Introduction
Gastric cancer (GC), as a common malignancy of
digestive tract carcinomas, is one of the leading causes of
cancer-associated mortality worldwide, and it has been previously
reported that from >950,000 new GC cases, 720,000 deaths
occurred in annually (1–3). Despite the early diagnosis and
effective therapies, GC is often diagnosed at advanced stage with
the characters of extensive invasion and distant metastasis,
leading to the low survival rate (4,5).
Additionally, as it is a major health threat, GC puts a large
economic burden on every patient's family (5). Therefore, there is an urgency to
identify novel biomarkers that can be detected early, and at the
same time they can block or slow down the malignancy progression in
order to improve GC prognosis. Previous studies have identified
various cellular products to be novel molecules or targets in the
progression of GC, including long non-coding RNAs (lncRNAs)
(6,7).
LncRNAs are identified as a class RNA molecules with
length of more than 200 nucleotides, that have no significant
protein-coding capacity, but occupy a majority of mammalian genome
production (8,9). LncRNAs are involved in various
biological processes including cell proliferation, apoptosis and
migration (10). Additionally,
lncRNAs have been reported to function as important genes in human
diseases, including lncRNA PVT1 was demonstrated to be associated
with prostate cancer and H19 was associated with breast cancer
(11–13). Recent studies have revealed that
lncRNAs also have critical roles in the progression and metastasis
of GC (14,15). However, the molecular mechanism of
lncRNAs in the progression and metastasis of GC remains to be
elucidated. Identification of novel biomarkers is required for the
diagnosis and treatment of patients with GC.
Small nucleolar RNA host gene 12 (SNHG12) is located
in chromosome 1 and 675 nucleotides in size. Previous studies
suggested that SNHG12 exerted its oncogenic function by promoting
cell growth and inhibiting cell apoptosis in various cancer types
(10,16). The present study aimed to
investigate the role of SNHG12 in GC. Therefore, SNHG12 expression
and its clinicopathological and prognostic significance for GC
patients was assessed. Subsequently, the function of SNHG12 on the
proliferation and invasion of GC cell lines was examined. The
findings of the current study suggested that SNHG12 may be a tumor
promoter by acting as a miR-320 sponge in GC, indicating that
SNHG12 may be a novel biomarker for GC progression.
Materials and methods
Human tissue specimens and cell
culture
GC tissues and adjacent normal gastric epithelial
tissues were obtained from 60 patients who underwent surgery at
Department of General Surgery, Second Affiliated Hospital of
Zhejiang University (Hangzhou, China) between May 2015 and June
2016. Specimens were immediately frozen in liquid nitrogen and
stored at −80°C. Informed consent was obtained from all patients.
The present study was approved by the Ethics Committee of the
Second Affiliated Hospital of Zhejiang University. The clinical
characteristics recorded included sex, age, size of carcinoma,
tumor-node-metastasis (TNM) stage, distant and lymphatic
metastasis. Overall survival and the disease-free survival rates
were recorded from the date of surgery to the mortality of the
patient or last contact.
The SGC7901, NCI-N87 and AGS human gastric cancer
cell lines and normal gastric epithelial cell line GES-1 were
obtained from American Type Culture Collection (Manassas, VA, USA).
Cells were cultured in Dulbecco's modified Eagle's medium (cat. no.
sc-224478; Santa Cruz Biotechnology, Inc., Dallas, TX, USA)
supplemented 10% fetal bovine serum (FBS), 100 U/ml penicillin
(cat. no. sc-391048; Santa Cruz Biotechnology, Inc.) and 100 U/ml
streptomycin (cat. no. sc-202821; Santa Cruz Biotechnology, Inc.)
at 37°C in a humidified 5% CO2 atmosphere.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from cells or tissues using
TRIzol (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA,
USA). cDNA synthesis was performed at 37°C for 15 min, then 85°C
for 5 sec using reverse transcriptase (Applied Biosystems; Thermo
Fisher Scientific, Inc.) following the manufacturer protocol. qPCR
was conducted with ABI 7500 system (Applied Biosystems; Thermo
Fisher Scientific, Inc.) using SYBR-Green (Takara Biotechnology
Co., Ltd., Dalian, China). PCR was performed for 25 cycles of: 10
sec at 98°C, 10 sec at 55°C and 20 sec at 72°C. The primer
sequences used were as follows: SNHG12: Forward (F),
5′-TCTGGTGATCGAGGACTTCC-3′ and reverse (R),
5′-ACCTCCTCAGTATCACACACT-3′; GAPDH F, 5′-ACACCCACTCCTCCACCTTT-3′
and R, 5′-TTACTCCTTGGAGGCCATGT-3′. LncRNA, mRNA and miRNA
expression levels were normalized using U6 small RNA. Fold-changes
were counted using the 2−ΔΔCq method (17).
Cell transfection
SNHG12 siRNA (si-SNHG12;
5′-GCAGTGGCGTGATCAAGGCTCATTGCAGCCT-3′) and control siRNA (si-ctrl;
5′-TTCCTGCGTGGGCCCAGCGCCGGGCACTGA-3′) were purchased from Guangzhou
RiboBio Co., Ltd. (Guangzhou, China). miR-320 and control miRNA
mimic/inhibitors were synthesized by GenePharma Co., Ltd.,
(Shanghai, China). Transfections were performed using a
Lipofectamine 2000 kit (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer instructions. The transfected cells
were harvested and assessed 48 h post-transfection.
