Introduction
Transplantation is currently facing a problem of
organ shortage (1). Therefore,
xenotransplantation has been intensively pursued as a promising
supplement to allotransplantation (2). Pigs have been identified as potential
donors as they possess a similar biochemical profile to that of
humans. However, the immunological barriers must be overcome prior
to the achievement of successful xenotransplantation. The biggest
hurdle in xenotransplantation is humoral rejection (1), which exists in two forms, namely
hyperacute rejection and delayed xenograft rejection. Hyperacute
rejection can be prevented by the knockout of the
α-1,3-galactosyltransferase gene (1), so delayed xenograft rejection appears
to be the most direct barrier.
Activation of transcription factor nuclear factor
(NF)-κB serves a significant function in delayed xenograft
rejection (3). Nuclear factor of κ
light polypeptide gene enhancer in B-cells inhibitor, α (IκBα) is
the crucial inhibitor of NF-κB (4). However, IκBα can be phosphorylated
and then degraded following stimulation with inflammatory agents
(5), leading to NF-κB activation.
To avoid the phosphorylation, a mutant version of IκBα lacking the
phosphorylation sites was designed (6). Since NF-κB signaling is crucial for
the growth and development of vertebrates (7), it is not feasible to inhibit this
pathway in the embryonic period of donor animals by simply
replacing IκBα with its mutant type. Therefore, it has been
suggested that conditional gene targeting at porcine IκBα
may be a proper choice: First the genomic IκBα locus could
be targeted with a construct consisting of the wild type and the
mutant version of IκBα, then the wild-type IκBα could
still be expressed at the period of development, but the mutant
could substitute for the wild type when the donor is mature. To
achieve the controlled expression of the two types of porcine
IκBα, a blueprint of the conditional gene targeting was
designed (see Fig. 1). The first
step of the process involved the engineering of a targeting vector
from the available plasmid pFPC-1 (8). The present study demonstrated how to
