Introduction
Hepatocellular carcinoma (HCC) is the fifth most
common malignancy and the second most common contributing factor to
cancer mortality rates worldwide (1–4).
Although progress has been made in diagnostic and therapeutic
strategies, the prognosis of patients with advanced HCC remains
unsatisfactory, primarily due to its frequent recurrence and
metastases (5–9). The 5-year survival rate has been
previously reported to be 25–39% following resection (10–13).
Therefore, it is necessary to investigate the underlying molecular
mechanisms in the development of HCC to identify novel diagnostic
markers and innovative therapeutic targets for patients with
HCC.
MicroRNAs (miRNAs), a class of small, endogenous,
single-stranded, non-coding RNAs, are 18–25 nucleotides in length
and can alter gene expression. The aberrant expression of miRNAs
can directly induce multiple diseases, including cancer. miRNAs
have been used as non-invasive tumor biomarkers as they are
involved in differentiation, proliferation and apoptosis in various
malignancies (14–17). In HCC, the abnormal expression of
miRNAs is essential during the progression of HCC (18–21).
As reported, several miRNAs with lower expression levels can act as
tumor suppressors in HCC tumorigenesis and growth (22–27),
whereas certain upregulated miRNAs may function as HCC activators
(28–31).
miR-223-3p is an miRNA, which has been identified in
human diseases in previous studies. In penile carcinoma, the
overexpression of miR-223-3p was considered a molecular classifier
to differentiate tumors from non-cancerous tissues (32). The upregulation of miR-223-3p may
be a potential diagnostic biomarker in esophageal squamous cell
carcinoma (33). In addition,
downregulated levels of miR-223-3p are closely associated with poor
prognosis in bladder cancer (34).
In HCC, cell proliferation is enhanced by low expression levels of
miR-223-3p (35). A number of
studies have also reported that downregulated miR-223-3p is a
potentially favorable biomarker for early-stage diagnosis of HCC
(36–38). However, certain studies have
revealed that miR-223-3p is preferentially expressed in HCC,
compared with healthy controls (39,40).
Although it is maintained that circulating miR-223-3p can provide a
certain degree of diagnostic accuracy in patients with HCC
(41,42), reports on the diagnostic value for
HCC in serum and tissues remain limited. Therefore, the present
study aimed to perform a more comprehensive meta-analysis of the
diagnostic value of miR-223-3p and precursor miR-223, based on data
extracted from the Gene Expression Omnibus (GEO), The Cancer Genome
Atlas (TCGA) and qualified literature.
miR-223-3p can directly target different genes and
signaling pathways in a diverse set of diseases. For example, in
non-small-cell lung cancer (NSCLC), an increased level of
miR-223-3p induces the downregulation of insulin-like growth factor
1 receptor (IGF1R), which may indicate a potential therapeutic
strategy for tyrosine kinase inhibitor-resistant NSCLC (43). The invasiveness of bladder
carcinoma can be suppressed by targeting nuclear receptor
coactivator 1 (NCOA1) by miR-223-3p (34). In addition, miR-223-3p can target
stathmin 1 (STMN1) and IGF1R in gastric carcinoma to inhibit tumor
progression (44). A previous
study also reported that miR-223-3p accelerated the development of
HCC through mediating the G2/M transition by targeting STMN1
(45), and miR-223-3p inhibits the
onset and progression of HCC by repressing the gene expression of
c-Myc (46). However, the
potential mechanisms underlying the activity of miR-223-3p in HCC
remain to be fully elucidated. Therefore, the specific molecular
mechanisms by which miR-223-3p is involved in the occurrence and
development of HCC requires investigation. The present study aimed
to clarify the role of miR-223-3p in HCC tumorigenesis. The use of
high-throughput data, including microarray and RNA-sequencing, and
bioinformatics may provide novel insights into how miR-223-3p
functions in HCC, and to provide a theoretical foundation for the
diagnosis and personalized therapy of patients with HCC.
Materials and methods
Clinical significance: GEO database
Mining expression profiles from
microarrays
The HCC microarray profiles were obtained from the
National Center for Biotechnology Information GEO (http://www.ncbi.nlm.nih.gov/geo/) based on the
following search strategy: ‘(miR or microRNA or miRNA) and
(malignan* or cancer or tumor or tumour or neoplas* or carcinoma)
and (hepatocellular or liver or hepatic OR HCC)’. The search
included all terms that begin with a word by entering the word
followed by an asterisk (*), the wildcard character. Data including
the expression level of miR-223-3p in human HCC and in
non-cancerous controls were collected.
Manufacturing receiver operating curve
(ROC) and scatter plots of the expression of miR-223-3p
Based on the expression level of miR-223-3p, ROC and
scatter plots were separately produced in GraphPad Prism 6.0
(GraphPad Software, Inc., La Jolla, CA, USA). The sensitivity and
specificity of miR-223-3p in HCC were also evaluated for
meta-analysis.
Statistical analysis
SPSS software (version 22.0; IBM Corp., Armonk, NY,
USA) was used to conduct statistical analysis. The expression
profiles from the GEO were normalized by log2-transformation. The
expression of miR-223-3p was calculated as the mean ± standard
deviation. An independent Student's t-test was performed to
calculate the difference between HCC cases and non-cancerous
tissues. P<0.05 was considered to indicate a statistically
significant difference.
Clinical significance: TCGA database
TCGA data
To further clarify the correlations between the
precursor miR-223 and HCC, the reads per million (RPM) miRNA mapped
data of miR-223 in 375 HCC and 50 non-cancerous liver tissues were
scanned and extracted from the TCGA database (http://cancergenome.nih.gov). Additionally, the raw
mRNA count (level 3) and the corresponding clinical information of
the 375 patients with HCC were obtained.
Analyzing the clinical features of
miR-223
For determining the miR-223-expressing conditions in
the different clinicopathological features of HCC, scatter plots
were constructed. The Kaplan-Meier curve was used to examine the
prognostic value of miR-223 in HCC.
Statistical analysis
The mRNA expression level values were normalized by
conversion to a log2 scale using the edgeR package of R software
(version 3.5; http://www.bioconductor.org/packages/release/bioc/html/edgeR.html),
in addition to the RPM data. The mRNAs with an absolute log2-fold
change >2 were identified as differentially expressed. The
differences in the levels of miR-223 between various clinical
features were assessed using an independent Student's t-test.
P<0.05 was considered to indicate a statistically significant
difference.
Identification of diagnostic accuracy
from eligible studies
A literature search (up to 09/05/2017) was performed
using the following databases: PubMed (https://www.ncbi.nlm.nih.gov/pubmed), EBSCO
(https://www.ebsco.com), Wiley Online Library
(http://onlinelibrary.wiley.com), Web of
Science (http://isiknowledge.com), Science Direct
(http://www.sciencedirect.com), Cochrane
Central Register of Controlled Trials (http://www.cochranelibrary.com), Google Scholar
(https://scholar.google.com), EMBASE
(https://www.embase.com), Ovid (http://ovidsp.ovid.com) and LILACS (http://lilacs.bvsalud.org), in addition to the
following Chinese databases: Chinese CNKI (http://www.cnki.net), Chinese Chong Qing VIP
(http://lib.cqvip.com), Chinese Wan Fang
(http://www.wanfangdata.com.cn) and China
Biology Medicine disc (http://www.sinomed.ac.cn). The search strategy was as
follows ‘(miR-223 or miRNA-223 or microRNA-223 or miR223 or
miRNA223 or microRNA223 or miR 223 or miRNA 223 or microRNA 223 or
miR-223 or miRNA-223-3p or microRNA-223-3p) and (malignan* or
cancer or tumor or tumour or neoplasm*, or carcinoma) and
(hepatocellular or liver or hepatic or HCC). The inclusion criteria
were as follows: i) Studies evaluating the diagnostic value of
downregulated miR-223-3p (or miR-223) in serum, plasma or HCC
tissues; ii) studies providing sufficient information for
calculating the sensitivity and specificity of miR-223-3p (or
miR-223) for diagnosing HCC; and iii) studies published in English
or Chinese. For eligible studies with data published more than
once, those with the largest patient sample size were included. In
addition, the following exclusion criteria were used: i) Studies
detecting miR-223-3p (or miR-223) in cell lines or animals; ii)
results published as an abstract, summary, case report, comment
letter, review or editorial; and iii) studies without sufficient
data for evaluating the diagnostic value of miR-223-3p (or miR-223)
in human HCC.
The quality assessment of literature was performed
independently by two authors (Ms. Li-jie Zhang and Ms. Mei-Ling
Yang) using the Quality Assessment of Diagnostic Accuracy Studies
(QUADAS)-2 Tool (www.bris.ac.uk/quadas). A discussion of consistency
was applied when divergence appeared. QUADAS-2 was used to judge
bias and applicability between ‘high’ and ‘low’ risks, which
consisted of patient selection, index test, reference standard, and
flow and timing (47).
