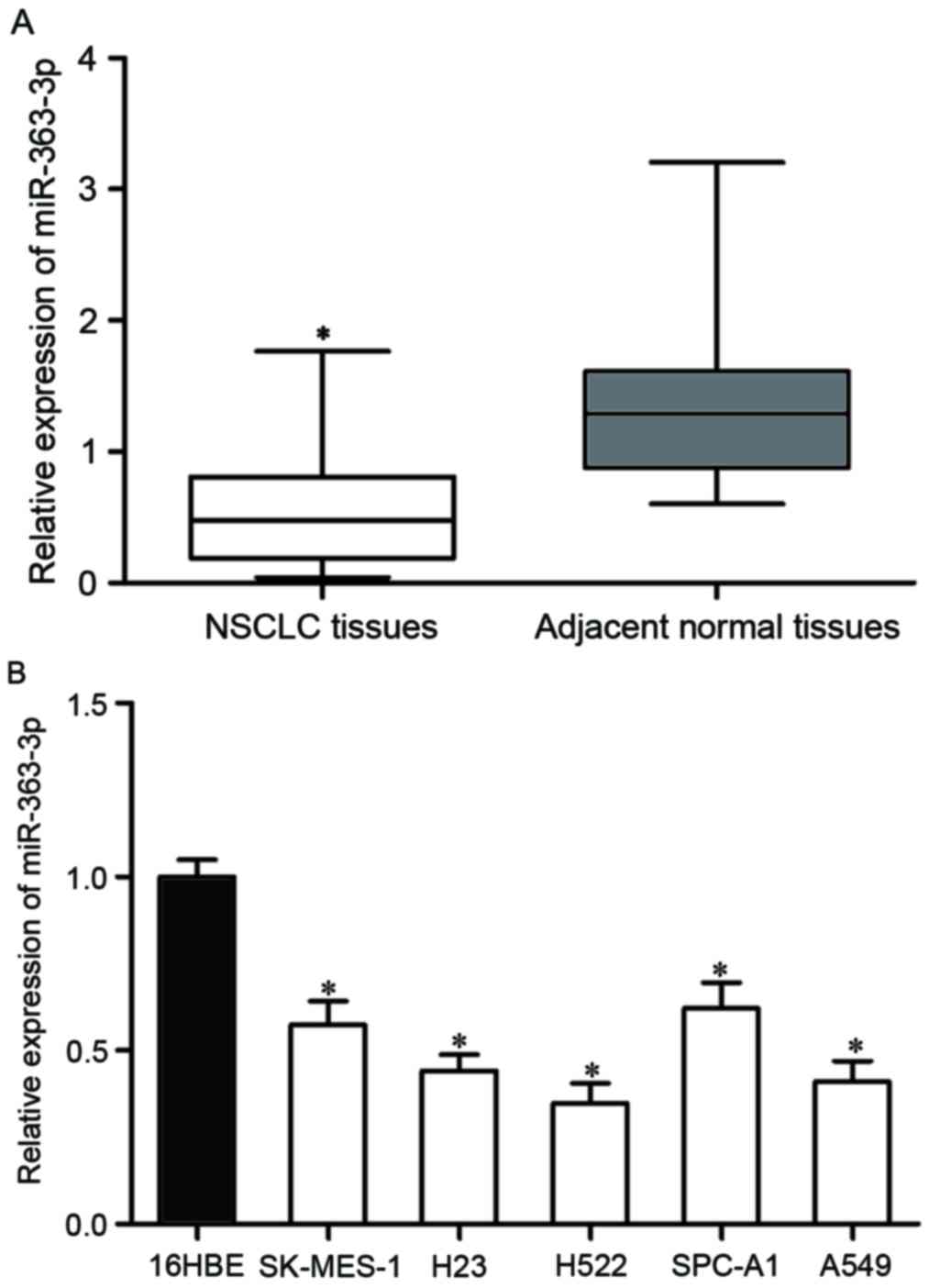
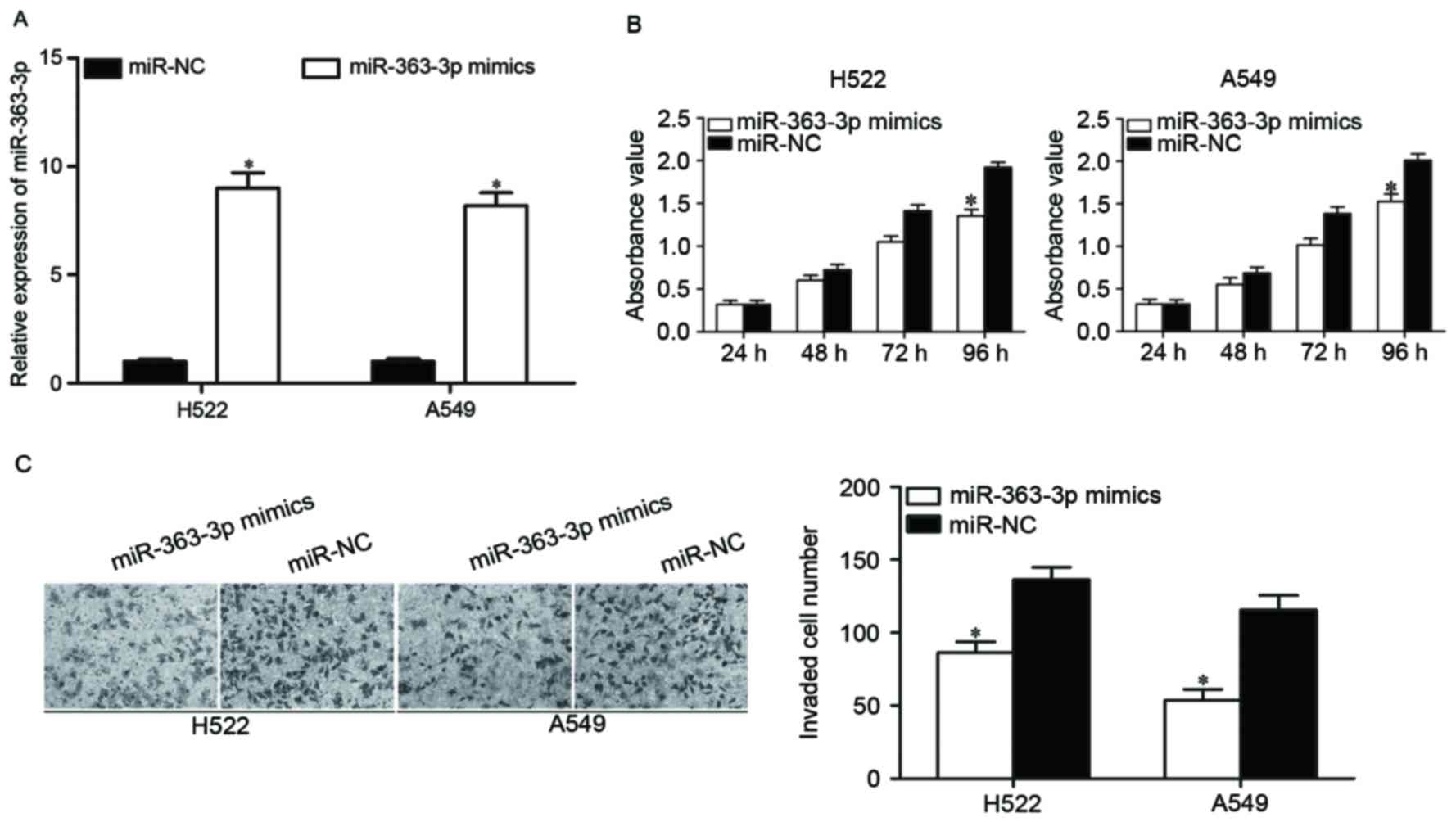
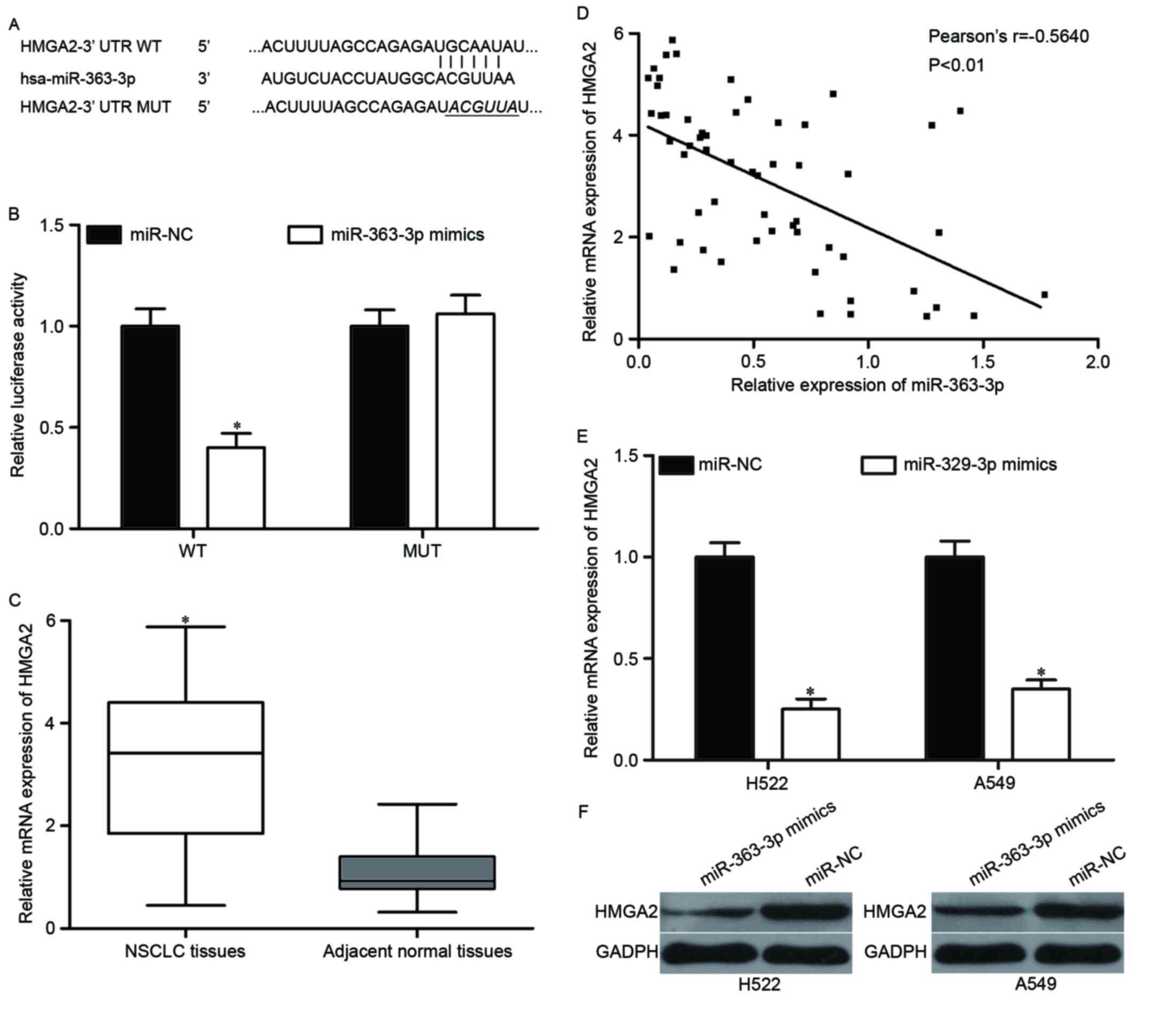
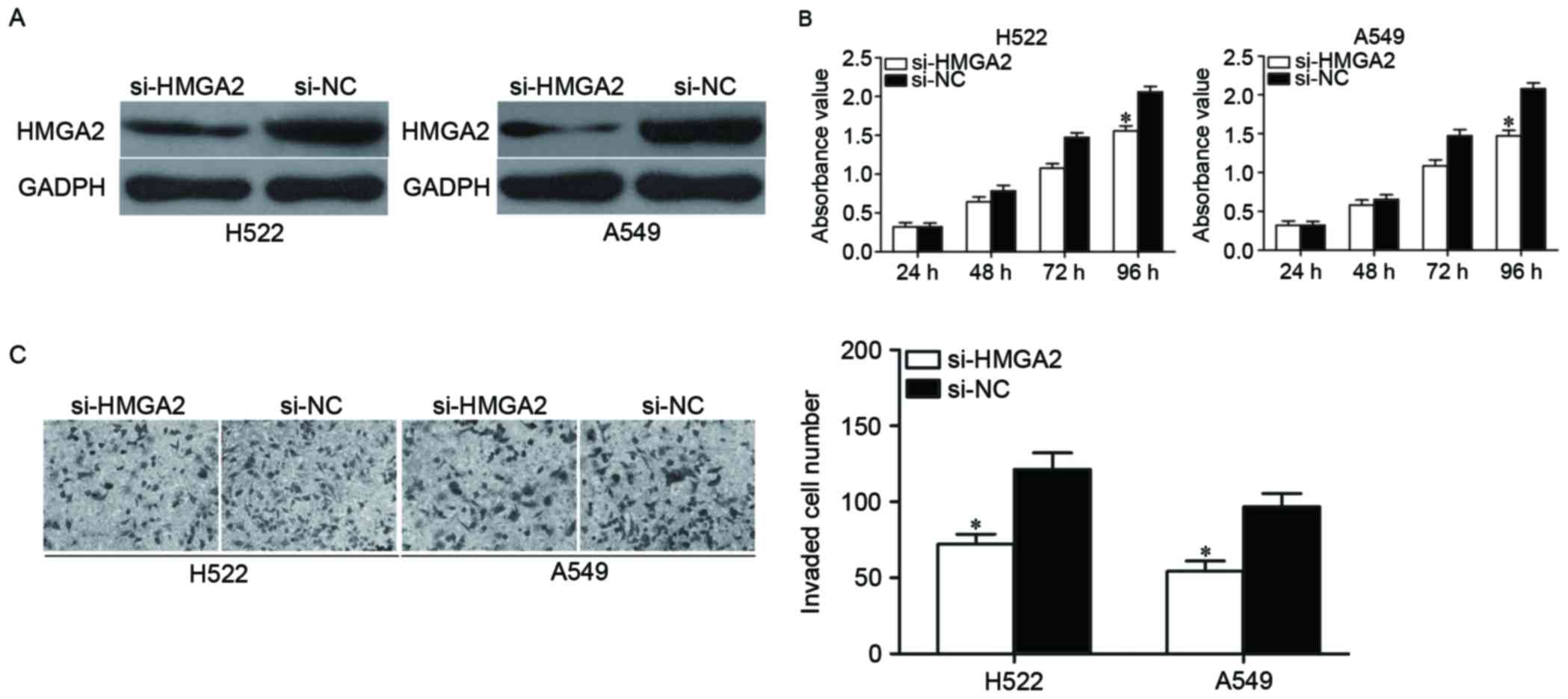
|
1
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Debevec L and Debeljak A:
Multidisciplinary management of lung cancer. J Thorac Oncol.
2:5772007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Spira A and Ettinger DS: Multidisciplinary