construct a highly complex targeting vector by a classic but useful
method.
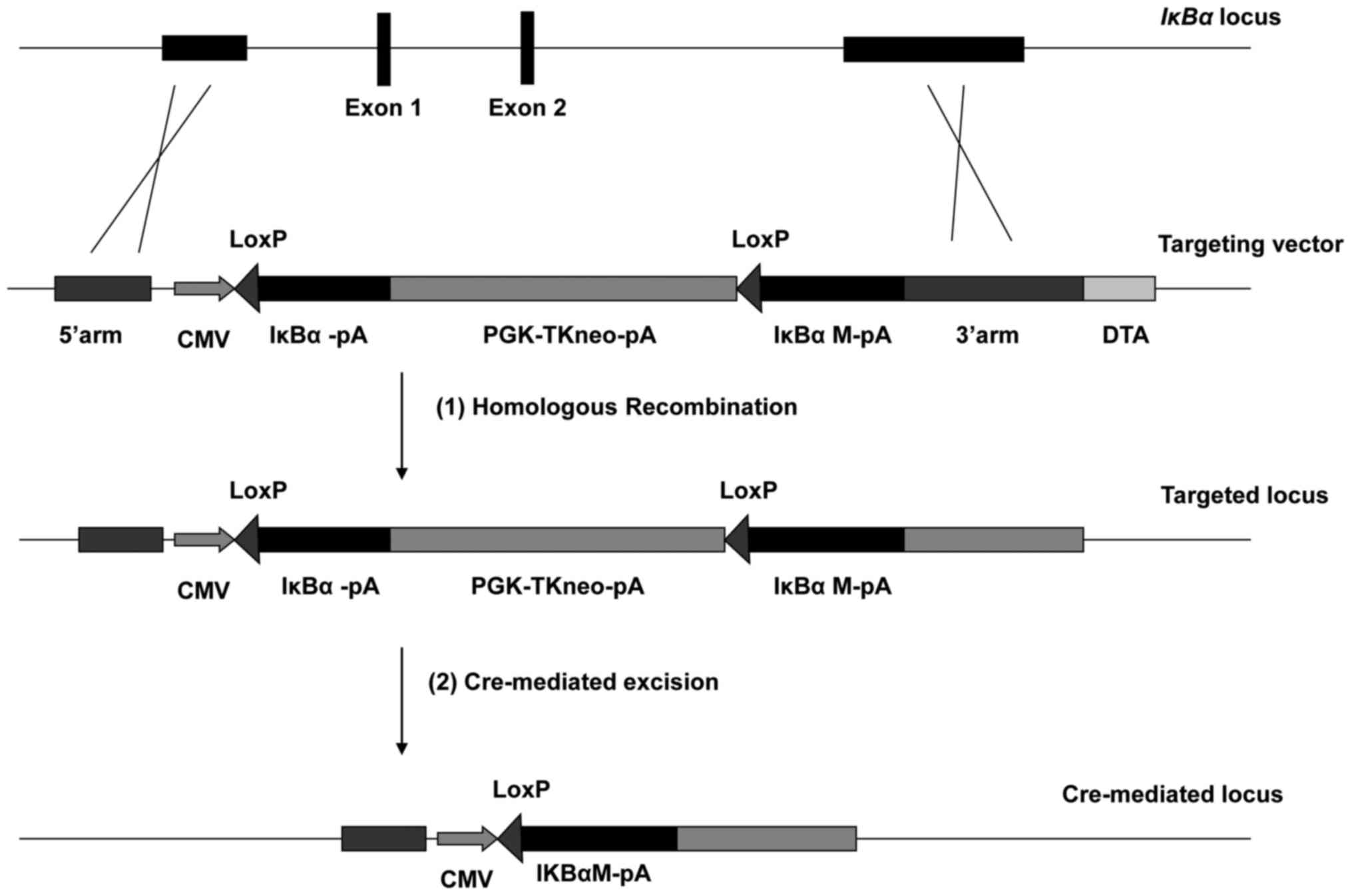 | Figure 1.Blueprint of conditional gene
targeting at porcine IκBα. The porcine IκBα gene is
composed of 6 exons, located on chromosome 7. The targeting process
consisted of two steps: i) First, targeting genomic IκBα
locus with a construct consisting of the wt IκBα and its
mutant IκBαM, to still be able to express the wt IκBα
during the development of the animal and ii) substituting the
mutant for the wt by Cre-mediated excision when the donor is
mature. CMV, human cytomegalovirus immediate-early
promoter/enhancer; PGK, promoter of mouse phosphoglycerate kinase-1
gene; TKneo, a fusion of thymidine kinase and neomycin resistance
genes; DTA, a gene coding for fragment A of diphtheria toxin; wt,
wild type; IκBα, nuclear factor of κ light polypeptide gene
enhancer in B-cells inhibitor, α. |
Materials and methods
Oligonucleotides
The sequences and uses of the oligonucleotides (PAGE
purified) (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA,
USA) are presented in Table I.
 | Table I.Oligonucleotides. |
Table I.
Oligonucleotides.
| Name | Sequence | Use |
|---|
| 5A-F |
5′-GGCGCGCCTGTTCGTGCTTTCTGATTTCC-3′ | Forward primer with
AscI site for 5′ arm of IκBα |
| 5A-R |
5′-TTAATTAATGAGGTCGTGTTCCTCCATT-3′ | Reverse primer with
PacI site for 5′ arm of IκBα |
| 3A-F |
5′-GTCTTCCTCCACTGTTCTTGCCTCCTTTGT-3′ | Forward primer for
3′ arm of IκBα |
| 3A-R |
5′-TGTCCCTCTTCTGTTGGTAGACTCCACTCC-3′ | Reverse primer for
3′ arm of IκBα |
| LoxP-A |
5′-CTAGCATAACTTCGTATAGCATACATTATACGAAGTTATCG-3′ | Annealed to LoxP-B
to yield the LoxP fragment |
| LoxP-B |
5′-GATCCGATAACTTCGTATAATGTATGCTATACGAAGTTATG-3′ | Annealed to LoxP-A
to yield the LoxP fragment |
| NTEC-A |
5′-AGCTGCGGCCGCACGCGTCTTAGAGCATGCTGGGGATGCGGTGGGCTCTATGG-3′ | Annealed to NTEC-B
to yield the NTEC fragment |
| NTEC-B |
5′-AATTCCATAGAGCCCACCGCATCCCCAGCATGCTCTAAGACGCGTGCGGCCGC-3′ | Annealed to NTEC-A
to yield the NTEC fragment |
| NHCL-A |
5′-AGCTTGCTAGCCTTAGAGCATGCTGGGGATGCGGTGGGCTCTATGGTATCGATG-3′ | Annealed to NHCL-B
to yield the NHCL fragment |
| NHCL-B |