Meta-analysis
Stata (version 12.0; StataCorp, College Station, TX,
USA) to conduct the meta-analysis. The ‘midas’ module of Stata 12.0
(StataCorp LP, College Station, TX, USA) was used to combine the
diagnostic data from GEO, TCGA and the literature. The
heterogeneity was calculated using a χ2 test and
I2, and I2>50% or/and P<0.05 for the Q
test were considered to be statistically significant in
heterogeneity. The fixed-effect model was used if there was
significant heterogeneity. Otherwise, the random effect model was
used (48). The summarized indices
included sensitivity, specificity, likelihood ratio and diagnostic
odds ratio with a 95% confidence interval (CI). Based on the
sensitivity and specificity, a summary receiver operating
characteristic curve (SROC) was used for evaluation. Publication
bias was measured by plotting Deek's funnel plot (49). P<0.05 (two-sided) was considered
to indicate a statistically significant difference
Bioinformatics evaluation
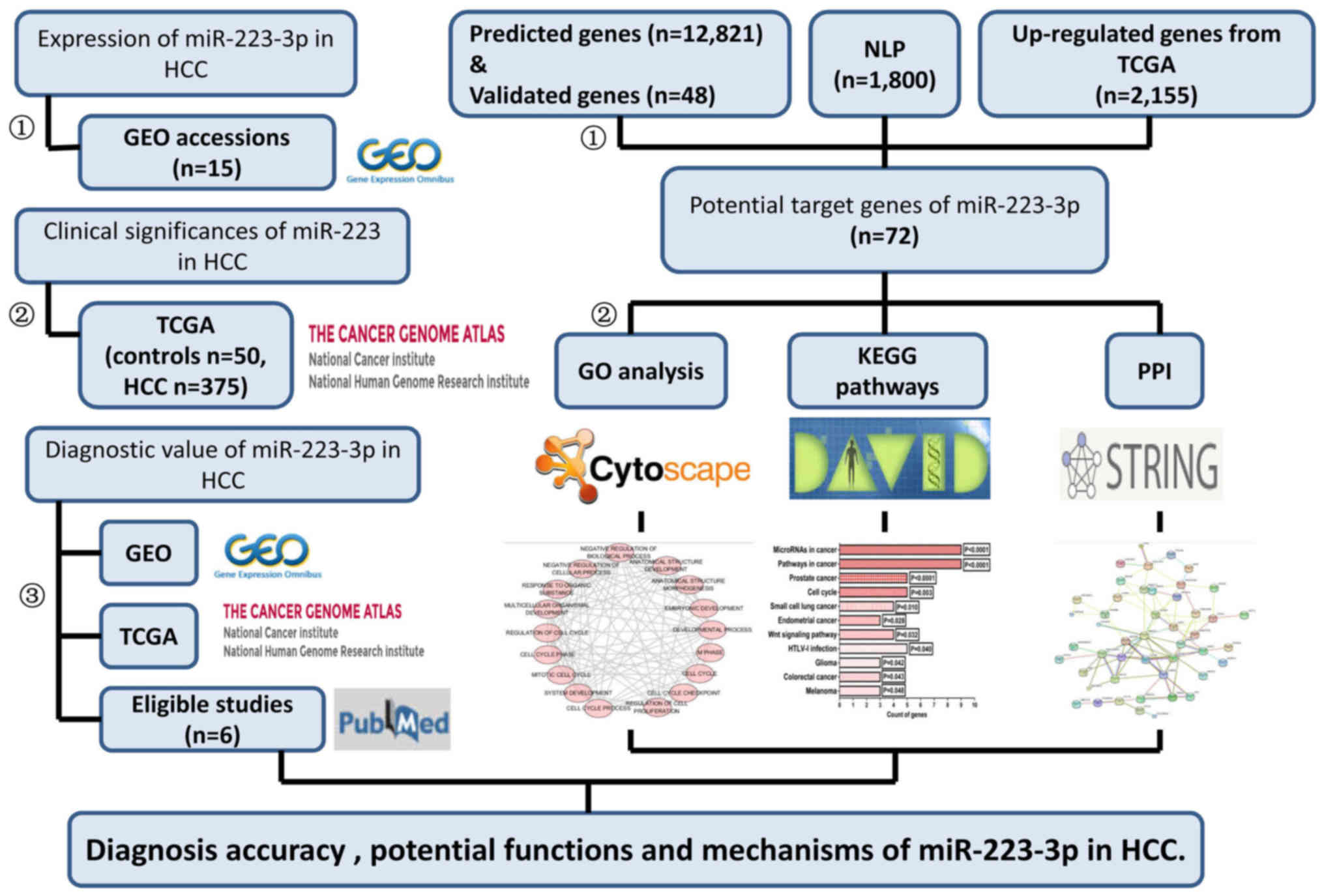
Selection of potential target genes. The targets of
miR-223-3p were selected from the intersection of 13 prediction
tools, natural language processing (NLP) of HCC, and differentially
expressed genes from TCGA. A total of 12 online tools were used for
predicting potential targets: miRWalk (http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2),
Micro T4 (http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=microtv4/index),
miRanda (http://www.microrna.org/microrna/getDownloads.do),
miRBridge (https://www.ncbi.nlm.nih.gov/pubmed/?term=20385095),
miRDB (http://mirdb.org/miRDB/download.html), miRMap
(http://mirmap.ezlab.org/downloads/mirmap201301e/),
miRNAMap (ftp://mirnamap.mbc.nctu.edu.tw/miRNAMap2), PICTAR2
(http://dorina.mdc-berlin.de/rbp_browser/download_hg19.html),
PITA (https://genie.weizmann.ac.il/pubs/mir07/mir07_exe.html),
RNA22 (https://cm.jefferson.edu/rna22/), RNAhybrid
(https://bibiserv.cebitec.uni-bielefeld.de/rnahybrid/dl_pre-page.html),
and TargetScan (http://www.targetscan.org/cgi-bin/targetscan/data_download.cgi?db=vert_61).
The genes found to overlap on at least three prediction programs
were identified as targets. The predicted targets of miR-223-3p in
the miTarBase program were confirmed using western blot analysis
and quantitative polymerase chain reaction reporter assays. Genes
expressed aberrantly in HCC in studies published between 01/01/1980
and 05/25/2015 were gathered with NLP techniques, as described in
our previous study (50).
Additionally, the extracted genes were added to a list using ABNER
(http://pages.cs.wisc.edu/~bsettles/abner/) (51). The correlations between hub genes
and miR-223-3p were assessed using Spearman's correlation.
Prospective mechanisms and
functions
The molecular mechanisms and biological functions of
potential miR-223-3p targets in HCC were identified by enrichment
analysis and pathway annotations. Gene Ontology (GO) analysis and
the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways
analysis were performed using the Database for Annotation,
Visualization and Integrated Discovery (DAVID; https://david.ncifcrf.gov/). P<0.05 was considered
to indicate a significant pathway. GO associations were also
visualized in Cytoscape (version 3.4.0; http://www.cytoscape.org/).
Protein-protein interaction (PPI)
network construction
The potential targets were mapped to the Search Tool
for the Retrieval of Interacting Genes database (http://www.string-db.org/) (52) to evaluate significant correlations
between different protein pairs. The overall design of the present
study is shown in the flow chart in Fig. 1.
Results
Expression of miR-223-3p in HCC in
GEO
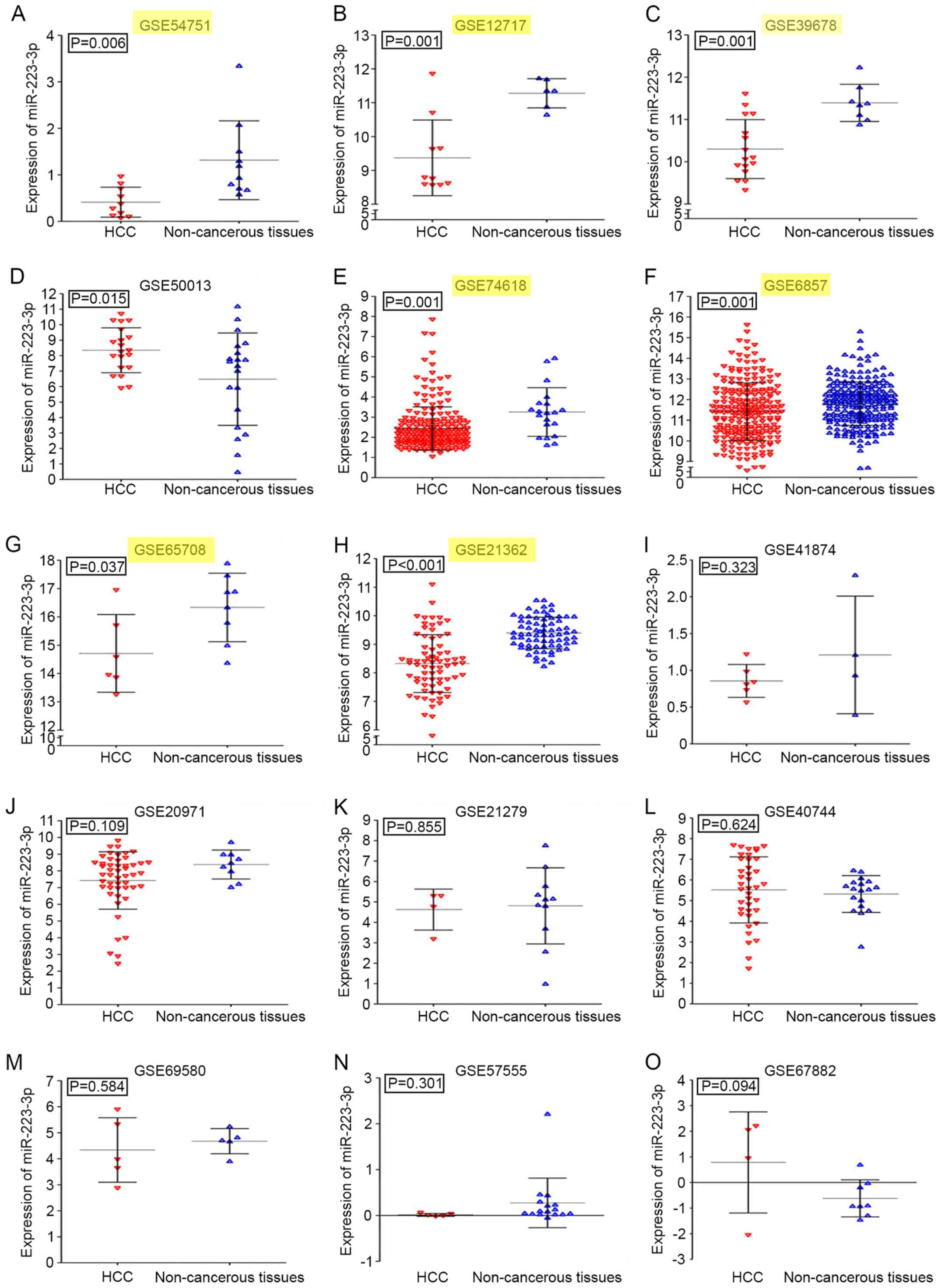
A total of 15 series (GSE) in the GEO database were
extracted for the present study, which consisted of 706 HCC samples
and 457 normal controls. In addition to GSE21279, detecting the
expression level of the precursor miR-223, the other 14 GEO
accessions were all identified as miR-223-3p expression profiles.
Detailed information regarding each GEO accession is shown in
Table I. The expression level of
miR-223-3p was significantly downregulated in patients with HCC in
seven of the 15 GEO datasets (Fig.
2A-O). Compared with the non-cancerous controls, the miR-223-3p
HCC expression profiles were significantly downregulated in
GSE54751, GSE12717, GSE39678, GSE74618, GSE6857, GSE65708 and
GSE21362 (P<0.05). By contrast, the expression level of
miR-223-3p in GSE50013 was upregulated in HCC groups (P=0.015). The
levels of miR-223-3p were also obtained based on TCGA expression
profiles. In liver HCC, the levels of miR-223-3p in cancer tissues
were also lower, compared with those in the normal controls
(Fig. 3A).