management of lung cancer. N Engl J Med. 350:379–392. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Boffetta P and Nyberg F: Contribution of
environmental factors to cancer risk. Br Med Bull. 68:71–94. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Didkowska J, Manczuk M, McNeill A, Powles
J and Zatonski W: Lung cancer mortality at ages 35–54 in the
European Union: Ecological study of evolving tobacco epidemics.
BMJ. 331:189–191. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ridge CA, McErlean AM and Ginsberg MS:
Epidemiology of lung cancer. Semin Intervent Radiol. 30:93–98.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Paliogiannis P, Attene F, Cossu A, Budroni
M, Cesaraccio R, Tanda F, Trignano M and Palmieri G: Lung cancer
epidemiology in North Sardinia, Italy. Multidiscip Respir Med.
8:452013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Søes S, Daugaard IL, Sørensen BS, Carus A,
Mattheisen M, Alsner J, Overgaard J, Hager H, Hansen LL and
Kristensen LS: Hypomethylation and increased expression of the
putative oncogene ELMO3 are associated with lung cancer development
and metastases formation. Oncoscience. 1:367–374. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Reungwetwattana T, Weroha SJ and Molina
JR: Oncogenic pathways, molecularly targeted therapies, and
highlighted clinical trials in non-small-cell lung cancer (NSCLC).