5′-AATTCATCGATACCATAGAGCCCACCGCATCCCCAGCATGCTCTAAGGCTAGCA-3′ | Annealed to NHCL-A
to yield the NHCL fragment |
| pUC1-A |
5′-GATCCATCGATGTCGACACGCGTTTTAAACGGCCGGGCCGGCCATGCAT-3′ | Annealed to pUC1-B
to yield the pUC1 fragment |
| pUC1-B |
5′-AGCTATGCATGGCCGGCCCGGCCGTTTAAAACGCGTGTCGACATCGATG-3′ | Annealed to pUC1-A
to yield the pUC1 fragment |
| pUC2-A |
5′-AATTGCGGCCGCGTTTAAACAAGCTTGAATTCGATATCGCTAGCG-3′ | Annealed to pUC2-B
to yield the pUC2 fragment |
| pUC2-B |
5′-GATCCGCTAGCGATATCGAATTCAAGCTTGTTTAAACGCGGCCGC-3′ | Annealed to pUC2-A
to yield the pUC2 fragment |
Cloning of the homologous arms
For cloning, 2.0- and 6.0-kb porcine genomic
fragments (5′ and 3′ arms, respectively) were amplified by
polymerase chain reaction (PCR) using the LA-Taq™ DNA
polymerase (Takara Biotechnology Co., Ltd., Dalian, China) and
genomic DNA from porcine iliac endothelial cells (PIEC; Shanghai
Cell Bank of Chinese Academy of Sciences, Shanghai, China) as a
template. The primers used were 5A-F and 5A-R for the 5′arm and
3A-F and 3A-R for the 3′arm. The thermocycling conditions for the
25 µl PCR reaction was as follows 95°C for 5 min, 30 cycles of 60°C
for 30 sec and 72°C for 2 min, and finally 72°C for 10 min. A total
of 50 ng genomic DNA was used as the template. The 5′arm with
AscI/PacI sites was subcloned into pMD18-T vector,
yielding pMD-5′arm. The 3′arm was subcloned into
pCR®−XL-TOPO® (Invitrogen; Thermo Fisher
Scientific, Inc.), yielding pCR-XL-TOPO-3′arm.
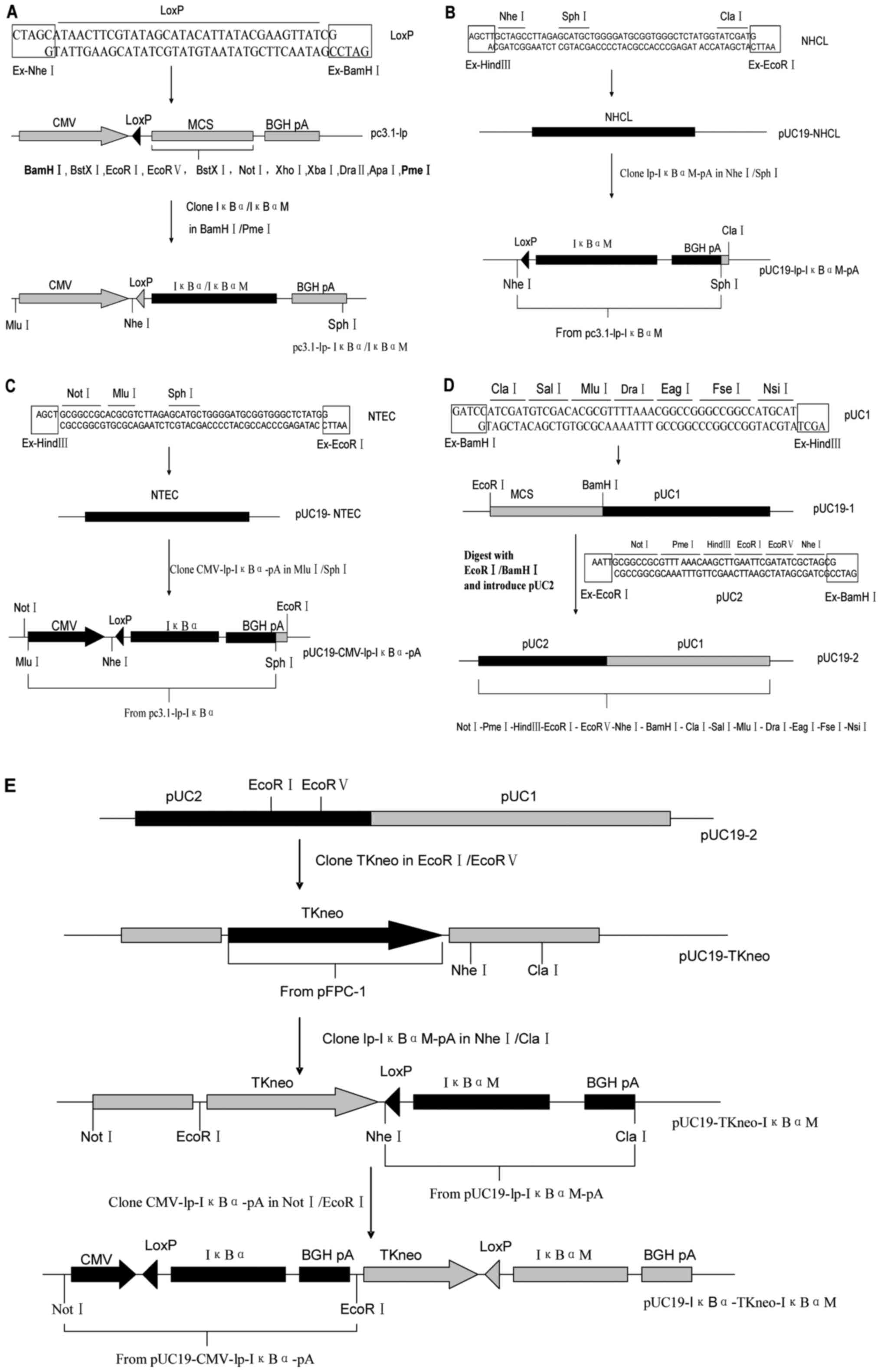
Construction of the Cre/LoxP
system
To generate a Cre/LoxP system
(pUC19-IκBα-TKneo-IκBαM; Fig. 2E)
for controlled expression of the two types of porcine IκBα,
an oligonucleotide-based method was deployed. There were three
steps to the whole process. All the plasmids were purchased from
Transgene (Beijing, China).
First, a series of suitable restriction sites were
designed and generated for assembling various DNA fragments. DNA
fragments (54- and 53-mer; NHCL and NTEC) were produced by
annealing oligonucleotides NHCL-A/NHCL-B and NTEC-A/NTEC-B
respectively (Table I). The
fragments NHCL and NTEC were then inserted at the