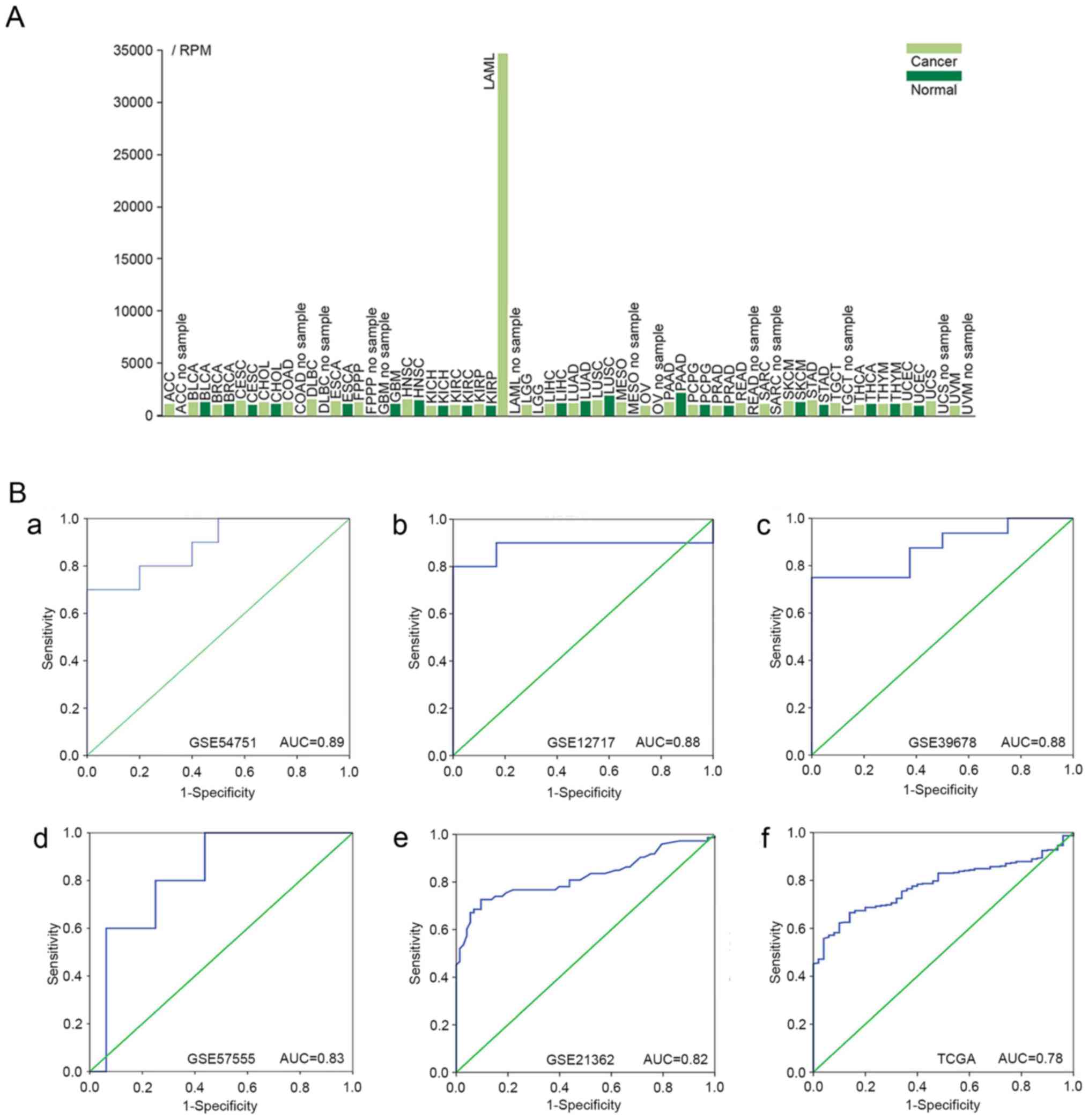 | Figure 3.Expression profiles. (A) TCGA
expression profiles of miR-223-3p in different diseases. The light
green bars represent cancer tissues. The dark green bars represent
normal tissues. ACC, adenoid cystic carcinoma; BLCA, bladder
urothelial carcinoma; BRCA, breast invasive carcinoma; CESC,
cervical squamous cell carcinoma and endocervical adenocarcinoma;
CHOL, cholangiocarcinoma; COAD, colon adenocarcinoma; DLBC, diffuse
large B-cell lymphoma; ESCA, esophageal carcinoma; FPPP,
Formalin-fixed Paraffin Pilot Project; GBM, glioblastoma
multiforme; HNSC, head and neck squamous cell carcinoma; KICH,
chromophobe renal cell carcinoma; KIRC, kidney renal clear cell
carcinoma; KIRP, kidney renal papillary cell carcinoma; LAML, acute
myeloid leukemia; LGG, brain lower grade glioma; LIHC, liver
hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung
squamous cell carcinoma; MESO, mesothelioma; OV, ovarian serous
cystadenocarcinoma; PAAD, pancreatic adenocarcinoma; PCPG,
pheochromocytoma and paraganglioma; PRAD, prostate adenocarcinoma;
READ, rectum adenocarcinoma; SARC, sarcoma; SKCM, skin cutaneous
melanoma; STAD, stomach adenocarcinoma; TGCT, testicular germ cell
cancer; THCA, papillary thyroid carcinoma; THYM, thymoma; UCEC,
uterine corpus endometrial carcinoma; UCS, uterine carcinosarcoma;
UVM, uveal melanoma; TCGA, The Cancer Genome Atlas; RPM, reads per
million. (B) miR-223-3p ROC curves in GEO and TCGA datasets. (a)
ROC of GSE54751; (b) ROC of GSE12717; (c) ROC of GSE39678; (d) ROC
of GSE57555; (e) ROC of GSE21362; (f) ROC of TCGA dataset. ROC,
receiver operating characteristic; GEO, Gene Expression Omnibus;
TCGA, The Cancer Genome Atlas. |
 | Table I.Fifteen gene expression omnibus
series datasets of HCC. |
Table I.
Fifteen gene expression omnibus
series datasets of HCC.
|
| HCC | Non-cancerous |
|
|---|
|
|
|
|
|
|---|
| ID | n | Mean ± SD | n | Mean ± SD | P-value |
|---|
| GSE57555 |
5 | 0.013±0.032 | 16 | 0.275±0.540 | 0.300 |
| GSE69580 |
5 | 4.337±1.241 |
5 | 4.677±0.483 | 0.951 |
| GSE54751 | 10 | 0.412±0.323 | 10 | 1.314±0.848 | 0.006 |
| GSE41874 |
6 | 0.855±0.224 |
4 | 1.209±0.800 | 0.323 |
| GSE50013 | 20 | 8.342±1.452 | 20 | 6.460±2.979 | 0.017 |
| GSE40744 | 39 | 5.518±1.600 | 18 | 5.318±0.891 | 0.549 |
| GSE20971 | 49 | 7.424±1.716 |
9 | 8.383±0.866 | 0.108 |
| GSE21362 | 73 | 8.324±1.013 | 73 | 9.396±0.553 | <0.001 |
| GSE12717 | 10 | 9.370±1.118 |
6 | 11.279±0.432 | 0.001 |
| GSE6857 | 241 | 11.417±1.396 | 241 | 11.802±1.054 | 0.006 |
| GSE67882 |
4 | 0.783±1.976 |
8 | −0.621±0.724 | 0.094 |
| GSE65708 |
6 | 14.711±1.372 |
8 | 16.335±1.210 | 0.037 |
| GSE74618 | 218 | 2.434±1.074 | 20 | 3.254±1.211 | 0.001 |
| GSE39678 | 16 | 10.300±0.697 |
8 | 11.393±0.440 | 0.001 |
| GSE21279 |
4 | 4.624±1.001 | 11 | 4.809±1.862 | 0.855 |
The ROC analysis revealed a significant diagnostic
value of miR-223-3p for HCC. Of the 15 GEO profiles, five with an
AUC >0.80 are shown in Fig.
3B-a-e (P<0.05).
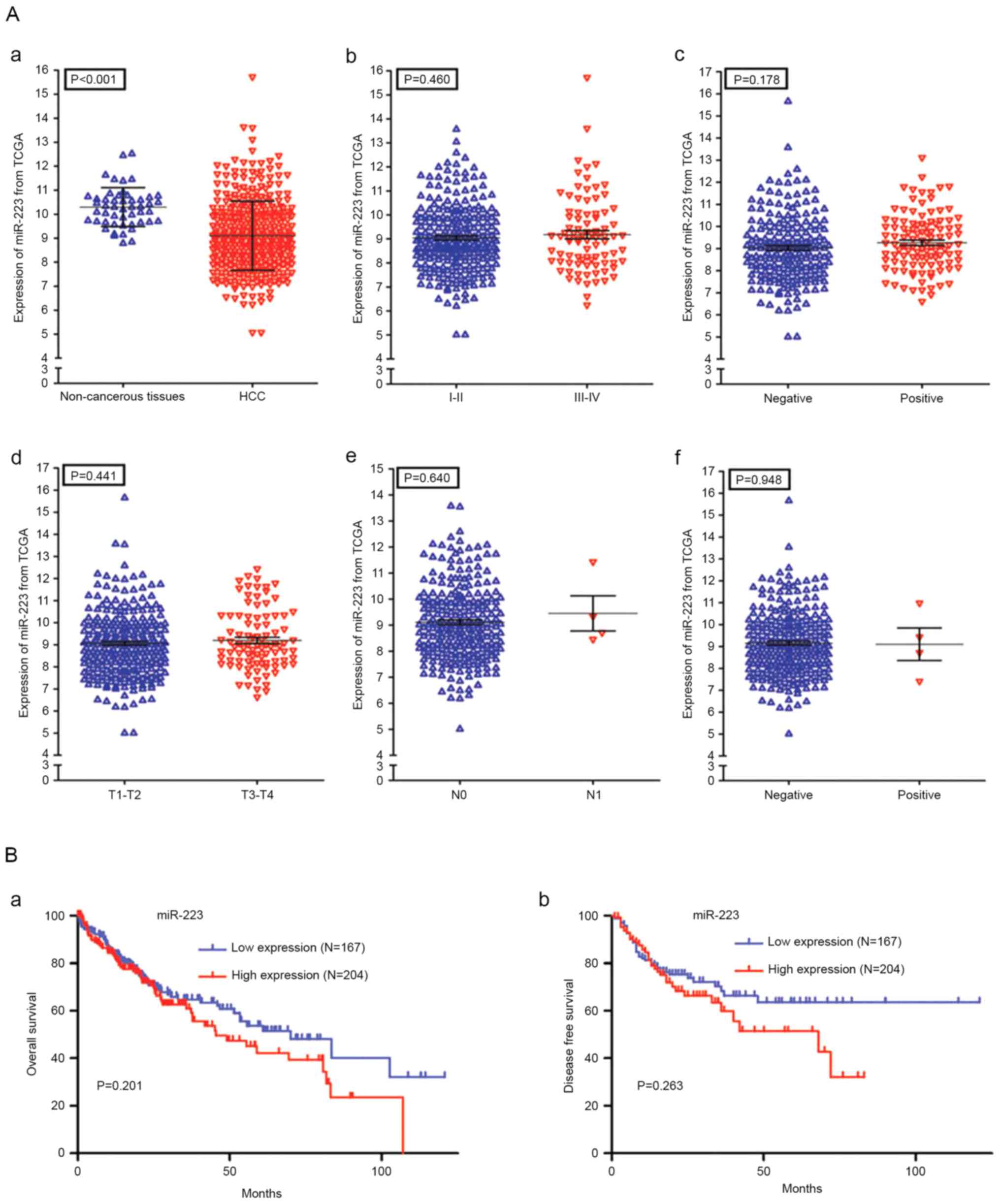
Expression of miR-223 in HCC in
TCGA
A total of 371 patients with HCC (252 men and 119
women) and 50 normal controls were derived from the TCGA dataset.