Cell proliferation assays
Cell viability was determined using Cell Counting
Kit-8 (CCK-8) and the colony formation assays. For the CCK-8 assay,
the transfected GC cells (2×103) were seeded into
96-well plates and incubated for 1–6 days at 37°C. CCK-8 (10 µl;
Beyotime Institute of Biotechnology, Shanghai, China) solution was
added to each well 1 h prior to the endpoint. Subsequently, the
plates were incubated at 37°C for 2 h. A Benchmark Microplate
Reader (Bio-Rad Laboratories, Inc., Hercules, CA, USA) was used to
determine the spectrophotometric absorbance value at 460 nm. For
the colony formation assay, the transfected cells (500 cells/well)
were placed into 6-well plates and cultured in RPMI 1640 medium
(Gibco; Thermo Fisher Scientific, Inc.) containing 10% FBS at 37°C
with 5% CO2 for 10 days. Rinsed with phosphate-buffered
saline (PBS), the cells were fixed with 4% paraformaldehyde for 15
min at room temperature and stained with crystal violet (1 mg/ml)
at 37°C for 10 min. A total of ten fields were selected at random
to examine and the number of colonies (≥50 cells) was detected with
a light microscope.
Immunohistochemistry (IHC)
analysis
IHC was conducted as previously described (18). Briefly, the transfected cells were
grown on coverslips and fixed with 4% paraformaldehyde for 30 min
at room temperature. Following being rinsed three times with 0.1 M
PBS (pH 7.4), the cells were incubated with a primary mouse
anti-Ki67 antibody (cat. no. 612254; 1:250; BD Biosciences,
Franklin Lakes, NJ, USA) overnight at 4°C. The next day, the cells
were incubated with rabbit horseradish peroxidase (HRP-conjugated
secondary antibody IgG (cat. no. sc-3749, 1:500; Santa Cruz
Biotechnology, Inc.) for 15 min at room temperature, followed by
3,3′-diaminobenzidine staining at room temperature for 4 min and
counterstained with hematoxylin. Finally, the cells were assessed
using a light microscope.
Invasion assay
Cell invasive capacity was determined using an
invasion assay. The transfected cells (2×105) were
seeded in the top chamber with Matrigel-coated membrane (24-well
insert; pore size: 8-µm; Corning Inc., Corning, NY, USA) of the
inserts. The RPMI 1640 media with 10% FBS was added to the bottom
chamber. The cells were incubated for 48 h, the cells on the upper
surface of the membrane were swabbed off gently, and cells on the
lower surface were fixed in 4% paraformaldehyde for 15 min at room
temperature and stained with 2% crystal violet at 37°C for 10 min
(Sigma-Aldrich; Merck Millipore, Darmstadt, Germany) staining.
Total number of invasive cells were counted using a light
microscope BX51 (Olympus America, Inc., Melville, NY, USA).
Western blot assay
Total protein (50 µg) was extracted using
radioimmunoprecipitation assay lysis buffer (Sigma-Aldrich; Merck
KGaA) and quantified by bicinchoninic acid assay kit (Beyotime
Institute of Biotechnology). Then the protein was separated using
10% SDS-PAGE and transferred onto polyvinylidene fluoride
membranes. The membranes were immunoblotted with 5% powdered milk
dissolved in PBS containing 0.1% Tween-20 at room temperature for 3
h. Then the membranes were incubated overnight at 4°C with primary
rabbit antibodies: CRK like proto-oncogene, adaptor protein (CRKL;
1:500; cat. no. sc-365471; Santa Cruz Biotechnology, Inc.),
phosphorylated (p)-AKT serine/threonine kinase 1 (Akt; 1:500; cat.
no. sc-24500; Santa Cruz Biotechnology, Inc.),
p-extracellular-signal regulated kinase (Erk; 1:1,000; cat. no.
4370; Cell Signaling Technology, Inc., Danvers, MA, USA) and GAPDH
(1:2,000; cat. no. sc-420485; Santa Cruz Biotechnology, Inc.).
Then, a 1:3,000 HRP-conjugated anti-rabbit secondary antibody (cat.
no. 1662,408; 1:3,000; Bio-Rad Laboratories Inc., Hercules, CA,
USA) was incubated with the membrane for 1 h at room temperature.
Signals were quantified using a ChemiDoc XRS system (Bio-Rad
Laboratories, Inc.).
Reporter vector construction and
luciferase reporter assay
A bioinformatics database (lncipedia.org) was used to search for potential
microRNAs that may bind to SNHG12. It was demonstrated that SNHG12
fragments contained the predicted binding site of miR-320.
Subsequently, the fragments were amplified using PCR (17) and cloned them into pGL3-control
vector (Promega Corporation, Madison, WI, USA) to form the reporter
vector SNHG12-wild-type (Wt SNHG12). To test the binding
specificity, the corresponding mutant fragments were cloned to form
the reporter vector SNHG12-mutant-type (Mut SNHG12). SGC7901 and
AGS cells (1.0×106 cells) were seeded into a 6-well dish
with reporter vector Wt SNHG12 or Mut SNHG12 (100 ng) and indicated
miRNAs (25 nM) using Lipofectamine 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). Each well had 800 ml serum-free medium added and
cells were incubated for 6 h in a CO2-incubator.
