|
1
|
Coventry LL, Finn J and Bremner AP: Sex
differences in symptom presentation in acute myocardial infarction:
A systematic review and meta-analysis. Heart. 40:477–491. 2011.
|
|
2
|
Valensi P1, Lorgis L and Cottin Y:
Prevalence, incidence, predictive factors and prognosis of silent
myocardial infarction: A review of the literature. Arch Cardiovasc
Dis. 104:178–188. 2011. View Article : Google Scholar
|
|
3
|
Panza JA: Myocardial ischemia and the
pains of the heart. N Engl J Med. 346:1934–1935. 2002. View Article : Google Scholar
|
|
4
|
Langenbrunner JC, Marquez PV and Wang S:
Toward a Healthy and Harmonious Life in China: Stemming the Rising
Tide of Non-Communicable DiseasesHuman Development Unit; East Asia
and Pacific region. Washington, DC: The World Bank; 2011,
http://documents.worldbank.org/curated/en/618431468012000892/Toward-a-healthy-and-harmonious-life-in-China-stemming-the-rising-tide-of-non-communicable-diseasesOctober
21–2016
|
|
5
|
Velagaleti RS, Pencina MJ, Murabito JM,
Wang TJ, Parikh NI, D'Agostino RB, Levy D, Kannel WB and Vasan RS:
Long-term trends in the incidence of heart failure after myocardial
infarction. Circulation. 118:2057–2062. 2008. View Article : Google Scholar :
|
|
6
|
Pfeffer MA and Braunwald E: Ventricular
remodeling after myocardial infarction. Experimental observations
and clinical implications. Circulation. 81:1161–1172. 1990.
View Article : Google Scholar
|
|
7
|
Go AS, Mozaffarian D, Roger VL, Benjamin
EJ, Berry JD, Borden WB, Bravata DM, Dai S, Ford ES, Fox CS, et al:
Heart disease and stroke statistics-2013 update: A report from the
American Heart Association. Circulation. 127:e6–e245. 2013.
View Article : Google Scholar
|
|
8
|
Rosenberg L, Kaufman DW, Helmrich SP,
Miller DR, Stolley PD and Shapiro S: Myocardial infarction and
cigarette smoking in women younger than 50 years of age. JAMA.
253:2965–2969. 1985. View Article : Google Scholar
|
|
9
|
Leong DP, Smyth A, Teo KK, McKee M,
Rangarajan S, Pais P, Liu L, Anand SS and Yusuf S: INTERHEART
Investigators: Patterns of alcohol consumption and myocardial
infarction risk: Observations from 52 Countries in the interheart
case-control study. Circulation. 130:390–398. 2014. View Article : Google Scholar
|
|
10
|
Acute myocardial infarction and combined
oral contraceptives: Results of an international multicentre
case-control study: WHO Collaborative Study of Cardiovascular
Disease and Steroid Hormone Contraception. Lancet. 349:1202–1209.
1997. View Article : Google Scholar
|
|
11
|
Cassidy A, Mukamal KJ, Liu L, Franz M,
Eliassen AH and Rimm EB: High anthocyanin intake is associated with
a reduced risk of myocardial infarction in young and middle-aged
women. Circulation. 127:188–196. 2013. View Article : Google Scholar :
|
|
12
|
Freiberg MS, Chang CC, Kuller LH,
Skanderson M, Lowy E, Kraemer KL, Butt AA, Goetz Bidwell M, Leaf D,
Oursler KA, et al: HIV infection and the risk of acute myocardial
infarction. JAMA Intern Med. 173:614–622. 2013. View Article : Google Scholar :
|
|
13
|
Helgadottir A, Manolescu A, Thorleifsson
G, Gretarsdottir S, Jonsdottir H, Thorsteinsdottir U, Samani NJ,
Gudmundsson G, Grant SF, Thorgeirsson G, et al: The gene encoding
5-lipoxygenase activating protein confers risk of myocardial
infarction and stroke. Nat Genet. 36:233–239. 2004. View Article : Google Scholar
|
|
14
|
Do R, Stitziel NO, Won HH, Jørgensen AB,
Duga S, Merlini Angelica P, Kiezun A, Farrall M, Goel A, Zuk O, et
al: Multiple rare alleles at LDLR and APOA5 confer risk for
early-onset myocardial infarction. Nature. 518:102–106. 2015.