The results from the ROC analysis for TCGA RNA-seq data are shown
in Fig. 3B-f (AUC, 0.78; 95% CI,
0.74–0.83; P<0.001). Additionally, the present study analyzed
the associations between the level of miR-223 and the clinical
features of patients with HCC. As is shown in Fig. 4A-a, there was a significant
difference in the expression of miR-223 between the HCC tissues
(9.112±1.436) and healthy controls (10.300±0.811; P<0.001).
However, no statistically significant correlations were found
between the level of miR-223 and various clinicopathological
features of the patients with HCC (Fig. 4A-b-f; Table II). The effect of the expression
of miR-223 on survival outcomes in HCC was also insignificant
(P=0.201 for overall survival, P=0.263 for disease free survival;
Fig. 4B).
 | Table II.Association between levels of
microRNA-223 and clinicopathological variables in patients with HCC
from The Cancer Genome Atlas database. |
Table II.
Association between levels of
microRNA-223 and clinicopathological variables in patients with HCC
from The Cancer Genome Atlas database.
| Parameter | n | Mean ± SD | T-value | P-value |
|---|
| Group |
|
|
| <0.001 |
|
HCC | 371 | 9.112±1.436 | 8.659 |
|
|
Normal | 50 | 10.300±0.811 |
|
|
| Sex |
|
|
| 0.736 |
|
Male | 252 |
9.095±1.426 | 0.337 |
|
|
Female | 119 | 9.149±1.462 |
|
|
| Tumor status |
|
|
| 0.625 |
| Tumor
present | 111 | 9.116±1.531 | 0.489 |
|
|
Tumor-free | 233 | 9.037±1.327 |
|
|
| Family history |
|
|
| 0.174 |
| + | 110 | 9.284±1.479 | 1.361 |
|
| − | 210 | 9.053±1.427 |
|
|
| Risk factors |
|
|
| 0.734 |
| + | 251 | 9.092±1.435 | 0.339 |
|
| − | 101 | 9.149±1.435 |
|
|
| Other
malignancy |
|
|
| 0.485 |
| + | 36 | 9.271±1.446 | 0.700 |
|
| − | 335 | 9.095±1.436 |
|
|
| Inflammation |
|
|
| 0.749 |
| + | 117 | 9.060±1.418 | 0.321 |
|
| − | 118 | 9.000±1.483 |
|
|
| Ethnicity |
|
|
| 0.650 |
|
Hispanic or Latino | 18 | 9.297±1.844 | 0.461 |
|
|
Non-Hispanic or Latino | 334 | 9.094±1.418 |
|
|
| T stage |
|
|
| 0.981 |
| G
I–II | 275 | 9.105±1.295 | 0.024 |
|
| G
III–IV | 93 | 9.109±1.812 |
|
|
| N stage |
|
|
| 0.894 |
| N1 |
4 | 9.190±1.300 | 0.133 |
|
|
NX-N0 | 254 | 9.093±1.446 |
|
|
| M stage |
|
|
| 0.286 |
| M1 |
4 | 8.324±0.844 | 1.069 |
|
|
MX-M0 | 269 | 9.087±1.423 |
|
|
| Pathologic
stage |
|
|
| 0.913 |
| Stage
I–II | 260 | 9.084±1.296 | 0.110 |
|
| Stage
III–IV | 87 | 9.108±1.875 |
|
|
| Vascular
invasion |
|
|
| 0.214 |
| + | 110 | 9.227±1.635 | 1.247 |
|
| − | 206 | 9.002±1.298 |
|
|
Inclusion of previous literature and
quality assessment
A total of 295 relevant studies were identified in
the literature search, and 100 were excluded due to duplicated
publications. In total, three publications, comprising six studies
with sufficient data, were eventually considered to be eligible for
analysis in the present study. The detailed characteristics are
shown in Table III. The results
of the quality assessment of the literature are shown in Table IV.
 | Table III.Characteristics of included
studies. |
Table III.
Characteristics of included
studies.
| Author/source,
year | Country | Male/female | Sample type | Type of
control | Detecting
method | (Refs.) |
|---|
| Khairy et
al, 2016 | Egypt | 59/19 | Serum | CLD | RT-PCR | (53) |
| Bhattacharya et
al, 2016 | USA | 42/14 | Serum | CLD | RT-PCR | (37) |
| Bhattacharya et
al, 2016 | USA | 39/14 | Serum | Normal | RT-PCR | (37) |
| Duo et al,
2015 | China | 80/40 | Serum | Normal | RT-PCR | (54) |
| Duo et al,
2015 | China | 82/38 | Serum | CLD | RT-PCR | (54) |
| Duo et al,
2015 | China | 93/57 | Serum | Unclear | RT-PCR | (54) |
| GSE12717, 2008 | USA | NA | Tissue | Unclear | SO | – |
| GSE20971, 2011 | France | NA | Tissue | Normal | SO | – |
| GSE21279, 2010 | China | NA | Tissue | CLD | HTS | – |
| GSE21362, 2011 | Japan | NA | Tissue | Unclear | SO | – |
| GSE39678, 2012 | South Korea | NA | Tissue | Normal | SO | – |
| GSE41874, 2013 | Japan | NA | Tissue | Unclear | SO | – |
| GSE54751, 2014 | USA | NA | Tissue | Unclear | RT-PCR | – |
| GSE57555, 2015 | Japan | NA | Tissue | Unclear | ISO | – |
| GSE65708, 2016 | China | NA | Serum | CLD | RT-PCR | – |
| GSE6857, 2008 | USA | NA | Tissue | Unclear | SO | – |
| GSE69580, 2015 | Taiwan | NA | Tissue | CLD | ISO | – |
| GSE74618, 2016 | Spain | NA | Tissue | Unclear | ISO | – |
| TCGA, 2016 | NR | NA | Tissue | Normal | HTS | – |
 | Table IV.Quality assessment of diagnostic
accuracy studies. |
Table IV.
Quality assessment of diagnostic
accuracy studies.
|
| Risk of bias |
|
| Applicability
concerns |
|
|
|---|
|
|
|
|
|
|
|
|
|---|
| Author, year | Patient | Index | Reference | Flow and
timing | Patient | Index | Reference | (Refs.) |
|---|
| Khairy et
al, 2016 | Unclear | High | Low | Unclear | Unclear | Low | Unclear | (53) |
| Bhattacharya et
al, 2016 | Unclear | Unclear | Low | Low | Unclear | Low | High | (37) |
| Duo et al,
2015 | Unclear | Unclear | Low | Unclear | Unclear | Low | High | (54) |
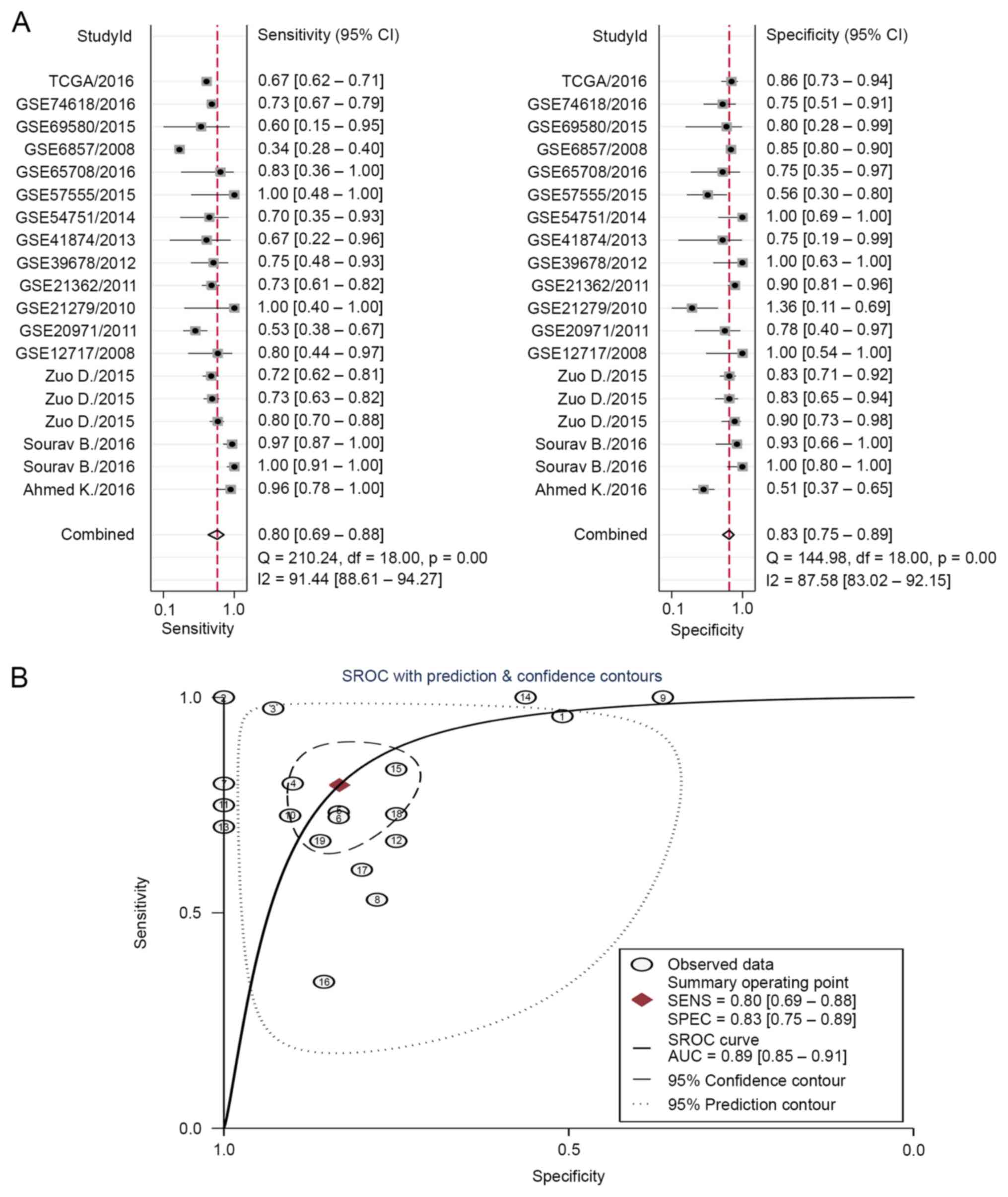
Meta-analysis for diagnostic value of
miR-223-3p in HCC
Pooled diagnostic performance
The diagnostic accuracy of miR-223-3p and miR-223
for HCC was pooled from 19 studies (Table V). The I2 value for the
AUC was 0.97 (95% CI, 0.95–0.99), therefore, a random model was
used in the following analysis. The overall pooled sensitivity was
0.80 with a 95% CI between 0.69 and 0.88, whereas the specificity
was 0.83 with a 95% CI between 0.75 and 0.89 (Fig. 5A). The positive and negative
likelihood ratios were 4.8 (95% CI, 3.1–7.2) and 0.24 (95% CI,
0.15–0.39), respectively. Overall, the diagnostic odds ratio was 19
(95% CI, 10.0–39.0). The diagnostic accuracy was further evaluated
by plotting the SROC, and by calculating the AUC, which was 0.89
(95% CI, 0.85–0.91), indicating a high diagnostic accuracy in
distinguishing patients with HCC from healthy controls (Fig. 5B).
 | Table V.Diagnostic value of mcroiRNA-223 in
19 individual studies. |
Table V.