Clin Lung Cancer. 13:252–266. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sánchez de Cos J, Sojo González MA,
Montero MV, Pérez Calvo MC, Vicente MJ and Valle MH: Non-small cell
lung cancer and silent brain metastasis. Survival and prognostic
factors. Lung Cancer. 63:140–145. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lai EC: Micro RNAs are complementary to
3′UTR sequence motifs that mediate negative post-transcriptional
regulation. Nat Genet. 30:363–364. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Iorio MV and Croce CM: MicroRNA
dysregulation in cancer: Diagnostics, monitoring and therapeutics.
A comprehensive review. EMBO Mol Med. 4:143–159. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Mitchell PS, Parkin RK, Kroh EM, Fritz BR,
Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant
KC, Allen A, et al: Circulating microRNAs as stable blood-based
markers for cancer detection. Proc Natl Acad Sci USA. 105:pp.
10513–10518. 2008; View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K,
Guo J, Zhang Y, Chen J, Guo X, et al: Characterization of microRNAs
in serum: A novel class of biomarkers for diagnosis of cancer and
other diseases. Cell Res. 18:997–1006. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Vychytilova-Faltejskova P, Radova L,
Sachlova M, Kosarova Z, Slaba K, Fabian P, Grolich T, Prochazka V,
Kala Z, Svoboda M, et al: Serum-based microRNA signatures in early
diagnosis and prognosis prediction of colon cancer. Carcinogenesis.
37:941–950. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li P, Liu H, Wang Z, He F, Wang H, Shi Z,
Yang A and Ye J: MicroRNAs in laryngeal cancer: Implications for
diagnosis, prognosis and therapy. Am J Transl Res. 8:1935–1944.
2016.PubMed/NCBI
|
|
19
|
Shin SS, Park SS, Hwang B, Moon B, Kim WT,
Kim WJ and Moon SK: MicroRNA-892b influences proliferation,
migration and invasion of bladder cancer cells by mediating the
p19ARF/cyclin D1/CDK6 and Sp-1/MMP-9 pathways. Oncol Rep.
36:2313–2320. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Qiu X, Zhu H, Liu S, Tao G, Jin J, Chu H,
Wang M, Tong N, Gong W, Zhao Q, et al: Expression and prognostic
value of microRNA-26a and microRNA-148a in gastric cancer. J
Gastroenterol Hepatol. 32:819–827. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chandrasekaran KS, Sathyanarayanan A and
Karunagaran D: MicroRNA-214 suppresses growth, migration and
invasion through a novel target, high mobility group AT-hook 1, in
human cervical and colorectal cancer cells. Br J Cancer.
115:741–751. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Feng F, Kuai D, Wang H, Li T, Miao W, Liu
Y and Fan Y: Reduced expression of microRNA-497 is associated with
greater angiogenesis and poor prognosis in human gliomas. Hum
Pathol. 58:47–53. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Huang T, She K, Peng G, Wang W, Huang J,
Li J, Wang Z and He J: MicroRNA-186 suppresses cell proliferation
and metastasis through targeting MAP3K2 in non-small cell lung
cancer. Int J Oncol. 49:1437–1444. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Esquela-Kerscher A and Slack FJ:
Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer.
6:259–269. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tsuji S, Kawasaki Y, Furukawa S, Taniue K,
Hayashi T, Okuno M, Hiyoshi M, Kitayama J and Akiyama T: The
miR-363-GATA6-Lgr5 pathway is critical for colorectal
tumourigenesis. Nat Commun. 5:31502014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hu F, Min J, Cao X, Liu L, Ge Z, Hu J and
Li X: MiR-363-3p inhibits the epithelial-to-mesenchymal transition
and suppresses metastasis in colorectal cancer by targeting Sox4.