View Article : Google Scholar
|
|
15
|
Han H, Wang H, Yin Z, Jiang H, Fang M and
Han J: Association of genetic polymorphisms in ADH and ALDH2 with
risk of coronary artery disease and myocardial infarction: A
meta-analysis. Gene. 526:134–141. 2013. View Article : Google Scholar
|
|
16
|
Valenta Z, Mazura I, Kolár M, Grünfeldová
H, Feglarová P, Peleška J, Tomecková M, Kalina J, Slovák D and
Zvárová J: Determinants of excess genetic risk of acute myocardial
infarction-a matched case-control study. Eur J Biomed Inform.
8:34–43. 2012.
|
|
17
|
Gautier L, Cope L, Bolstad BM and Irizarry
RA: affy-analysis of Affymetrix GeneChip data at the probe level.
Bioinformatics. 20:307–315. 2004. View Article : Google Scholar
|
|
18
|
Liu Q, Wong L and Li J: Z-score biological
significance of binding hot spots of protein interfaces by using
crystal packing as the reference state. Biochim Biophys Acta.
1824:1457–1467. 2012. View Article : Google Scholar
|
|
19
|
Paic F, Igwe JC, Nori R, Kronenberg MS,
Franceschetti T, Harrington P, Kuo L, Shin DG, Rowe DW, Harris SE
and Kalajzic I: Identification of differentially expressed genes
between osteoblasts and osteocytes. Bone. 45:682–692. 2009.
View Article : Google Scholar :
|
|
20
|
Liu C and Xuan Z: Prioritization of
cancer-related genomic variants by SNP association network. Cancer
Inform. 14 Suppl 2:S57–S70. 2015.
|
|
21
|
Yu F, Yang Z, Hu X, Sun Y, Lin H and Wang
J: Protein complex detection in PPI networks based on data
integration and supervised learning method. BMC Bioinformatics. 16
Suppl 12:S32015. View Article : Google Scholar :
|
|
22
|