Diagnostic value of mcroiRNA-223 in
19 individual studies.
| ID | Author/source,
year | HCC (n) | Controls (n) | AUC | Sensitivity | Specificity | TP (n) | FP (n) | FN (n) | TN (n) | (Refs.) |
|---|
| 1 | Khairy et
al, 2016 | 23 | 55 | 0.680 | 0.960 | 0.509 | 22 | 27 |
1 | 28 | (53) |
| 2 | Bhattacharya et
al, 2016 | 39 | 17 | 0.997 | 0.972 | 0.941 | 39 |
0 |
0 | 17 | (37) |
| 3 | Bhattacharya et
al, 2016 | 39 | 14 | 1.000 | 1.000 | 1.000 | 38 |
1 |
1 | 13 | (37) |
| 4 | Duo et al,
2015 | 90 | 30 | NA | 0.800 | 0.900 | 72 |
3 | 18 | 27 | (54) |
| 5 | Duo et al,
2015 | 90 | 30 | NA | 0.730 | 0.833 | 66 |
5 | 24 | 25 | (54) |
| 6 | Duo et al,
2015 | 90 | 60 | NA | 0.719 | 0.833 | 65 | 10 | 25 | 50 | (54) |
| 7 | GSE12717, 2008 | 10 |
6 | 0.883 | 0.800 | 1.000 |
8 |
0 |
2 |
6 | – |
| 8 | GSE20971, 2011 | 49 |
9 | 0.673 | 0.531 | 0.778 | 26 |
2 | 23 |
7 | – |
| 9 | GSE21279, 2010 |
4 | 11 | 0.568 | 1.000 | 0.364 |
4 |
7 |
0 |
4 | – |
| 10 | GSE21362, 2011 | 73 | 73 | 0.824 | 0.726 | 0.904 | 53 |
7 | 20 | 66 | – |
| 11 | GSE39678, 2012 | 16 |
8 | 0.875 | 0.750 | 1.000 | 12 |
0 |
4 |
8 | – |
| 12 | GSE41874, 2013 |
6 |
4 | 0.625 | 0.667 | 0.750 |
4 |
1 |
2 |
3 | – |
| 13 | GSE54751, 2014 | 10 | 10 | 0.890 | 0.700 | 1.000 |
7 |
0 |
3 | 10 | – |
| 14 | GSE57555, 2015 |
5 | 16 | 0.825 | 1.000 | 0.562 |
5 |
7 |
0 |
9 | – |
| 15 | GSE65708, 2016 |
6 |
8 | 0.813 | 0.833 | 0.750 |
5 |
2 |
1 |
6 | – |
| 16 | GSE6857, 2008 | 241 | 241 | 0.597 | 0.340 | 0.855 | 82 | 35 | 159 | 206 | – |
| 17 | GSE69580, 2015 |
5 |
5 | 0.560 | 0.600 | 0.800 |
3 |
1 |
2 |
4 | – |
| 18 | GSE74618, 2016 | 218 | 20 | 0.731 | 0.729 | 0.750 | 159 |
5 | 59 | 15 | – |
| 19 | TCGA, 2016 | 375 | 50 | 0.782 | 0.667 | 0.860 | 250 |
7 | 125 | 43 | – |
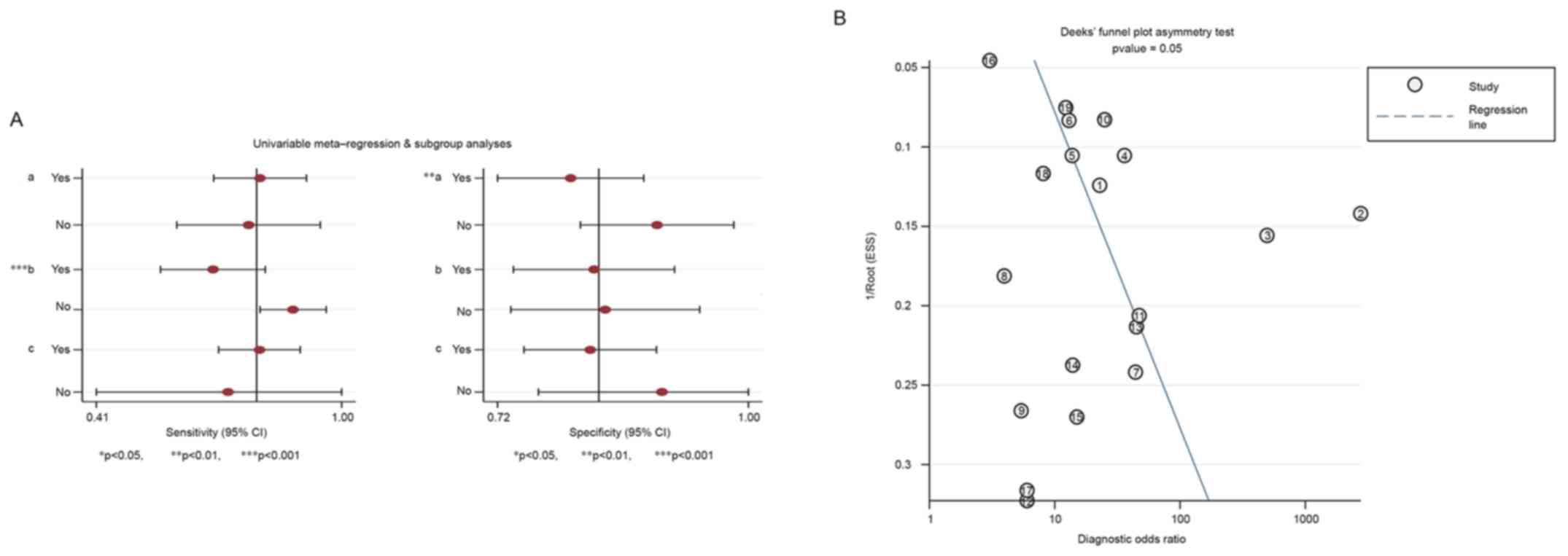
Univariate regression and subgroup
analysis
As the threshold effect accounted for only 5% of the
heterogeneity across a total of 19 studies, univariate regression
and subgroup analysis was used to further investigate the sources
of heterogeneity. The sample type (serum or tissues) and
histological type for the controls may contribute to heterogeneity.
The sample type was ultimately revealed to be a statistically
significant source of heterogeneity (P<0.001; Fig. 6A).
Publication bias
Publication bias in the present study was determined
using Deek's funnel plot, and the P-value (P=0.054), indicated no
statistical significance in publication bias (Fig. 6B).
Potential mechanisms and functions of
miR-223-3p in HCC
Potential target genes of
miR-223-3p
Based on the 12 online predictive tools mentioned
above, a total of 12,821 genes, which appeared on three or more
platforms, were screened, and 48 validated genes were obtained from
miTarBase. In addition, 1,800 genes associated with HCC were
identified from 64,577 studies in the processing of NLP, and 2,155
upregulated genes in HCC were identified from TCGA in the present
study. Finally, 72 genes, which intersected in the three
aforementioned methods, were included in further analysis.
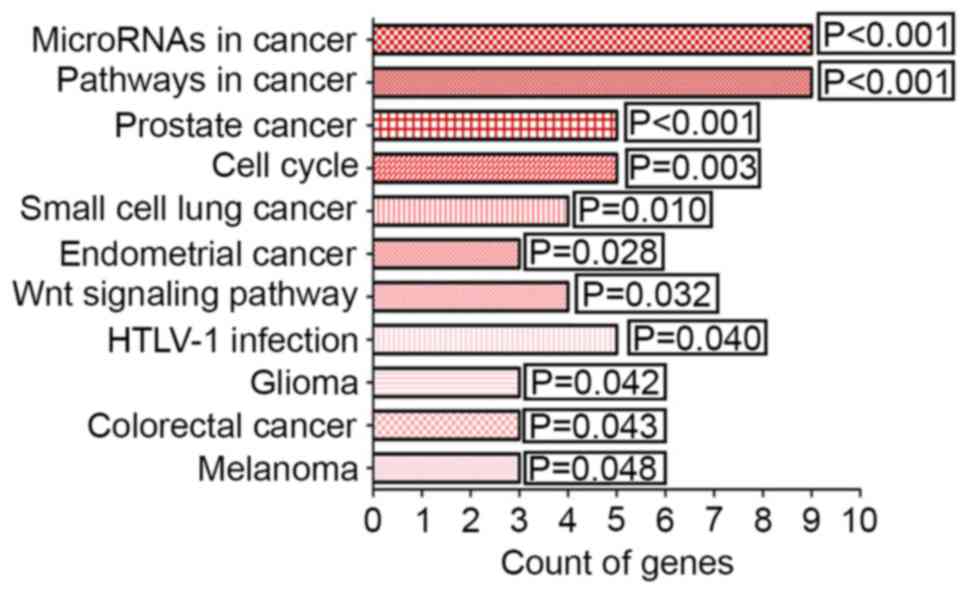
Pathway annotation of KEGG
From the KEGG pathway annotation, a total of 11
pathways were identified (listed in Table VI). The pathways with
significantly different gene counts are shown in Fig. 7 (P<0.05). The highest degree of
significance was in ‘microRNAs in cancer’ (P<0.001). The genes
DNA methyltransferase 1 (DNMT1), kinesin family member 23 (KIF23),
STMN1 and others are involved in this pathway. Additionally,
several potential target genes were closely associated with KEGG
pathways, including the terms ‘pathways in cancer’ (P<0.001),
‘prostate cancer’ (P<0.001) and ‘cell cycle’ (P=0.003).
 | Table VI.Biological pathways enriched in KEGG
of HCC. |
Table VI.