Luciferase activity was detected 48 h after co-transfection using a
dual-luciferase reporter assay system normalized by Renilla
luciferase (Promega Corporation, Madison, WI, USA) following the
manufacturer's protocol.
Statistical analysis
Data are presented as the mean ± standard deviation
and statistical analysis was performed using SPSS version 18.0
(SPSS, Inc., Chicago, IL, USA). Differences between groups were
determined using Student's t-test and one-way analysis of variance
(ANOVA). Tukey's test was used when it was necessary following a
one-way ANOVA. Survival curves were analyzed using the Kaplan-Meier
method and calculated using the log-rank test. The relationship of
SNHG12 and miR-320 expression was detected using Pearson
correlation. P<0.05 was considered to indicate a statistically
significant difference.
Results
SNHG12 is upregulated in GC tissues
and cell lines
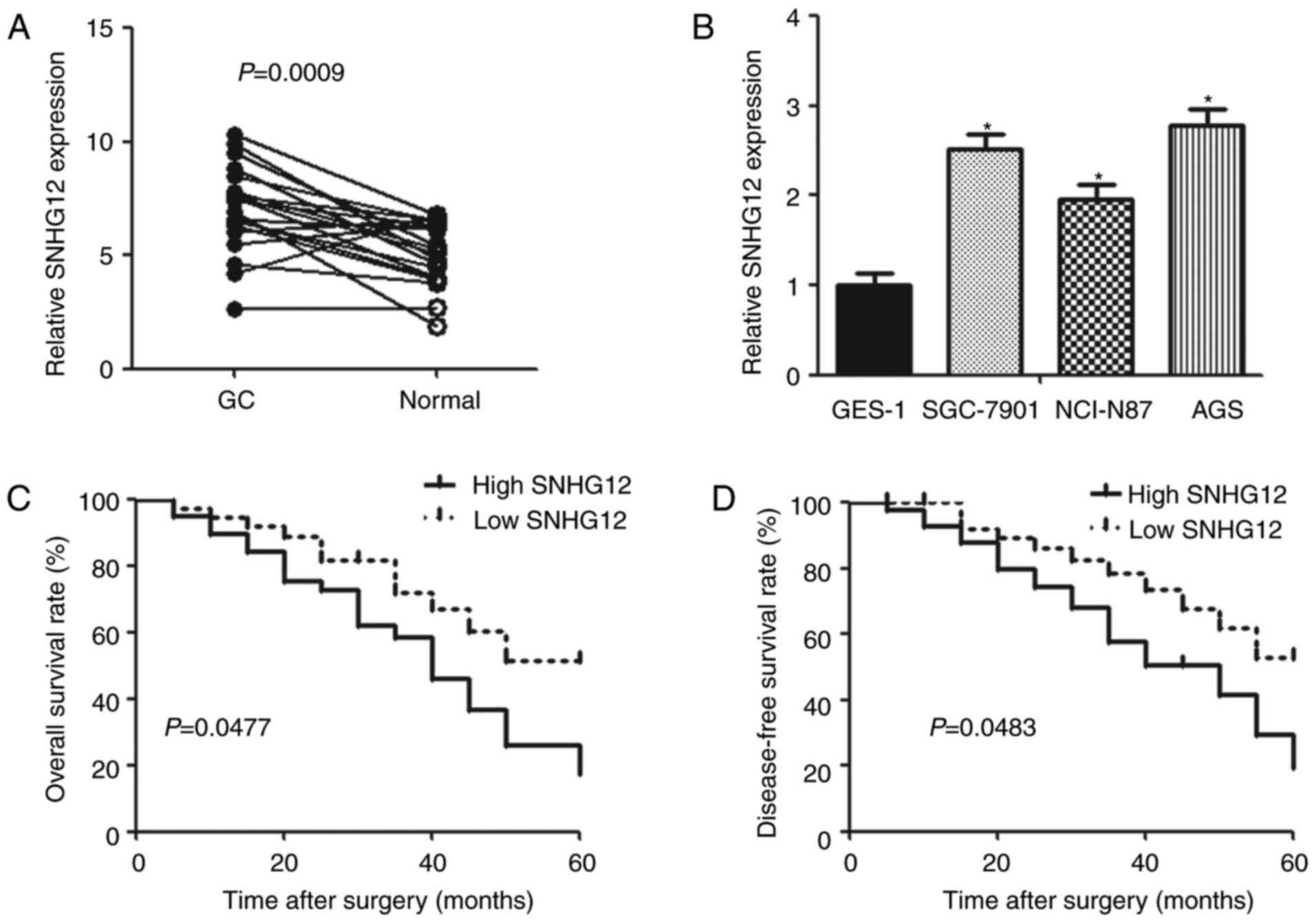
The expression of SNHG12 was examined in GC tissues
and adjacent normal tissues. According to the RT-qPCR analysis, the
expression of SNHG12 was higher in GC tissues compared with normal
tissues (Fig. 1A). SNHG12
expression increased significantly in gastric cancer cell lines,
including SGC7901, NCI-N87 and AGS compared with normal gastric
epithelial cells GES-1 (Fig.
1B).
Correlations between SNHG12 expression
in patients with GC and clinical characteristics
To investigate the relationship of clinical
characteristics and SNHG12 expression in gastric cancer, a cut-off
value (median expression value) of SNHG12 expression was used to
divide all 60 patients into two groups. As presented in Table I, patients with SNHG12 level higher
than the cut-off value were placed in the high expression group,
and patients with lower SNHG12 expression were assigned to the low
expression group. The expression of SNHG12 was positively
associated with tumor size (P=0.0043), TNM stage (P=0.0384),
distant metastasis (P=0.0018) and lymphatic metastasis (P=0.0045).