Liu AN, Wang LL, Li HP, Gong J and Liu XH:
Correlation between posttraumatic growth and posttraumatic stress
disorder symptoms based on pearson correlation coefficient: A
meta-analysis. J Nerv Ment Dis. 205:380–389. 2017. View Article : Google Scholar
|
|
23
|
Malta DC, Bernal RI, Almeida MC, Ishitani
LH, Girodo AM, Paixão LM, Oliveira MT, Junior Pimenta FG and Júnior
Silva JB: Inequities in intraurban areas in the distribution of
risk factors for non communicable diseases, Belo Horizonte, 2010.
Rev Bras Epidemiol. 17:629–641. 2014.(In English, Portuguese).
View Article : Google Scholar
|
|
24
|
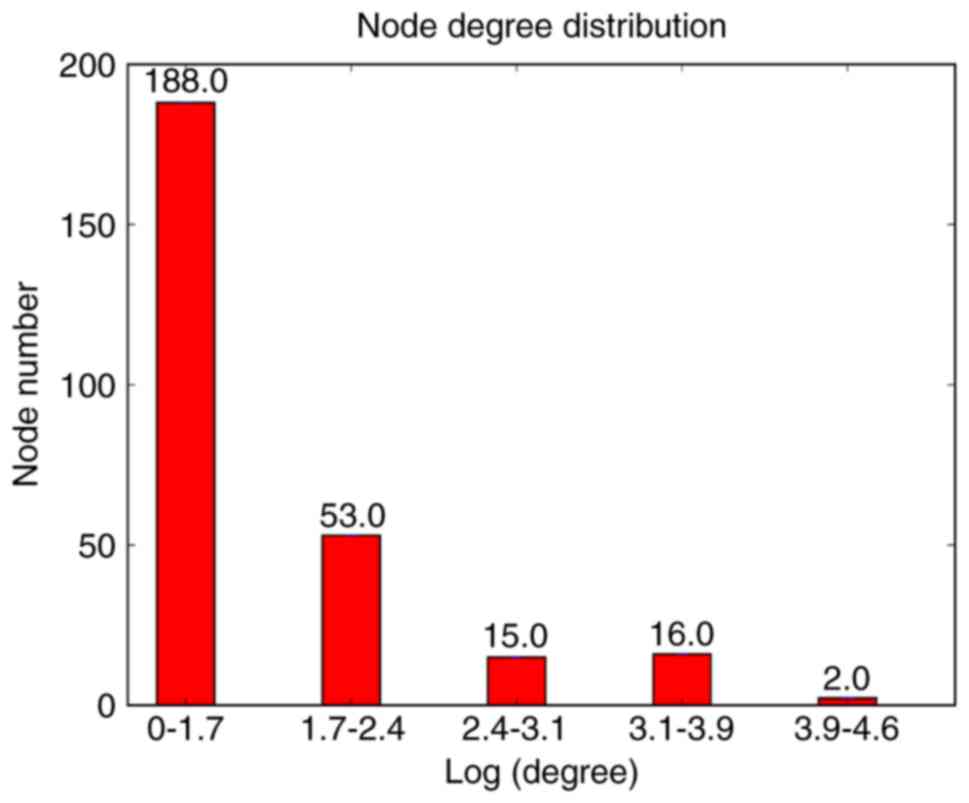
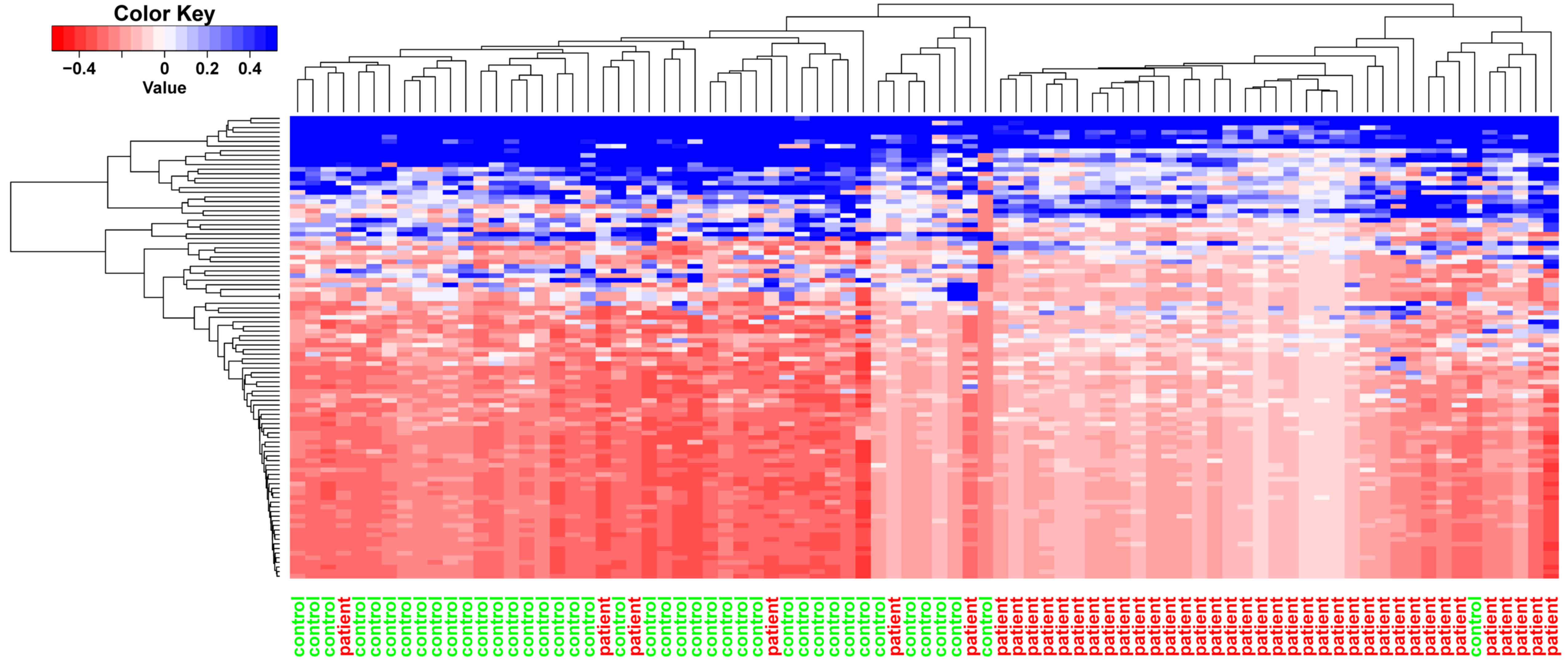
Metsalu T and Vilo J: ClustVis: A web tool
for visualizing clustering of multivariate data using principal
component analysis and heatmap. Nucleic Acids Res. 43:W566–W570.
2015. View Article : Google Scholar :
|
|
25
|
Qureshi MN, Min B, Jo HJ and Lee B:
Multiclass classification for the differential diagnosis on the
ADHD subtypes using recursive feature elimination and hierarchical
extreme learning machine: Structural MRI study. PLoS One.
11:e01606972016. View Article : Google Scholar :
|
|
26
|
Matwin S and Sazonova V: Direct comparison
between support vector machine and multinomial naive Bayes
algorithms for medical abstract classification. J Am Med Inform
Assoc. 19:9172012. View Article : Google Scholar :
|
|
27
|
Park HJ, Noh JH, Eun JW, Koh YS, Seo SM,
Park WS, Lee JY, Chang K, Seung KB, Kim PJ and Nam SW: Assessment
and diagnostic relevance of novel serum biomarkers for early
decision of ST-elevation myocardial infarction. Oncotarget.