Biological pathways enriched in KEGG
of HCC.
| KEGG pathway | Genes | P-value | FDR-value |
|---|
| MicroRNAs in
cancer | KIF23, E2F1, CCNE2,
DNMT3A, PDGFRB, IGF2BP1, DNMT1, STMN1, HMGA2 | <0.001 |
0.037 |
| Pathways in
cancer | E2F1, CCNE2,
CDKN2B, ITGA6, PDGFRB, LEF1, BIRC5, EGF, AXIN2 | <0.001 |
0.634 |
| Prostate
cancer | E2F1, CCNE2,
PDGFRB, LEF1, EGF | <0.001 |
1.031 |
| Cell cycle | E2F1, CCNE2,
CDKN2B, CHEK1, MCM6 |
0.003 |
3.661 |
| Small cell lung
cancer | E2F1, CCNE2,
CDKN2B, ITGA6 |
0.010 | 10.687 |
| Endometrial
cancer | LEF1, EGF,
AXIN2 |
0.028 | 26.228 |
| Wnt signaling
pathway | DKK1, SFRP4, LEF1,
AXIN2 |
0.032 | 29.532 |
| HTLV–I
infection | E2F1, CDKN2B,
PDGFRB, CHEK1, TCF3 |
0.040 | 35.953 |
| Glioma | E2F1, PDGFRB,
EGF |
0.042 | 36.973 |
| Colorectal
cancer | LEF1, BIRC5,
AXIN2 |
0.043 | 37.802 |
| Melanoma | E2F1, PDGFRB,
EGF |
0.048 | 41.102 |
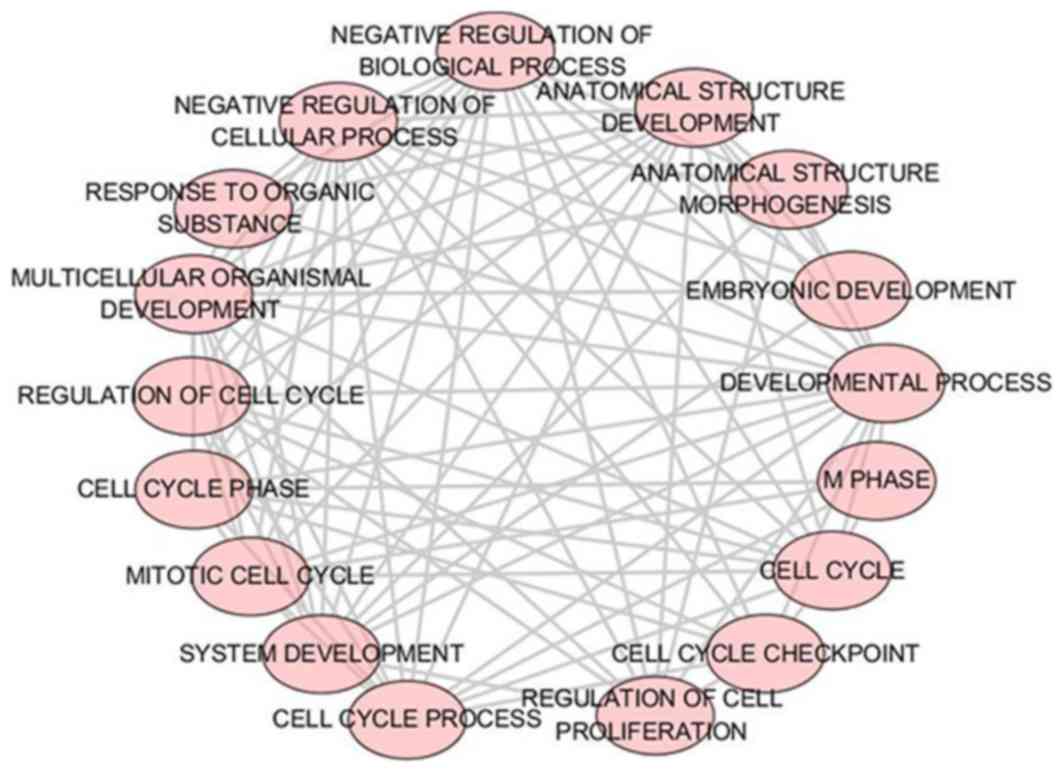
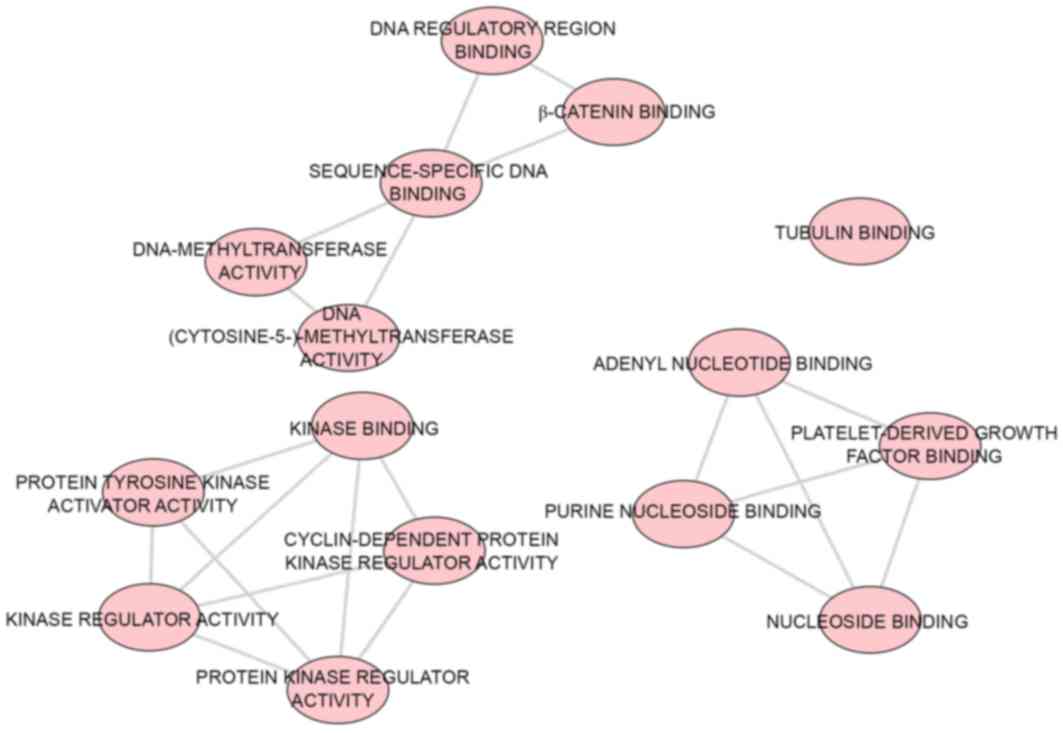
Biological functional analysis of
GO
A total of 43 significant GO categories were
screened out using DAVID, including 29 biological processes (BPs),
nine cellular components (CCs) and five molecular functions (MFs).
The top 10 significant analyses of BPs are shown in Table VII, in addition to the nine CCs
and five MFs (P<0.05). The results showed that, in the BP
category, potential targets of miR-223-3p were concentrated
predominantly on the terms ‘mitotic cytokinesis’ (P<0.001),
‘positive regulation of cytokinesis’ (P<0.001) and ‘negative
regulation of transcription from RNA polymerase II promoter’
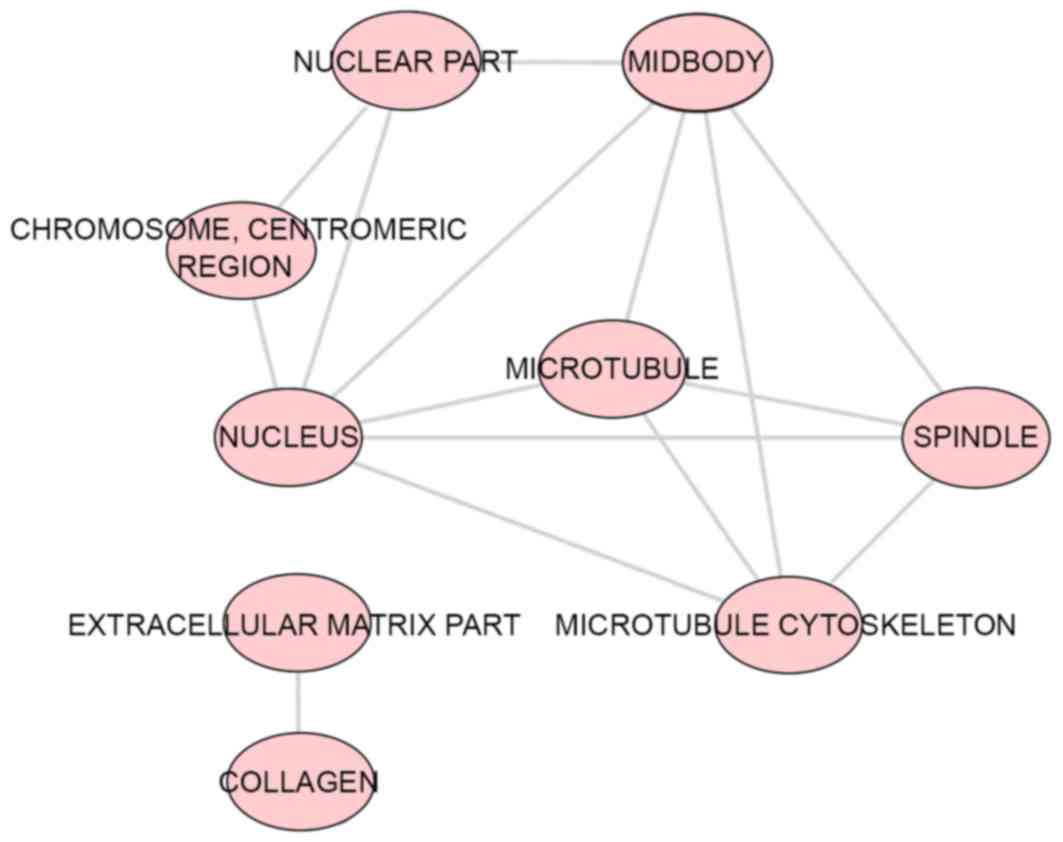
(P=0.003; Fig. 8). For the CCs,
the target genes were predominantly enriched in the terms
‘centralspindlin complex’ (P<0.001), ‘microtubule’ (P=0.001) and
‘midbody’ (P=0.006; Fig. 9). In
the MF category ATP binding’ was the most enriched term (P=0.003),
followed by ‘chromatin binding’ (P=0.008) and ‘armadillo repeat
domain binding’ (P=0.02; Fig.
10).
 | Table VII.Top GO terms significantly enriched
with high potential target gene count. |
Table VII.