However, no significant difference was observed between SNHG12
level and sex or age. Patients with high SNHG12 expression
presented lower overall survival rate and disease-free survival
rate compared with those with low SNHG12 expression according to
the Kaplan-Meier analysis (P=0.0477, P=0.0483, respectively;
Fig. 1C and D).
 | Table I.Correlations between SNHG12 expression
in patients with gastric cancer and their clinical
characteristics. |
Table I.
Correlations between SNHG12 expression
in patients with gastric cancer and their clinical
characteristics.
|
|
| Relative SNHG12
expression |
|
|---|
|
|
|
|
|
|---|
| Group | Number of
patients | Low | High | P-value |
|---|
| Sex |
|
|
| 0.7952 |
| Male | 34 | 17 | 16 |
|
|
Female | 26 | 13 | 14 |
|
| Age |
|
|
| 0.7906 |
|
>50 | 38 | 11 | 12 |
|
| ≤50 | 22 | 19 | 18 |
|
| Tumor size (cm) |
|
|
| 0.0043 |
|
>5 | 19 | 11 | 22 |
|
| ≤5 | 41 | 19 | 8 |
|
| TNM stage |
|
|
| 0.0384 |
| I–II | 28 | 18 | 10 |
|
|
III–IV | 32 | 12 | 20 |
|
| Distant
metastasis |
|
|
| 0.0018 |
|
Negative | 27 | 19 | 7 |
|
|
Positive | 33 | 11 | 23 |
|
| Lymphatic
metastasis |
|
|
| 0.0045 |
| No | 36 | 21 | 10 |
|
|
Yes | 24 | 9 | 20 |
|
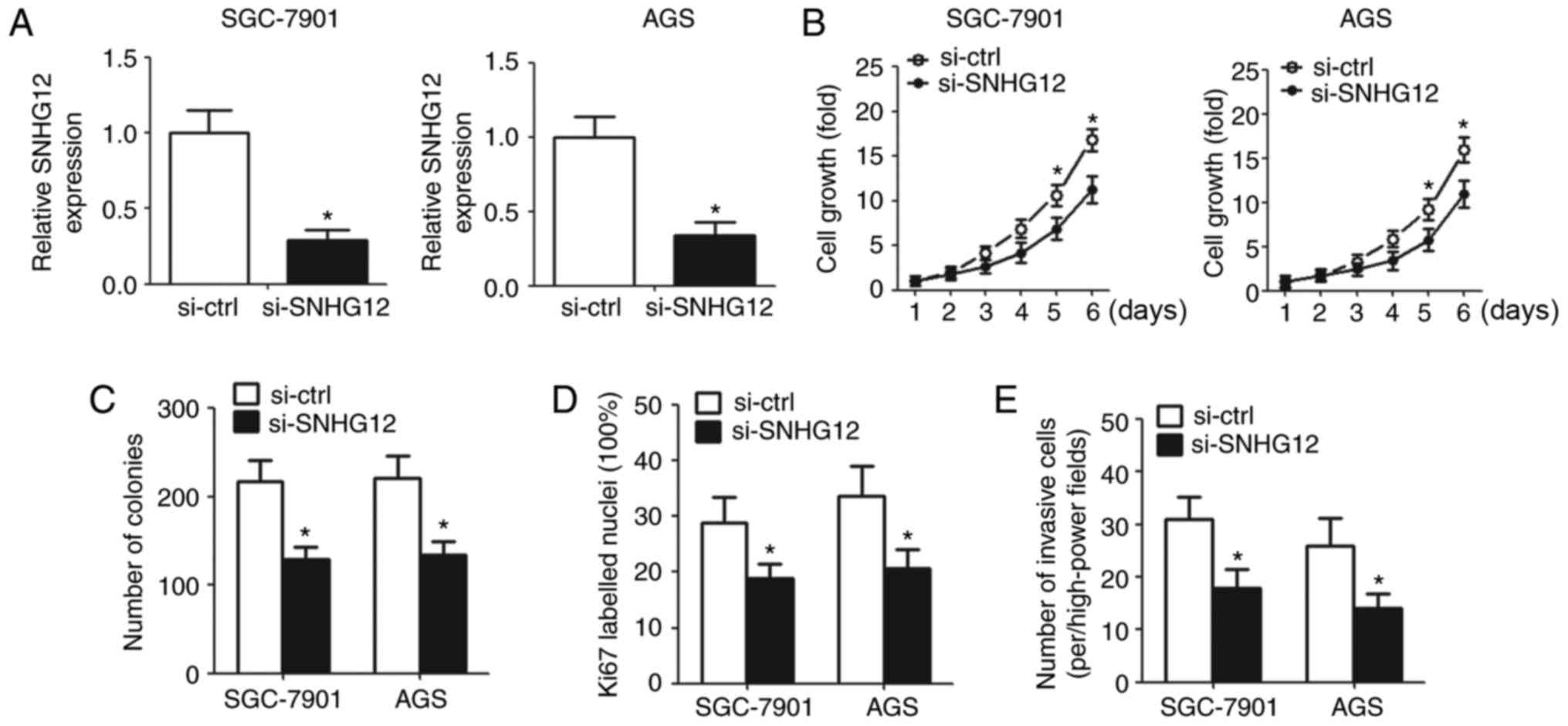
Knockdown of SNHG12 inhibits cell
proliferation and invasion in vitro
To elucidate the biological function of SNHG12, the
present study downregulated SNHG12 in GC cells by transfection with
SNHG12 siRNA. RT-qPCR was used to detect SNHG12 expression in
transfected cells (Fig. 2A). CCK-8
assay demonstrated that downregulation of SNHG12 suppressed cell
growth (Fig. 2B). The colony
formation assay revealed that the number of colonies decreased
significantly in the SNHG12 knockdown group compared with the
control group (Fig. 2C).