HindIII/EcoRI sites of pUC19 to respectively get
pUC19-NHCL (Fig. 2B) and
pUC19-NTEC (Fig. 2C). Then 49- and
45-mer DNA fragments (pUC1 and pUC2) were constructed by annealing
oligonucleotides pUC1-A/pUC1-B and pUC2-A/pUC2-B respectively. The
fragments pUC1 and pUC2 were sequentially inserted at the
BamHI/HindIII and EcoRI/BamHI sites of
pUC19, yielding pUC19-2 (Fig. 2D).
The intermediate product, pUC19-1, can also be used to subclone the
3′arm (as mentioned below).
Meanwhile, the fragments lp-IκBαM-pA and
CMV-lp-IκBα-pA were constructed (Fig.
2A) on the basis of pcDNA3.1(+) (Invitrogen; Thermo Fisher
Scientific, Inc.). To introduce a LoxP site in front of IκBα
or IκBαM (mutant IκBα) cDNA, a 41-merDNA fragment
(LoxP), assembled by annealing oligonucleotides LoxP-A and LoxP-B,
was inserted at the NheI/BamHI sites of pcDNA3.1(+)
to get a plasmid pc3.1-lp. Second, the IκBα/IκBαM cDNA
fragment was excised by the restriction enzymes NheI and
PmeI and subcloned into pc3.1-lp, yielding
pc3.1-lp-IκBα/IκBαM.
The second step was to prepare respectively the
three major fragments (CMV-lp-IκBα-pA, TKneo and lp-IκBαM-pA)
flanked by suitable restriction sites according to the distribution
of restriction sites in the MCS of pUC19-2. For this purpose, TKneo
fragment was excised by the endonucleases EcoRI/ScaI
from the plasmid pFPC-1, and then inserted at the
EcoRI/EcoRV sites of pUC19-2, yielding pUC19-TKneo
(Fig. 2E). Meanwhile, fragments
lp-IκBαM-pA and CMV-lp-IκBα-pA were excised by the endonucleases
NheI/SphI and MluI/SphI respectively
from the plasmids pc3.1-lp-IκBαM and pc3.1-lp-IκBα, and then
subcloned into the vectors pUC19-NHCL and pUC19-NTEC, yielding
pUC19-lp-IκBαM-pA (Fig. 2B) and
pUC19-CMV-lp-IκBα-pA (Fig.
2C).
The third step was to assemble the three major
fragments (CMV-lp-IκBα-pA, TKneo and lp-IκBαM-pA) together to get
the expected Cre/LoxP system (Fig.
2E). The fragments lp-IκBαM-pA and CMV-lp-IκBα-pA were
respectively excised by the endonucleases NheI/ClaI
and NotI/EcoRI from the vectors pUC19-lp-IκBαM-pA and
pUC19-CMV-lp-IκBα-pA, and then sequentially inserted into
pUC19-TKneo, yielding pUC19-IκBα-TKneo-IκBαM.