Top GO terms significantly enriched
with high potential target gene count.
| Category | Term | Genes | P-value | FDR-value |
|---|
|
GOTERM_BP_DIRECT | GO:0000281-mitotic
cytokinesis | KIF23, CKAP2,
STMN1, RACGAP1 | <0.001 |
0.303 |
|
GOTERM_BP_DIRECT | GO:0032467-positive
regulation of cytokinesis | KIF23, KIF14,
RACGAP1, ECT2 | <0.001 |
0.537 |
|
GOTERM_BP_DIRECT | GO:0000122-negative
regulation of transcription from RNA polymerase II promoter | SHOX2, E2F1,
DNMT3A, DKK1, SIX1, LEF1, WHSC1, SOX9 |
0.003 |
3.933 |
|
GOTERM_BP_DIRECT | GO:0034504-protein
localization to nucleus | SIX1, COL1A1,
SOX9 |
0.007 |
9.137 |
|
GOTERM_BP_DIRECT | GO:0060325-face
morphogenesis | DKK1, LEF1,
COL1A1 |
0.007 |
9.833 |
|
GOTERM_BP_DIRECT |
GO:0030326-embryonic limb
morphogenesis | FRAS1, DKK1,
LEF1 |
0.008 | 11.281 |
|
GOTERM_BP_DIRECT | GO:0030199-collagen
fibril organization | COL1A1, LOX,
MMP11 |
0.008 | 11.281 |
|
GOTERM_BP_DIRECT | GO:0010718-positive
regulation of epithelial to mesenchymal transition | LEF1, COL1A1,
AXIN2 |
0.008 | 11.281 |
|
GOTERM_BP_DIRECT | GO:0001942-hair
follicle development | DKK1, SOX9,
LGR5 |
0.009 | 12.801 |
|
GOTERM_BP_DIRECT |
GO:0000915-actomyosin contractile ring
assembly | KIF23, RACGAP1 |
0.010 | 13.331 |
|
GOTERM_CC_DIRECT |
GO:0097149-centralspindlin complex | KIF23, RACGAP1,
ECT2 | <0.001 |
0.060 |
|
GOTERM_CC_DIRECT |
GO:0005874-microtubule | KIF23, KIF14,
ARHGEF2, NDRG1, CCT3 |
0.001 |
1.397 |
|
GOTERM_CC_DIRECT |
GO:0030496-midbody | KIF23, KIF14,
RACGAP1, ECT2 |
0.006 |
6.867 |
|
GOTERM_CC_DIRECT |
GO:0005813-centrosome | KIF23, CKAP2, DTL,
CHEK1, NDRG1, AXIN2 |
0.010 | 10.162 |
|
GOTERM_CC_DIRECT | GO:0005886-plasma
membrane | KIF14, CCT3, MMP14,
CDH12, DKK1, CNR1, SULF1, RGS5, SORT1, PDGFRB, NDRG1, EGF,
AXIN2 |
0.019 | 19.542 |
|
GOTERM_CC_DIRECT |
GO:0005667-transcription factor
complex | E2F1, SIX1, LEF1,
TCF3 |
0.031 | 29.825 |
|
GOTERM_CC_DIRECT |
GO:0048471-perinuclear region of
cytoplasm | NOX4, XRCC3, STC2,
SORT1, NDRG1 |
0.044 | 39.035 |
|
GOTERM_CC_DIRECT |
GO:0005634-nucleus | E2F1, NOX4, CCNF,
ATAD2, WHSC1, PBK, HMGA2, SOX9, ECT2, MCM6, CCNE2, SHOX2, GLUL,
HEY1, SIX1, TXNRD1, NDRG1, TCF3 |
0.044 | 39.179 |
|
GOTERM_CC_DIRECT |
GO:0005654-nucleoplasm | KIF23, DNMT3A,
XRCC3, CDKN2B, DTL, ATAD2, CHEK1, RACGAP1, AXIN2, SOX9, MCM6 |
0.044 | 39.550 |
|
GOTERM_MF_DIRECT | GO:0005524-ATP
binding | KIF23, KIF14, GLUL,
XRCC3, MSH2, PFKFB2, ABCC4, PDGFRB, ATAD2, CHEK1, PBK, CCT3,
MCM6 |
0.003 |
3.662 |
|
GOTERM_MF_DIRECT |
GO:0003682-chromatin binding | DNMT3A, SIX1, LEF1,
ATAD2, WHSC1, SOX9 |
0.008 |
8.509 |
|
GOTERM_MF_DIRECT |
GO:0070016-armadillo repeat domain
binding | LEF1, AXIN2 |
0.021 | 21.262 |
|
GOTERM_MF_DIRECT | GO:0035326-enhancer
binding | LEF1, SOX9 |
0.030 | 28.444 |
|
GOTERM_MF_DIRECT | GO:0005096-GTPase
activator activity | RGS5, RACGAP1,
AXIN2, ECT2 |
0.039 | 35.353 |
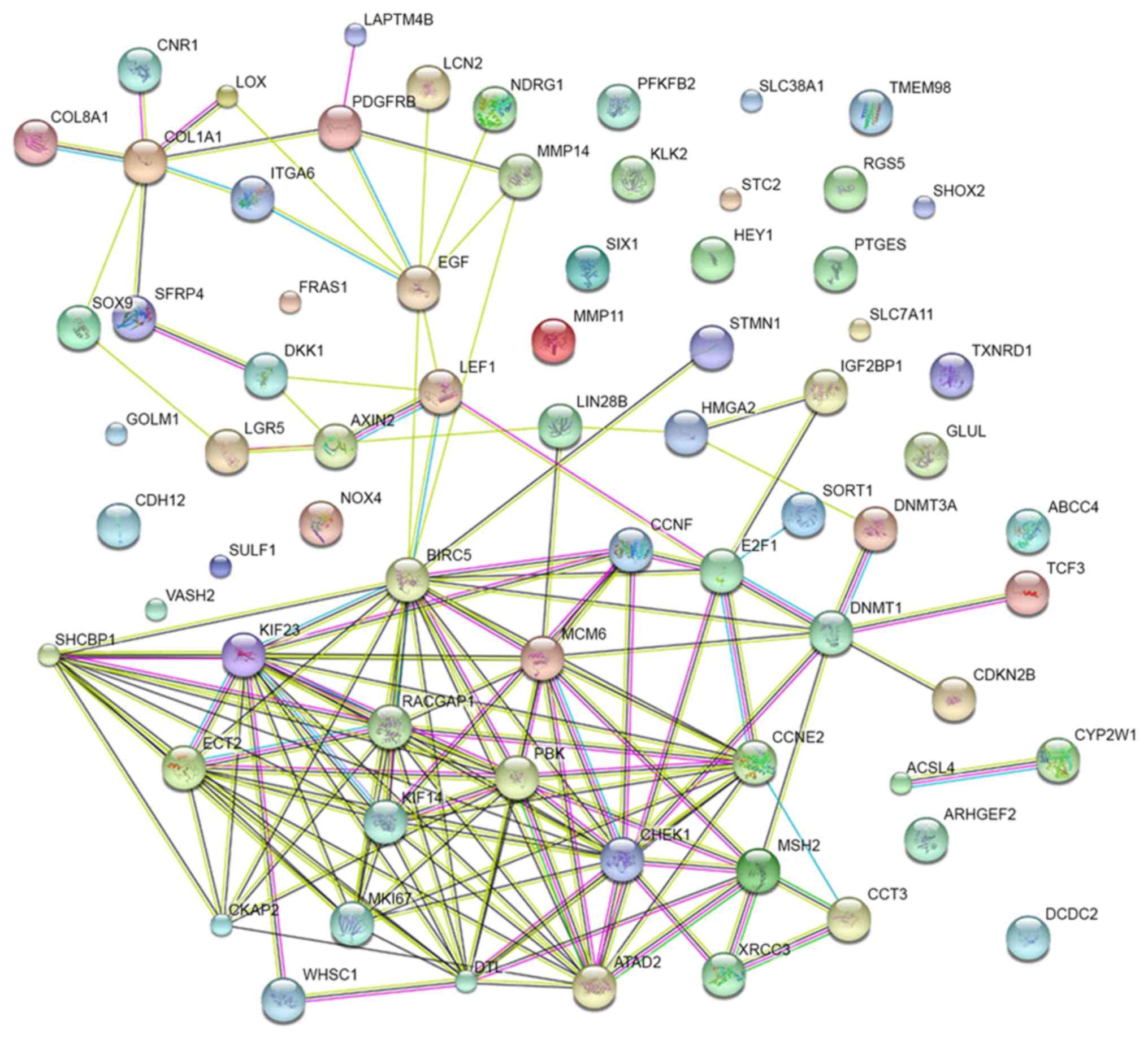
PPI network construction
A PPI network of the 72 potential targets of
miR-223-3p was mapped (Fig. 11).
The 10 protein pairs with the highest combined scores are listed in
Table VIII. The five genes,
which interacted >5 times between different protein pairs were
identified as the hub genes of miR-223-3p, including checkpoint
kinase 1 (CHEK1), DNA methyltransferase 1 (DNMT1), baculoviral IAP
repeat containing 5 (BIRC5), KIF23 and collagen, type I, α1
(COL1A1). In TCGA, compared with the normal controls, all five of
these targets were significantly upregulated in HCC (P<0.05).