Ki67-labeled nuclei were reduced in cells transfected with
si-SNHG12 compared with cells transfected with si-ctrl according to
IHC analysis (Fig. 2D). In
addition, cell invasion was markedly inhibited in cells with SNHG12
knockdown compared with cells transfected with si-ctrl (Fig. 2E).
SNHG12 promotes cell proliferation and
invasion by targeting miR-320 directly
A previous study identified lncRNAs as competitive
endogenous RNA (ceRNA) for miRNAs or naturally occurring miRNA
sponges, which have a critical function in the progression of gene
expression and signaling pathways (19). The present study used LNCipedia
(lncipedia.org) to predict putative miRNA, which
is modulated by SNHG12 and determined that miR-320 was the
potential target of SNHG12. To clarify whether there were direct
interactions between SNHG12 and miR-320, the correlation between
the expression of SNHG12 and miR-320 in GC tissue specimens was
investigated. A negative correlation was observed between SNHG12
and miR-320 (Fig. 3A). The
relative miR-320 expression increased significantly after the
knockdown of SNHG12 in GC cell lines (Fig. 3B). Additionally, western blot
analysis demonstrated that the expression of target genes (CRKL,
p-Akt and p-Erk) of miR-320 was downregulated after the knockdown
of SNHG12 (Fig. 3C).
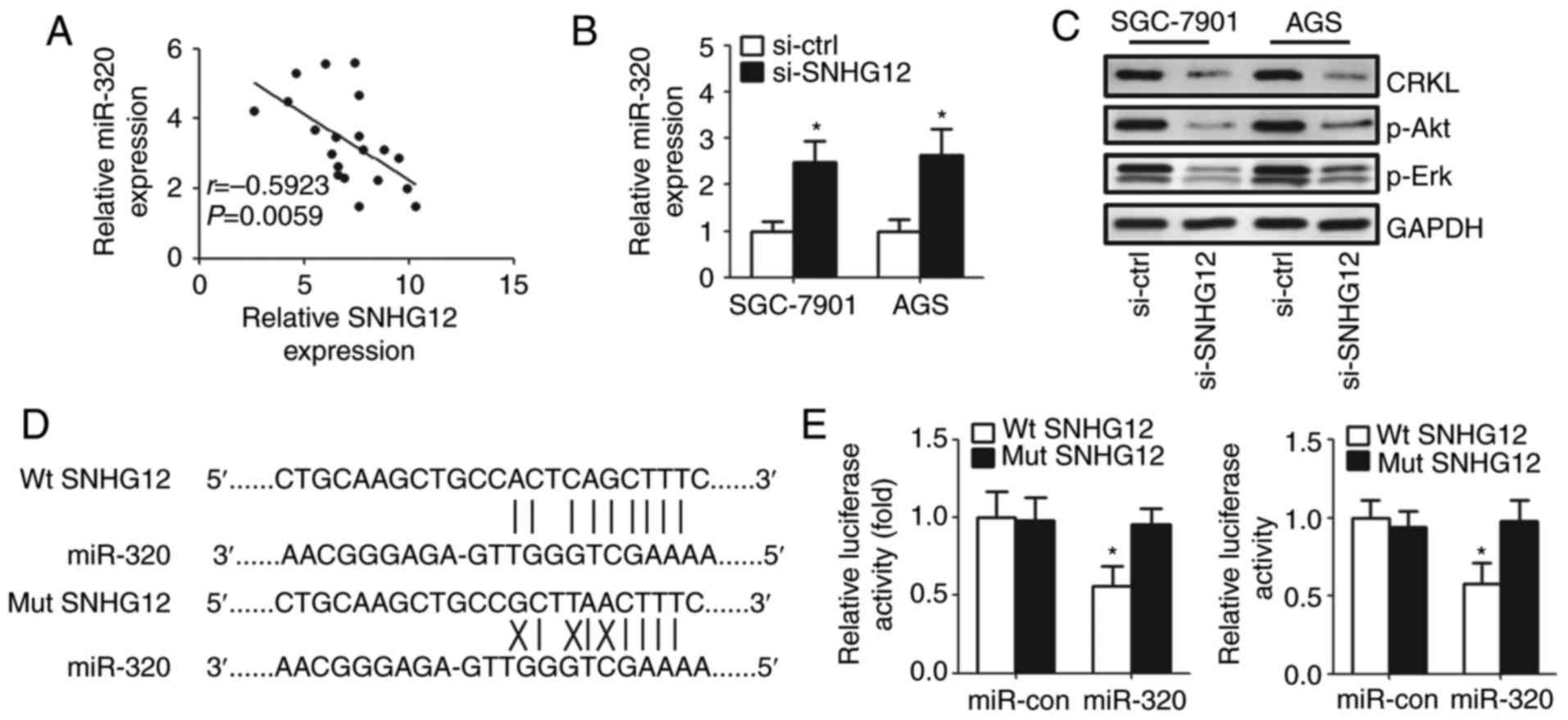 | Figure 3.SNHG12 may directly target miR-320.