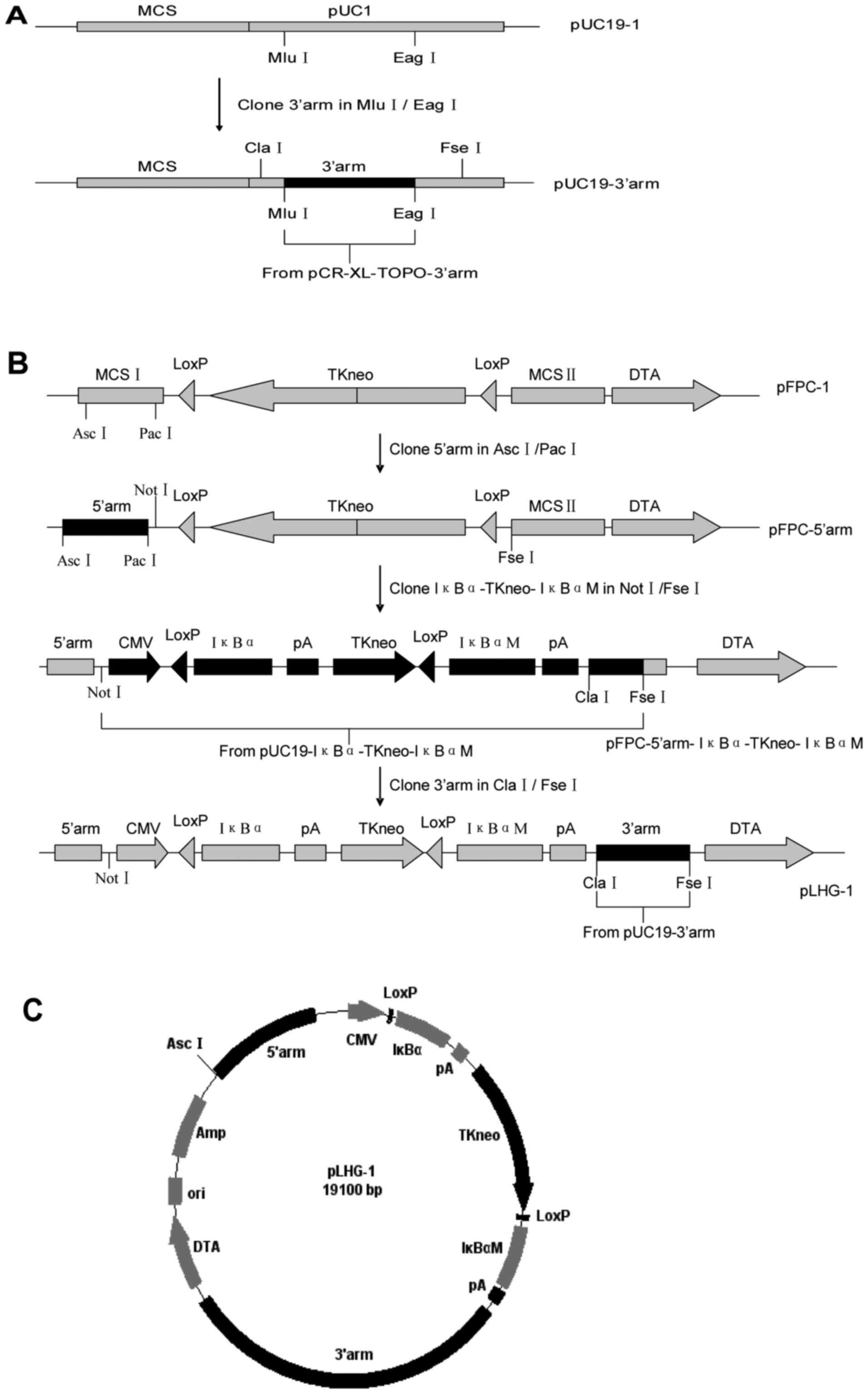
Construction of the conditional
targeting vector
A fragment containing the3′arm was excised by the
endonucleases MluI and EagI from pCR-XL-TOPO-3′arm,
and then inserted into pUC19-1, yielding pUC19-3′arm (Fig. 3A).
To generate the targeting vector (Fig. 3B), 5′arm, the fragment containing
the Cre/LoxP system (6.4kb), and 3′arm were respectively excised by
the endonucleases AscI/PacI, NotI/FseI
and ClaI/FseI from the vectors pMD-5′arm,
pUC19-IκBα-TKneo-IκBαM and pUC19-3′arm, and then sequentially
inserted into the plasmid pFPC-1, yielding pLHG-1 (19.1kb; Fig. 3C).
