population-based metagenomics analysis
reveals markers for gut microbiome composition and diversity.
Science. 352:565–569. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Derrien M and van Hylckama Vlieg JE: Fate,
activity, and impact of ingested bacteria within the human gut
microbiota. Trends Microbiol. 23:354–360. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mimee M, Citorik RJ and Lu TK: Microbiome
therapeutics-Advances and challenges. Adv Drug Deliver Revi.
105:44–54. 2016. View Article : Google Scholar
|
|
8
|
Chen T, Shi Y, Wang X, Wang X, Meng F,
Yang S, Yang J and Xin H: High-throughput sequencing analyses of
oral microbial diversity in healthy people and patients with dental
caries and periodontal disease. Mol Med Rep. 16:127–132. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Fang X, Wang X, Yang S, Meng F, Wang X,
Wei H and Chen T: Evaluation of the microbial diversity in
amyotrophic lateral sclerosis using high-throughput sequencing.
Front Microbiol. 7:14792016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang X, Hu X, Deng K, Cheng X, Wei J,
Jiang M, Wang X and Chen T: High-throughput sequencing of microbial
diversity in implant-associated infection. Infect Genet Evol.
43:307–311. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Planý M, Šoltys K, Budiš J, Mader A,
Szemes T, Siekel P and Kuchtaa T: Potential of high-throughput
sequencing for broad-range detection of pathogenic bacteria in
spices and herbs. Food Control. 70:360–370. 2016.
|
|
12
|
Ishaq SL and Wright AD: High-throughput
DNA sequencing of the ruminal bacteria from moose (Alces alces) in
Vermont, Alaska, and Norway. Microb Ecol. 68:185–195. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hamilton MJ, Weingarden AR, Unno T,
Khoruts A and Sadowsky MJ: High-throughput DNA sequence analysis
reveals stable engraftment of gut microbiota following
transplantation of previously frozen fecal bacteria. Gut Microbes.
4:125–135. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Moorthie S, Mattocks CJ and Wright CF:
Review of massively parallel DNA sequencing technologies. Hugo J.
5:1–12. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kircher M, Heyn P and Kelso J: Addressing
challenges in the production and analysis of illumina sequencing
data. BMC Genomics. 12:3822011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kircher M: Understanding and improving
high-throughput sequencing data production and analysis: 225.
2011.
|
|
17
|
Hall N: Advanced sequencing technologies
and their wider impact in microbiology. J Exp Biol. 210:1518–1525.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xu J, Lian F, Zhao L, Zhao Y, Chen X,
Zhang X, Guo Y, Zhang C, Zhou Q, Xue Z, et al: Structural
modulation of gut microbiota during alleviation of type 2 diabetes
with a Chinese herbal formula. Isme J. 9:552–562. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Schloss PD, Westcott SL, Ryabin T, Hall
JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH,
Robinson CJ, et al: Introducing mothur: Open-source,
platform-independent, community-supported software for describing
and comparing microbial communities. Appl Environ Microb.
75:7537–7541. 2009. View Article : Google Scholar
|
|
20
|
Caporaso JG, Kuczynski J, Stombaugh J,
Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich
JK, Gordon JI, et al: QIIME allows analysis of high-throughput
community sequencing data. Nat Methods. 7:335–336. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Davenport M, Poles J, Leung JM, Wolff MJ,
Abidi WM, Ullman T, Mayer L, Cho I and Loke P: Metabolic
alterations to the mucosal microbiota in inflammatory bowel
disease. Inflamm Bowel Dis. 20:723–731. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Haas BJ, Gevers D, Earl AM, Feldgarden M,
Ward DV, Giannoukos G, Ciulla D, Tabbaa D, Highlander SK, Sodergren
E, et al: Chimeric 16S rRNA sequence formation and detection in
Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 21:494–504.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lu K, Ryan PA, Schlieper KA, Graffam ME,
Levine S, Wishnok JS, Swenberg JA, Tannenbaum SR and Fox JG:
Arsenic exposure perturbs the gut microbiome and its metabolic
profile in mice: An integrated metagenomics and metabolomics
analysis. Environ Health Perspect. 122:2842014.PubMed/NCBI
|
|
24
|
Boers SA, Jansen R and Hays JP: Suddenly
everyone is a microbiota specialist. Clin Microbiol Infec.
22:581–582. 2016. View Article : Google Scholar
|
|
25
|
Di Bella JM, Bao Y, Gloor GB, Burton JP
and Reid G: High throughput sequencing methods and analysis for
microbiome research. J Microbiol Meth. 95:401–414. 2013. View Article : Google Scholar
|
|
26
|
Van Vliet AH: Next generation sequencing
of microbial transcriptomes: Challenges and opportunities. Fems
Microbiol Lett. 302:1–7. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kennedy NA, Walker AW, Berry SH, Duncan
SH, Farquarson FM, Louis P, Thomson JM UK IBD Genetics Consortium,
Satsangi J, Flint HJ, et al: The impact of different DNA extraction
kits and laboratories upon the assessment of human gut microbiota
composition by 16S rRNA gene sequencing. PLoS One. 9:e889822014.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Boers SA, Hays JP and Jansen R: Micelle
PCR reduces chimera formation in 16S rRNA profiling of complex
microbial DNA mixtures. Sci Rep. 5:141812015. View Article : Google Scholar : PubMed/NCBI
|