(A) Inverse relationship between the SNHG12 and miR-320 expression
levels was identified in GC tissues. (B) Upregulation of miR-320
was found in GC cells with the knockdown of SNHG12 by reverse
transcription-quantitative polymerase chain reaction. (C) Western
blot analysis of CRKL, p-Akt and p-Erk expression in SGC-7901 and
AGS cells 48 h post-transfection. (D) Putative SNHG12 target
sequences in miR-320 are presented. (E) SGC-7901 and AGS cells were
co-transfected with the luciferase reporter plasmid carrying the
Wt/Mut sequences of SNHG12 and control or miR-320 mimics. A
dual-luciferase reporter system analysis was performed. Data were
expressed as the mean ± standard deviation, n=5. *P<0.05 vs.
control group. miR-320, microRNA-320; miR-con, microRNA control;
SNHG12, small nucleolar RNA host gene 12; si-SNHG12, SNHG12 small
interfering RNA; si-ctrl, control small interfering RNA; Wt,
wild-type; Mut, mutant; CRKL, CRK like proto-oncogene, adaptor
protein; p-Akt, phosphorylated-AKT serine/threonine kinase 1;
p-Erk, phosphorylated-extracellular-signal regulated kinase. |
To determine whether SNHG12 may directly bind
miR-320 in GC cells, the present study cloned the Wt and Mut SNHG12
sequences, which contained the potential binding site of miR-320
into a luciferase reporter gene system (Fig. 3D). As expected, miR-320 in both GC
cell lines led to a significantly reduced luciferase activity of
SNHG12, containing a Wt sequence without suppressing the activity
of SNHG12 with a mutant sequence (Fig.
3E).
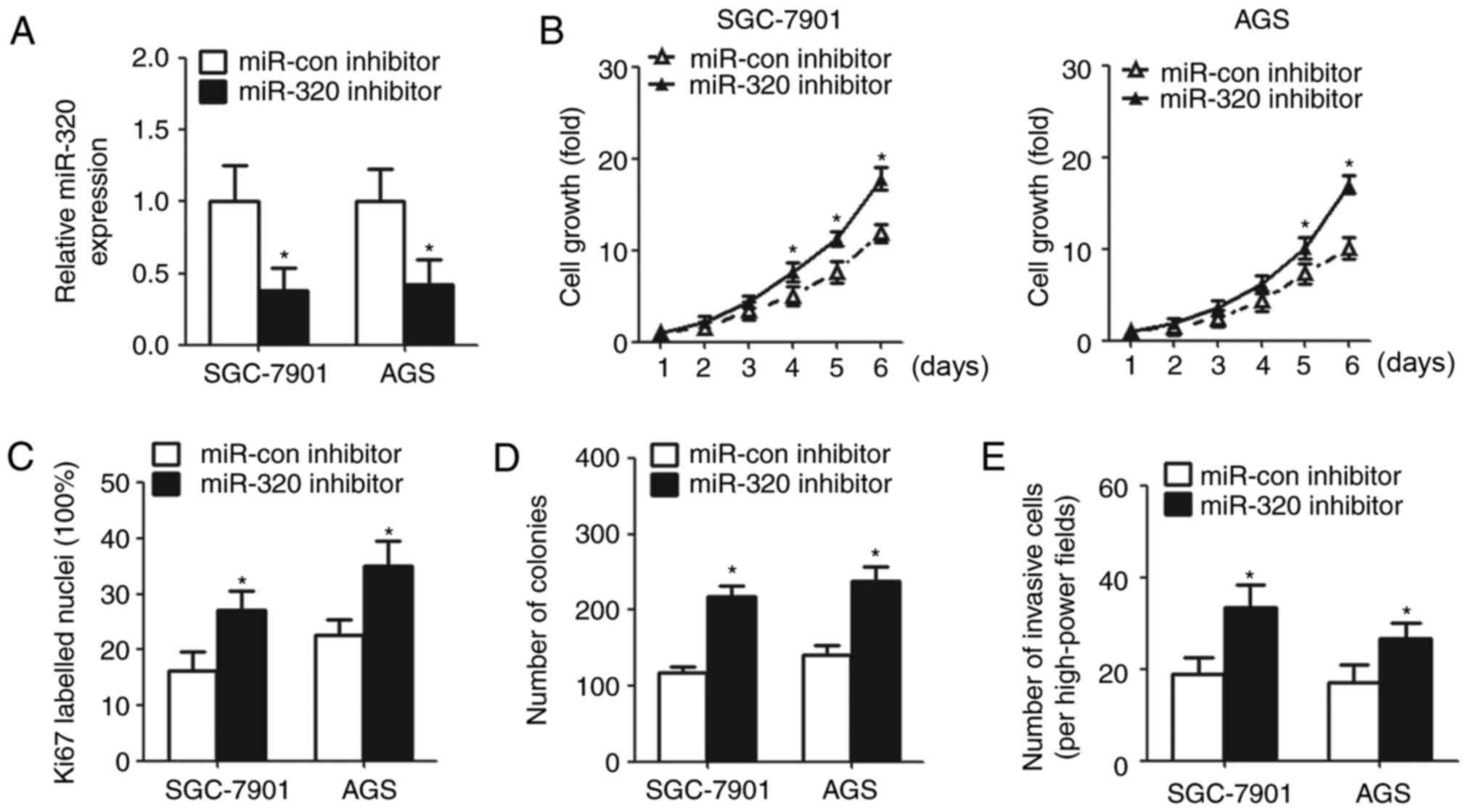
Inhibition of miR-320 reverses
SNHG12-induced effects on GC cells
In order to determine whether SNHG12 exerts its
function by sponging miR-320, the SNHG12-downregulated GC cells
were transfected with miR-320/miR-con inhibitor (Fig. 4A). The CCK-8 assay demonstrated
that cell growth was significantly promoted in the miR-320
inhibitor group (Fig. 4B).