Results and Discussion
A possible solution to delayed
xenograft rejection
There are numerous examples of how to inhibit NF-κB
signaling in cells (6,9,10)
and animals (11–14) by means of overexpressing different
types of IκBα. A point mutant of IκBα in which
serines 32 and 36 are substituted by alanine residues is no longer
phosphorylated in response to diverse stimuli (6). This mutant behaves as a potent
dominant negative IκB protein (6,11),
inhibiting NF-κB activation. The present study used the cDNA of
such a mutant IκBα (IκBαM) to construct a vector. As
NF-κB signaling is crucial for the development process of mammals
(7), a Cre/LoxP system was
designed and generated (see Fig.
2E), aimed at controlling expression of the two types of
porcine IκBα, the wild and the mutant types. This Cre/LoxP
system may provide a solution to delayed xenograft rejection.
As illustrated in Fig.
2E, the Cre/LoxP system consists of three major expression
cassettes of IκBα gene, TKneo (a fusion of the
thymidine kinase and neomycin resistance genes) and IκBαM
cDNA (followed by a poly A sequence). A marked difference between
this Cre/LoxP system and others (8,15,16)
is that not only the positive selectable marker but also the
IκBα cDNA is floxed (flagged by two LoxP sites). This is a
prerequisite to achieve controlled expression of the two types of
porcine IκBα. If the genomic IκBα locus was to be
replaced in vivo by the Cre/LoxP system, wild type
IκBα may be expressed during the developmental process. The
IκBα cDNA may be ablated by Cre recombinase when the donor
is mature, and as a result, a new expression cassette, for
IκBαM, would be generated. The expression of the mutant
IκBα may result in an almost complete inhibition of NF-κB
signaling (11) and subsequently a
way of preventing delayed xenograft rejection. Prior to producing
gene-targeted animals, it will be necessary to perform the
controlled expression described above in a porcine vascular
endothelial cell line as the vascular endothelium is the target of
delayed xenograft rejection (3).
The analysis of the phenotype of the targeted PIEC could, to a
certain extent, be a good model of what would occur in an in
vivo system.
In the last 20 years, gene targeting has been used
as a powerful tool for studying gene function (15,17,18).
In addition, the stable and site-specific modification of mammalian
genomes has a variety of applications in biomedicine and
biotechnology (19). Site-specific
integration, rather than random integration, is the best way of
achieving the stable long-term expression of an introduced gene
(19). The final purpose of the
present study was to apply the results of gene function studies to
NF-κB signaling to inhibit delayed xenograft rejection, not simply
to achieve the knockout of IκBα. Controlled expression of
the wild type and the mutant of IκBα may provide an example
of how specific biomedicine challenges may be overcome by gene
targeting.
An oligonucleotide based method for
vector construction
As illustrated in Figs.
2 and 3, the vector
construction method is based on conventional ligation reactions and
5 pairs of oligonucleotides facilitate the 7 rounds of assembly of
various fragments. This complicated targeting vector (pLHG-1;
Fig. 3C) was constructed within 3
weeks. Unlike ordinary targeting vectors, this vector is used to
introduce the wild type and the mutant of the same gene into the
target genomic locus. Thus, the targeting vector should contain 7
extra fragments (CMV promoter, IκBα cDNA, IκBαM cDNA,
2 copies of BGH pA and 2 copies of LoxP), apart from the homologous
arms and the selectable markers. This increases the difficulty of
vector construction. In order to eliminate errors, maps and
sequence files were generated for every cloning step designed for
the targeting vector prior to starting bench work (16). The vector was constructed using 5
short double-stranded DNA fragments made by annealing
oligonucleotides. The vector pLHG-1 was confirmed by various
endonucleases (NotI, EcoRI, BamHI and
HindIII; data not shown) digestion and sequencing with 5
primer sequences (Table II). An
alternative plasmid containing His-tagged IκBαM cDNA was
also constructed as described above (pLHG-2; data not shown).
 | Table II.Sequencing primers of pLHG-1. |
Table II.
Sequencing primers of pLHG-1.
| Name | Sequence |
|---|
| 3′arm-DTA |
5′TGGCCTGGGAGCTTCTG3′ |
| 3′arm-lpmIKBpA |
5′GAATGGACTTTAGTAAGGCATC3′ |
| Tkneo-lpmIKBpA |
5′CCATCACGAGATTTCGATTCCACC3′ |
|
5′arm-CMVlpIKBpA |
5′GAGACCTGGACATGGTGAACCT3′ |
| 5′arm-Amp |
5′GGGAACTGGTGGTCTTTATTC3′ |
To avoid unwanted mutations, the use of PCR
amplification in the assembly of various fragments was eliminated.