6:12970–12983. 2015. View Article : Google Scholar :
|
|
28
|
Kapiloff MS, Schillace RV, Westphal AM and
Scott JD: mAKAP: An A-kinase anchoring protein targeted to the
nuclear membrane of differentiated myocytes. J Cell Sci.
112:2725–2736. 1999.
|
|
29
|
Redden JM and Dodge-Kafka KL: AKAP
phosphatase complexes in the heart. J Cardiovasc Pharmacol.
58:354–362. 2011. View Article : Google Scholar :
|
|
30
|
Perino A, Ghigo A, Scott JD and Hirsch E:
Anchoring proteins as regulators of signaling pathways. Circ Res.
111:482–492. 2012. View Article : Google Scholar :
|
|
31
|
Sumandea CA, Garcia-Cazarin ML, Bozio CH,
Sievert GA, Balke CW and Sumandea MP: Cardiac troponin T, a
sarcomeric AKAP, tethers protein kinase A at the myofilaments. J
Biol Chem. 286:530–541. 2011. View Article : Google Scholar
|
|
32
|
Lehnart SE, Wehrens XH, Reiken S, Warrier
S, Belevych AE, Harvey RD, Richter W, Jin SL, Conti M and Marks AR:
Phosphodiesterase 4D deficiency in the ryanodine-receptor complex
promotes heart failure and arrhythmias. Cell. 123:25–35. 2005.
View Article : Google Scholar :
|
|
33
|
Li L, Li J, Drum BM, Chen Y, Yin H, Guo X,
Luckey SW, Gilbert ML, McKnight GS, Scott JD, et al: Loss of
AKAP150 promotes pathological remodelling and heart failure
propensity by disrupting calcium cycling and contractile reserve.
Cardiovasc Res. 113:147–159. 2017. View Article : Google Scholar
|
|
34
|
Nieves-Cintrón M, Hirenallur-Shanthappa D,
Nygren PJ, Hinke SA, Dell'Acqua ML, Langeberg LK, Navedo M, Santana
LF and Scott JD: AKAP150 participates in calcineurin/NFAT
activation during the down-regulation of voltage-gated K(+)
currents in ventricular myocytes following myocardial infarction.
Cell Signal. 28:733–740. 2016. View Article : Google Scholar
|
|
35
|
Bockus LB and Humphries KM: cAMP-dependent
protein kinase (PKA) signaling is impaired in the diabetic heart. J
Biol Chem. 290:29250–29258. 2015. View Article : Google Scholar :
|
|
36
|
Nelson PJ, Moissoglu K, Vargas J Jr,
Klotman PE and Gelman IH: Involvement of the protein kinase C
substrate, SSeCKS, in the actin-based stellate morphology of
mesangial cells. J Cell Sci. 112:361–370. 1999.
|
|
37
|
Hehnly H, Canton D, Bucko P, Langeberg LK,
Ogier L, Gelman I, Santana LF, Wordeman L and Scott JD: A mitotic
kinase scaffold depleted in testicular seminomas impacts spindle
orientation in germ line stem cells. Elife. 4:e093842015.
View Article : Google Scholar :
|
|
38
|
Yan X, Walkiewicz M, Carlson J, Leiphon L
and Grove B: Gravin dynamics regulates the subcellular distribution
of PKA. Exp Cell Res. 315:1247–1259. 2009. View Article : Google Scholar :
|
|
39
|
Petrat F, Boengler K, Schulz R and de
Groot H: Glycine, a simple physiological compound protecting by yet
puzzling mechanism(s) against ischaemia-reperfusion injury: Current
knowledge. Br J Pharmacol. 165:2059–2072. 2012. View Article : Google Scholar :
|
|
40
|
Lu Y, Zhang J, Ma B, Li K, Li X, Bai H,
Yang Q, Zhu X, Ben J and Chen Q: Glycine attenuates cerebral
ischemia/reperfusion injury by inhibiting neuronal apoptosis in
mice. Neurochem Int. 61:649–658. 2012. View Article : Google Scholar
|
|
41
|
Qi RB, Zhang JY, Lu DX, Wang HD, Wang HH
and Li CJ: Glycine receptors contribute to cytoprotection of
glycine in myocardial cells. Chin Med J (Engl). 120:915–921.
2007.
|
|
42
|
Pan C, Bai X, Fan L, Ji Y, Li X and Chen
Q: Cytoprotection by glycine against ATP-depletion-induced injury
is mediated by glycine receptor in renal cells. Biochem J.
390:447–453. 2005. View Article : Google Scholar :
|