Similarly, IHC analysis revealed that transfection with the miR-320
inhibitor increased Ki67-labeled nuclei in SNHG12-downregulated GC
cells (Fig. 4C). The number of
colonies and invasive cells were also upregulated in the miR-320
knockdown cells (Fig. 4D and
E).
Discussion
GC is a common disease with difficult complications
that include metastasis and recurrence, which seriously affect the
survival rate of patients with GC (20). A previous study revealed novel
functions of lncRNAs in the development and metastasis of human
cancers (21). SNHG12, being one
of the lncRNAs, with the small nucleolus RNA-encoding sequences in
its introns has been reported to contribute to the progression of
various tumors, such as hepatocellular carcinoma and osteosarcoma
(16,22). To the best of the authors'
knowledge; however, the function of SNHG12 in GC has not been
previously explored.
The present study determined that the expression
level of SNHG12 was significantly upregulated in GC tissues and
SGC-7901, NCI-N87 and AGS GC cell lines. Additionally, the
expression of SNHG12 was closely associated with tumor size, TNM
stage, distant and lymphatic metastasis. In addition, patients with
high SNHG12 expression had a lower overall survival and
disease-free survival rates according to the 60-month follow-up
survival survey. Additionally, silencing of SNHG12 in GC cells
inhibited cell proliferation and invasion. These findings suggested
that SNHG12 may be involved in the carcinogenesis and progression
of GC and it may be a prognostic marker for predicting overall and
disease-free survival of patients with GC. These findings were
consistent with previous studies demonstrating that SNHG12 acts as
a promoter in carcinoma progression and development (23,24).
Additionally, the present study validated miR-320 as
a direct-target gene of SNHG12, and identified a negative
correlation between SNHG12 and miR-320. Based on the bioinformatics
analysis, the putative binding sites between SNHG12 and miR-320
were revealed. Dual luciferase assay determined that miR-320 was a
direct downstream target of SNHG12. These findings suggested that
miR-320, a member of the miR-200 family, may have an essential role
in the progression of GC. This finding is consistent with previous
studies (25,26). miR-320 has been previously reported
to be implicated in many cancer-associated biological processes,
such as cell proliferation, invasion and metastasis (27–29).
A previous study indicated that miR-320 may inhibit proliferation
and invasion by targeting CRKL through the ERK and AKT pathway
(30). SNHG12 was observed to
upregulate the CRKL (a target gene of miR-320) expression in the
present study, which subsequently affected AKT and ERK signaling.
In addition, it was determined that the inhibition of miR-320
reversed the pro-tumor effects of SNHG12 on cell proliferation,
colony formation and invasion of GC cells.
In conclusion, the present study demonstrated that
SNHG12 may act as a promoter of tumorigenesis and development of GC
and become a potential prognostic biomarker for the diagnosis and
prognosis of GC. Additionally, SNHG12 was confirmed to act as a
sponge for miR-320 to modulate the CRKL expression, and further
affected the downstream AKT and ERK pathways.
Acknowledgements
The present study was supported by Zhejiang Public
Welfare Technology Research and Social Development Project (grant
no. 2014C33187).
References
|
1
|
Gu J, Li Y, Fan L, Zhao Q, Tan B, Hua K
and Wu G: Identification of aberrantly expressed long non-coding
RNAs in stomach adenocarcinoma. Oncotarget. 8:49201–49216.
2017.PubMed/NCBI
|
|
2
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang G, Zhao X, Li J, Yuan Y, Wen M, Hao
X, Li P and Zhang A: Racial disparities in stage-specific gastric
cancer: Analysis of results from the Surveillance Epidemiology and
End Results (SEER) program database. J Investig Med. 65:991–998.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen DL, Ju HQ, Lu YX, Chen LZ, Zeng ZL,
Zhang DS, Luo HY, Wang F, Qiu MZ, Wang DS, et al: Long non-coding
RNA XIST regulates gastric cancer progression by acting as a
molecular sponge of miR-101 to modulate EZH2 expression. J Exp Clin
Cancer Res. 35:1422016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liu Z, Chen Z, Fan R, Jiang B, Chen X,
Chen Q, Nie F, Lu K and Sun M: Over-expressed long noncoding RNA
HOXA11-AS promotes cell cycle progression and metastasis in gastric
cancer. Mol Cancer. 16:822017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li J, Gao J, Tian W, Li Y and Zhang J:
Long non-coding RNA MALAT1 drives gastric cancer progression by
regulating HMGB2 modulating the miR-1297. Cancer Cell Int.