With the help of the oligonucleotides, no n-directional cloning was
also avoided when assembling the different elements of the vector.
By making full use of directional cloning, satisfactory subcloning
efficiency was obtained throughout the whole process of vector
construction. In the last step of the assembly the efficiency of
the subclone step was confirmed to be high. This result is
comparable to the high efficiency of gateway system recombination
reported previously (15,20). In addition, methylation effects
should be avoided by proper design of the oligonucleotides when
dam-/dcm-methylation-deficient competent E. coli cells were
not used for plasmid transformation. To obtain high digestion
efficiency, moderate guanine-cytosine content (40–70%) of the
oligonucleotides is preferred.
Previously, various methods and technologies,
particularly recombination-based methods including recombineering
and gateway recombination, have been developed to facilitate and
simplify targeting vector construction. For recombineering, Zhang
et al (21) combined
genomic library screening and gene-targeting vector construction in
a single step, then Liu et al (22) and Cotta-de-Almeida et al
(23) managed to avoid the need to
construct or screen genomic libraries by manipulating bacterial
artificial chromosomes (BACs). Subsequently, recombineering was
employed more and more extensively in targeting vector
construction. For gateway recombination, Iiizumi et al
(15) improved the commercially
available Multi Site Gateway system and use it to facilitate
targeting vector construction. Nyabi et al (24) also applied Multi Site Gateway
system to build targeting vectors for transgenesis. To take
advantage of these two recombination methods, Ikeya et al
(20) tested a strategy of
generating gene-targeting vectors by combining in vivo
recombination (recombineering) with in vitro recombination
(gateway system). Wu et al (16) described a module cloning protocol
for constructing gene targeting vectors by combining the two
recombination methods. This ‘combined’ strategy may possess
potential in constructing gene targeting vectors, particularly
complicated ones. However, the two recombination methods
(recombineering and gateway recombination) exhibit their own
shortcomings in constructing targeting vectors which are as
complicated as the one designed in the present study. One of the
problems of using recombineering is the difficulty of getting
isogenic DNA for the cell line PIEC from commercially available BAC
clones, as isogenic DNA is preferred to improving targeting
efficiency (18). Additionally,
repetitive sequences in the original vector pFPC-1 would lead to
aberrant recombination, usually causing the failure to obtain
consistent results (22). For
gateway recombination, the destination vectors should be modified
by restriction enzyme-based cloning to be compatible with targeting
vector construction, and it is necessary to use restriction
enzyme-based cloning during the process of constructing the desired
targeting vector. The module cloning protocol (16) contains this conventional cloning
method. Furthermore, PCR amplification, which may introduce
unwanted mutations, has to be extensively used in the gateway
method. In addition, a MultiSite Gateway system remains too
expensive to be used for normal laboratory practice. For the
reasons described above, it was decided to construct the desired
targeting vector by restriction enzyme-based cloning. The present
study provided an example of constructing complicated targeting
vector with restriction enzyme-based cloning methods. It is
suggested that this strategy maybe employed in any normal
laboratory. The description here of an additional method to
efficiently construct targeting vectors will introduce more
flexibility in the field, therefore helping to meet the different
requirements of researchers.
Acknowledgements
The authors would like to thank Professor Yanxiu Liu
and Dr Chunmei Wang for their critical reading of the manuscript.
The present study was funded by the National Natural Science
Foundation of China (grants no. 31301936 and 31572383) and Project
of Qingdao People's Livelihood Science and Technology (grant no.
14-2-3-45-nsh).
References
|
1
|
Le Bas-Bernardet S, Anegon I and Blancho
G: Progress and prospects: Genetic engineering in
xenotransplantation. Gene Ther. 15:1247–1256. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mohiuddin MM: Clinical xenotransplantation
of organs: Why aren't we there yet? PLoS Med. 4:e752007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Auchincloss H Jr and Sachs DH: Xenogeneic
transplantation. Annu Rev Immunol. 16:433–470. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hayden MS and Ghosh S: Shared principles
in NF-kappaB signaling. Cell. 132:344–362. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cheong R, Hoffmann A and Levchenko A:
Understanding NF-kappaB signaling via mathematical modeling. Mol
Syst Biol. 4:1922008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Traenckner EB, Pahl HL, Henkel T, Schmidt
KN, Wilk S and Baeuerle PA: Phosphorylation of human I kappa
B-alpha on serines 32 and 36 controls I kappa B-alpha proteolysis
and NF-kappa B activation in response to diverse stimuli. EMBO J.