Biochem Biophys Res Commun. 474:35–42. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhou P, Huang G, Zhao Y, Zhong D, Xu Z,
Zeng Y, Zhang Y, Li S and He F: MicroRNA-363-mediated
downregulation of S1PR1 suppresses the proliferation of
hepatocellular carcinoma cells. Cell Signal. 26:1347–1354. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hou Y, Zhen J, Xu X, Zhen K, Zhu B, Pan R
and Zhao C: miR-215 functions as a tumor suppressor and directly
targets ZEB2 in human non-small cell lung cancer. Oncol Lett.
10:1985–1992. 2015.PubMed/NCBI
|
|
30
|
Ou Y, Zhai D, Wu N and Li X:
Downregulation of miR-363 increases drug resistance in
cisplatin-treated HepG2 by dysregulating Mcl-1. Gene. 572:116–122.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li X, Liu X, Fang J, Li H and Chen J:
microRNA-363 plays a tumor suppressive role in osteosarcoma by
directly targeting MAP2K4. Int J Clin Exp Med. 8:20157–20167.
2015.PubMed/NCBI
|
|
32
|
Conti A, Romeo SG, Cama A, La Torre D,
Barresi V, Pezzino G, Tomasello C, Cardali S, Angileri FF, Polito
F, et al: MiRNA expression profiling in human gliomas: Upregulated
miR-363 increases cell survival and proliferation. Tumour Biol.
37:14035–14048. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chen Y, Lu X, Wu B, Su Y, Li J and Wang H:
MicroRNA 363 mediated positive regulation of c-myc translation
affect prostate cancer development and progress. Neoplasma.
62:191–198. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hsu KW, Wang AM, Ping YH, Huang KH, Huang
TT, Lee HC, Lo SS, Chi CW and Yeh TS: Downregulation of tumor
suppressor MBP-1 by microRNA-363 in gastric carcinogenesis.
Carcinogenesis. 35:208–217. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhang PF, Sheng LL, Wang G, Tian M, Zhu
LY, Zhang R, Zhang J and Zhu JS: miR-363 promotes proliferation and
chemo-resistance of human gastric cancer via targeting of FBW7
ubiquitin ligase expression. Oncotarget. 7:35284–35292. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhuo HC, Song YF, Ye J, Lai GX and Liu DL:
MicroRNA-154 functions as a tumor suppressor and directly targets
HMGA2 in human non-small cell lung cancer. Genet Mol Res. 15:2016.
View Article : Google Scholar
|
|
37
|
Yang GL, Zhang LH, Bo JJ, Hou KL, Cai X,
Chen YY, Li H, Liu DM and Huang YR: Overexpression of HMGA2 in
bladder cancer and its association with clinicopathologic features
and prognosis HMGA2 as a prognostic marker of bladder cancer. Eur J
Surg Oncol. 37:265–271. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Sun M, Song CX, Huang H, Frankenberger CA,
Sankarasharma D, Gomes S, Chen P, Chen J, Chada KK, He C and Rosner
MR: HMGA2/TET1/HOXA9 signaling pathway regulates breast cancer
growth and metastasis. Proc Natl Acad Sci USA. 110:pp. 9920–9925.
2013; View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang X, Liu X, Li AY, Chen L, Lai L, Lin
HH, Hu S, Yao L, Peng J, Loera S, et al: Overexpression of HMGA2
promotes metastasis and impacts survival of colorectal cancers.
Clin Cancer Res. 17:2570–2580. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Winkler S, Murua Escobar H, Meyer B, Simon
D, Eberle N, Baumgartner W, Loeschke S, Nolte I and Bullerdiek J:
HMGA2 expression in a canine model of prostate cancer. Cancer Genet
Cytogenet. 177:98–102. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wu Y, Song Y and Liu H: Expression and its
clinical significance of HMGA2 in the patients with non-small cell
lung cancer. Zhongguo Fei Ai Za Zhi. 11:377–381. 2008.(In Chinese).
PubMed/NCBI
|
|
42
|
Kumar MS, Armenteros-Monterroso E, East P,
Chakravorty P, Matthews N, Winslow MM and Downward J: HMGA2
functions as a competing endogenous RNA to promote lung cancer
progression. Nature. 505:212–217. 2014. View Article : Google Scholar : PubMed/NCBI
|