17:442017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang KC and Chang HY: Molecular mechanisms
of long noncoding RNAs. Mol Cell. 43:904–914. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wapinski O and Chang HY: Long noncoding
RNAs and human disease. Trends Cell Biol. 21:354–361. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang O, Yang F, Liu Y, Lv L, Ma R, Chen C,
Wang J, Tan Q, Cheng Y, Xia E, et al: C-MYC-induced upregulation of
lncRNA SNHG12 regulates cell proliferation, apoptosis and migration
in triple-negative breast cancer. Am J Transl Res. 9:533–545.
2017.PubMed/NCBI
|
|
11
|
Troy A and Sharpless NE: Genetic ‘lnc’-age
of noncoding RNAs to human disease. J Clin Invest. 122:3837–3840.
2012. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang Y, Xu Y, Feng L, Li F, Sun Z, Wu T,
Shi X, Li J and Li X: Comprehensive characterization of lncRNA-mRNA
related ceRNA network across 12 major cancers. Oncotarget.
7:64148–64167. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ning S, Gao Y, Wang P and Li X, Zhi H,
Zhang Y, Liu Y, Zhang J, Guo M, Han D and Li X: Construction of a
lncRNA-mediated feed-forward loop network reveals global
topological features and prognostic motifs in human cancers.
Oncotarget. 7:45937–45947. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang F, Zhang L and Zhang C: Long
noncoding RNAs and tumorigenesis: Genetic associations, molecular
mechanisms, and therapeutic strategies. Tumour Biol. 37:163–175.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Huang B, Song JH, Cheng Y, Abraham JM,
Ibrahim S, Sun Z, Ke X and Meltzer SJ: Long non-coding antisense
RNA KRT7-AS is activated in gastric cancers and supports cancer
cell progression by increasing KRT7 expression. Oncogene.
35:4927–4936. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang JZ, Xu CL, Wu H and Shen SJ: LncRNA
SNHG12 promotes cell growth and inhibits cell apoptosis in
colorectal cancer cells. Braz J Med Biol Res. 50:e60792017.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Singh RP, Deep G, Chittezhath M, Kaur M,
Dwyer-Nield LD, Malkinson AM and Agarwal R: Effect of silibinin on
the growth and progression of primary lung tumors in mice. J Natl
Cancer Inst. 98:846–855. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Esteller M: Non-coding RNAs in human
disease. Nat Rev Genet. 12:861–874. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sun M, Nie FQ, Wang ZX and De W:
Involvement of lncRNA dysregulation in gastric cancer. Histol
Histopathol. 31:33–39. 2016.PubMed/NCBI
|
|
21
|
Rogoyski OM, Pueyo JI, Couso JP and
Newbury SF: Functions of long non-coding RNAs in human disease and
their conservation in Drosophila development. Biochem Soc Trans.
45:895–904. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ruan W, Wang P, Feng S, Xue Y and Li Y:
Long non-coding RNA small nucleolar RNA host gene 12 (SNHG12)
promotes cell proliferation and migration by upregulating
angiomotin gene expression in human osteosarcoma cells. Tumour
Biol. 37:4065–4073. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lan T, Ma W, Hong Z, Wu L, Chen X and Yuan
Y: Long non-coding RNA small nucleolar RNA host gene 12 (SNHG12)
promotes tumorigenesis and metastasis by targeting miR-199a/b-5p in
hepatocellular carcinoma. J Exp Clin Cancer Res. 36:112017.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang JZ, Xu CL, Wu H and Shen SJ: LncRNA
SNHG12 promotes cell growth and inhibits cell apoptosis in
colorectal cancer cells. Braz J Med Biol Res. 50:e60792017.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhu H, Jiang X, Zhou X, Dong X, Xie K,
Yang C, Jiang H, Sun X and Lu J: Neuropilin-1 regulated by miR-320
contributes to the growth and metastasis of cholangiocarcinoma
cells. Liver Int. Jun 15–2017.(Epub ahead of print). View Article : Google Scholar
|
|
26
|
Zhang T, Zou P, Wang T, Xiang J, Cheng J,
Chen D and Zhou J: Down-regulation of miR-320 associated with
cancer progression and cell apoptosis via targeting Mcl-1 in
cervical cancer. Tumour Biol. 37:8931–8940. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sun JY, Xiao WZ, Wang F, Wang YQ, Zhu YH,
Wu YF, Miao ZL and Lin YC: MicroRNA-320 inhibits cell proliferation
in glioma by targeting E2F1. Mol Med Rep. 12:2355–2359. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hsieh IS, Chang KC, Tsai YT, Ke JY, Lu PJ,
Lee KH, Yeh SD, Hong TM and Chen YL: MicroRNA-320 suppresses the
stem cell-like characteristics of prostate cancer cells by
downregulating the Wnt/beta-catenin signaling pathway.
Carcinogenesis. 34:530–538. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang W, Yang J, Xiang YY, Pi J and Bian J:
Overexpression of Hsa-miR-320 Is associated with invasion and
metastasis of ovarian cancer. J Cell Biochem. 118:3654–3661. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhao Y, Dong Q and Wang E: MicroRNA-320
inhibits invasion and induces apoptosis by targeting CRKL and
inhibiting ERK and AKT signaling in gastric cancer cells. Onco
Targets Ther. 10:1049–1058. 2017. View Article : Google Scholar : PubMed/NCBI
|