14:2876–2883. 1995.PubMed/NCBI
|
|
7
|
Gerondakis S, Grumont R, Gugasyan R, Wong
L, Isomura I, Ho W and Banerjee A: Unravelling the complexities of
the NF-kappaB signalling pathway using mouse knockout and
transgenic models. Oncogene. 25:6781–6799. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Perez-Campo FM, Spencer HL, Elder RH,
Stern PL and Ward CM: Novel vectors for homologous recombination
strategies in mouse embryonic stem cells: An ES cell line
expressing EGFP under control of the 5T4 promoter. Exp Cell Res.
313:3604–3615. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Goodman DJ, von Albertini MA, McShea A,
Wrighton CJ and Bach FH: Adenoviral-mediated overexpression of
I(kappa)B(alpha) in endothelial cells inhibits natural killer
cell-mediated endothelial cell activation. Transplantation.
62:967–972. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wrighton CJ, Hofer-Warbinek R, Moll T,
Eytner R, Bach FH and de Martin R: Inhibition of endothelial cell
activation by adenovirus-mediated expression of I kappa B alpha, an
inhibitor of the transcription factor NF-kappa B. J Exp Med.
183:1013–1022. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gareus R, Kotsaki E, Xanthoulea S, van der
Made I, Gijbels MJ, Kardakaris R, Polykratis A, Kollias G, de
Winther MP and Pasparakis M: Endothelial cell-specific NF-kappaB
inhibition protects mice from atherosclerosis. Cell Metab.
8:372–383. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Henke N, Schmidt-Ullrich R, Dechend R,
Park JK, Qadri F, Wellner M, Obst M, Gross V, Dietz R, Luft FC, et
al: Vascular endothelial cell-specific NF-kappaB suppression
attenuates hypertension-induced renal damage. Circ Res.
101:268–276. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kisseleva T, Song L, Vorontchikhina M,
Feirt N, Kitajewski J and Schindler C: NF-kappaB regulation of
endothelial cell function during LPS-induced toxemia and cancer. J
Clin Invest. 116:2955–2963. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ye X, Ding J, Zhou X, Chen G and Liu SF:
Divergent roles of endothelial NF-kappaB in multiple organ injury
and bacterial clearance in mouse models of sepsis. J Exp Med.
205:1303–1315. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Iiizumi S, Nomura Y, So S, Uegaki K, Aoki
K, Shibahara K, Adachi N and Koyama H: Simple one-week method to
construct gene-targeting vectors: Application to production of
human knockout cell lines. Biotechniques. 41:311–316. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wu S, Ying G, Wu Q and Capecchi MR: A
protocol for constructing gene targeting vectors: Generating
knockout mice for the cadherin family and beyond. Nat Protoc.
3:1056–1076. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Capecchi MR: Altering the genome by
homologous recombination. Science. 244:1288–1292. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Vasquez KM, Marburger K, Intody Z and
Wilson JH: Manipulating the mammalian genome by homologous
recombination. Proc Natl Acad Sci USA. 98:8403–8410. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sorrell DA and Kolb AF: Targeted
modification of mammalian genomes. Biotechnol Adv. 23:431–469.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ikeya M, Kawada M, Nakazawa Y, Sakuragi M,
Sasai N, Ueno M, Kiyonari H, Nakao K and Sasai Y: Gene
disruption/knock-in analysis of mONT3: Vector construction by
employing both in vivo and in vitro recombinations. Int J Dev Biol.
49:807–823. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang P, Li MZ and Elledge SJ: Towards
genetic genome projects: Genomic library screening and
gene-targeting vector construction in a single step. Nat Genet.
30:31–39. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu P, Jenkins NA and Copeland NG: A
highly efficient recombineering-based method for generating
conditional knockout mutations. Genome Res. 13:476–484. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cotta-de-Almeida V, Schonhoff S, Shibata
T, Leiter A and Snapper SB: A new method for rapidly generating
gene-targeting vectors by engineering BACs through homologous
recombination in bacteria. Genome Res. 13:2190–2194. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Nyabi O, Naessens M, Haigh K, Gembarska A,
Goossens S, Maetens M, De Clercq S, Drogat B, Haenebalcke L,
Bartunkova S, et al: Efficient mouse transgenesis using
Gateway-compatible ROSA26 locus targeting vectors and F1 hybrid ES
cells. Nucleic Acids Res. 37:e552009. View Article : Google Scholar : PubMed/NCBI
|