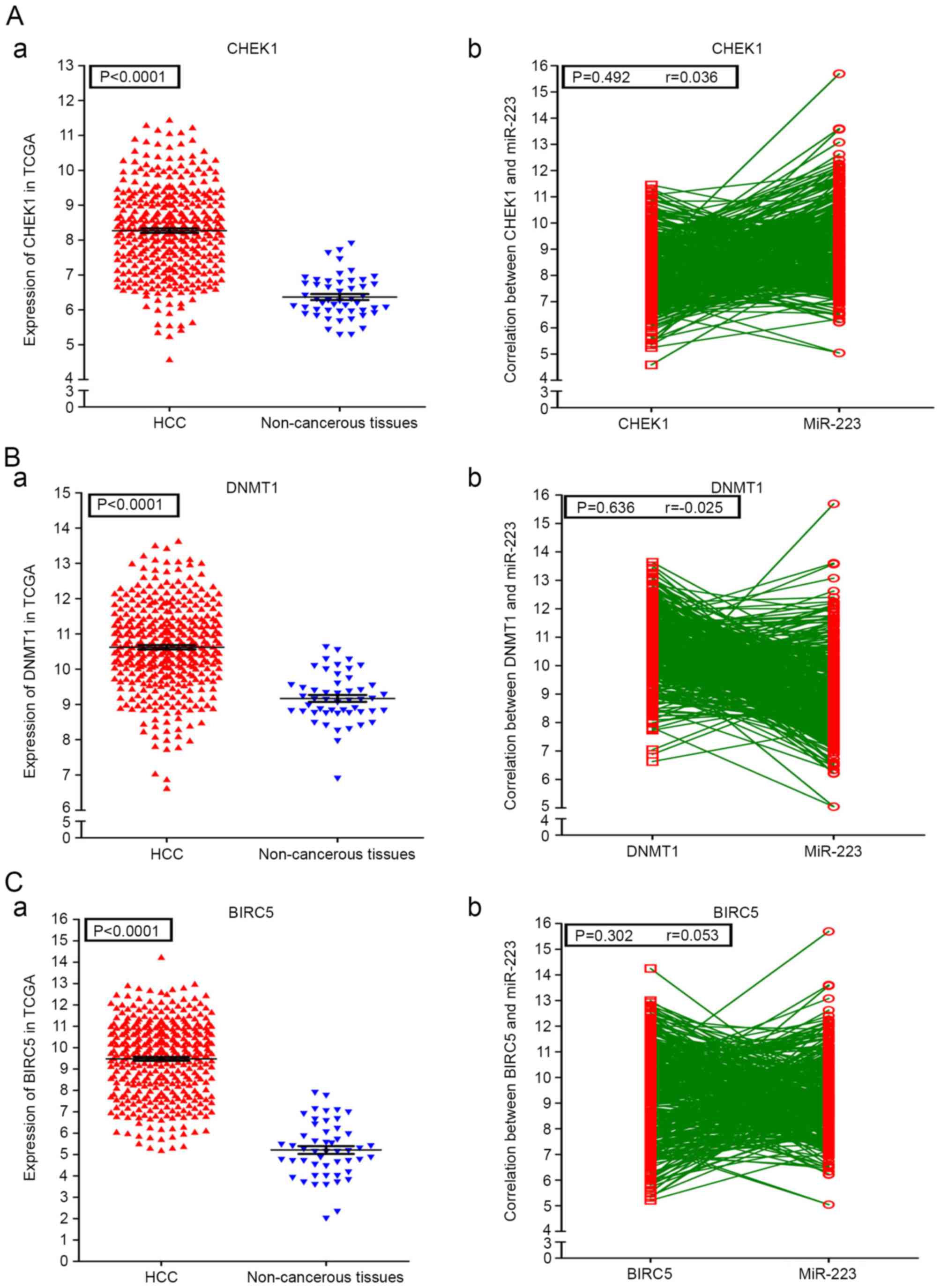
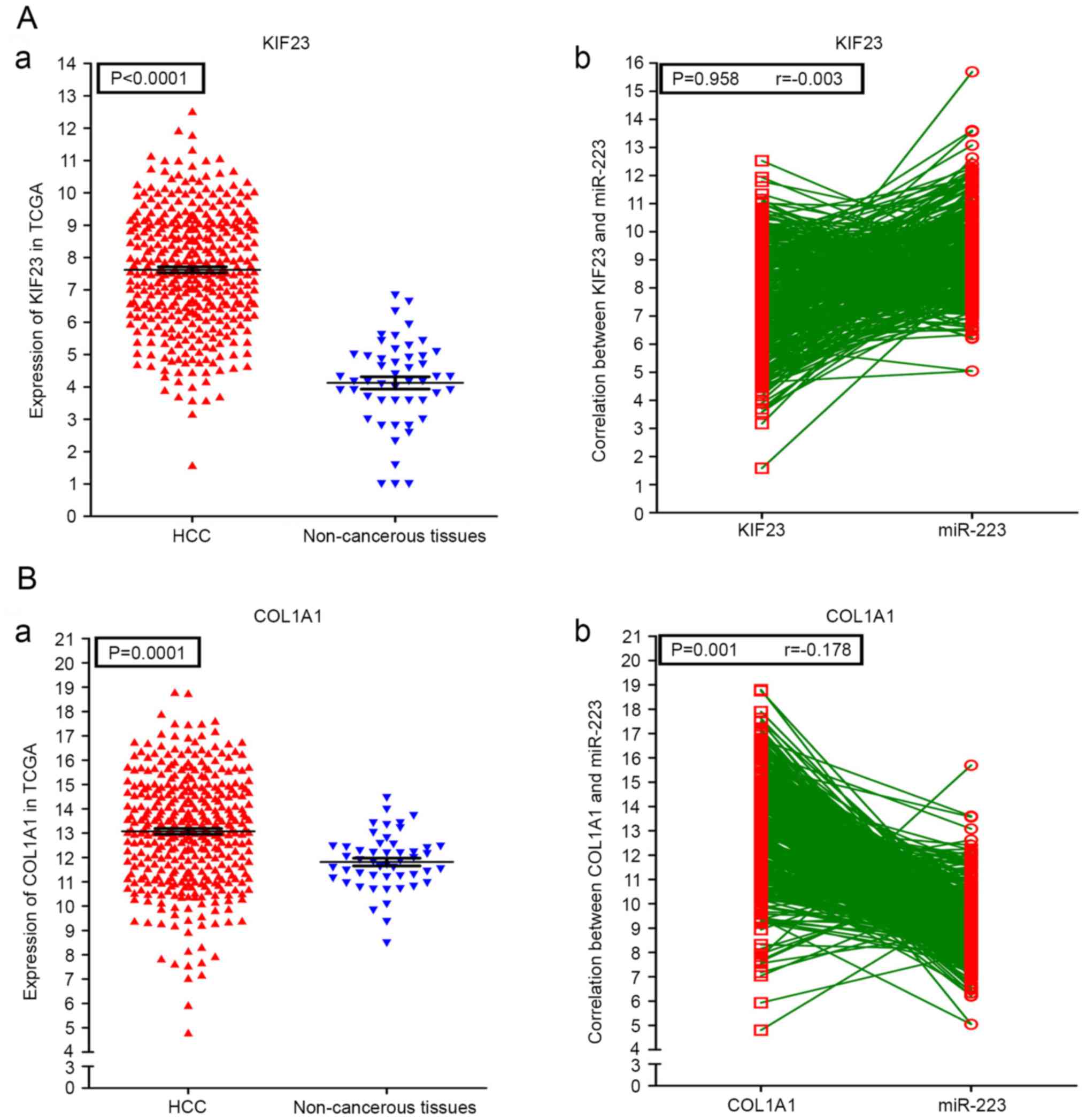
Additionally, correlations of the hub genes and miR-223 were also
shown in Figs. 12 and 13. High expression levels of the gene
COL1A1 were significantly correlated with a lower level of miR-223
in HCC (P=0.001; Fig. 13B-b).
 | Table VIII.Ten prominent protein-protein
interaction nodes according to the Search Tool for the Retrieval of
Interacting Genes. |
Table VIII.
Ten prominent protein-protein
interaction nodes according to the Search Tool for the Retrieval of
Interacting Genes.
| Node 1 | Node 2 | Co-expression | Experimentally
determined interaction | Database
annotated | Automated
textmining | Combined score |
|---|
| DNMT1 | DNMT3A | 0 | 0.798 | 0.9 | 0.987 | 0.999 |
| RACGAP1 | ECT2 | 0.288 | 0.914 | 0.9 | 0.907 | 0.999 |
| RACGAP1 | KIF23 | 0.578 | 0.996 | 0.9 | 0.979 | 0.999 |
| KIF23 | ECT2 | 0.245 | 0.564 | 0.9 | 0.966 | 0.998 |
| E2F1 | CCNE2 | 0 | 0.363 | 0.9 | 0.735 | 0.981 |
| BIRC5 | KIF23 | 0.161 | 0 | 0.9 | 0.738 | 0.976 |
| AXIN2 | LEF1 | 0 | 0.110 | 0.9 | 0.751 | 0.976 |
| RACGAP1 | BIRC5 | 0.173 | 0 | 0.9 | 0.714 | 0.974 |
| CHEK1 | MCM6 | 0.093 | 0.207 | 0.9 | 0.562 | 0.964 |
| DNMT1 | E2F1 | 0.070 | 0.564 | 0.9 | 0.116 | 0.959 |
Discussion
The results of the present study indicated that
miR-223-3p may be a tumor suppressor and a preferable diagnostic
biomarker for HCC. A decreased frequency of miR-223-3p was shown in
HCC samples in the GEO datasets, and the precursor miR-223 was
downregulated in HCC samples in the TCGA database. The AUC of the
SROC for miR-223-3p in diagnosing HCC was 0.89 (95% CI 0.85–0.91).
From the PPI network, five potential targets were identified as the
hub genes of miR-223-3p, including CHEK1, DNMT1, BIRC5, KIF23 and
COL1A1.
Previous studies have found that miR-223-3p can act
as an important novel biomarker for cancer screening in peripheral
circulating blood and tissues. However, whether the miR-223-3p is
downregulated in tumorigenesis or not remains controversial. The
reduced expression of miR-223-3p in plasma may be a novel
non-invasive detector in esophageal squamous cell carcinoma
(33), and abnormalities in the
expression of miR-223-3p in patients with HCC have enabled
discrimination from normal controls. Compared with normal controls,
the marked decrease in circulating miR-223-3p in HCC may serve as a
biomarker (36,38,55,56).
By contrast, in ovarian cancer tissues, miR-233-3p is expressed at
high levels, particularly in recurrent cancer tissues (57). In gingival squamous cell carcinoma,
significantly upregulated plasma levels of miR-223-3p have been
identified as a diagnostic biomarker (58). miR-223-3p was also found to be at a
higher level in the serum of patients with chronic
hepatitis-related HCC (39). As
reports on the expression of miR-223-3p in HCC have been
controversial, the present study used GEO and TCGA datasets to
perform a detailed analysis of the expression profiles of
miR-223-3p or its processor miR-223 in HCC samples, compared with
non-cancerous controls. A significant downregulation of miR-223-3p
in HCC was reported in the majority of the studies included. With a
total of 1,170 HCC samples in the present study, further
large-sample analyses are required.
As miRNAs exist stably in the circulation, including
the serum, plasma and urine, miR-223-3p may also be used as a
blood-based tumor marker for diagnosing HCC in its early stages. In
the present study, a comprehensive diagnostic meta-analysis was
performed, which focused on circulating miR-223-3p and on
miR-223-3p expression profiles in HCC tissues. The AUC of SROC was
0.89, which represents moderate accuracy for a diagnosis of HCC
(59). In accordance with the
present study, Li et al found that circulating miR-223-3p in
HCC was an informative biomarker, with the AUC of SROC being 0.8597
(41). The limitation of the study
by Li et al was that only one study with only 101 HCC serum
samples had been included, whereas 19 studies comprising 1,170 HCC
samples were used in the present study. The levels of miR-223-3p
were closely correlated with HCC in fresh tissues and
formalin-fixed paraffin-embedded (FFPE) samples. The expression of
miR-223-3p was deregulated in FFPE HCC samples, with a sensitivity
of 78.6% and a specificity of 72.7%. Additionally, downregulated
miR-223-3p may act as a biomarker for HCC prognosis following
orthotopic liver transplantation (60). Therefore, it may be possible to
diagnose HCC by detecting miR-223-3p in tissue biopsies. Additional
large-sample clinical studies are required to clarify the clinical
significance of the association between miR-223-3p and HCC.
Previous studies have revealed that miR-223-3p is
associated with different diseases. In prostate cancer, miR-223-3p
targeted septin 6 to promote cell proliferation, cell apoptosis,
cell invasion and other processes (61). NCOA1 and zinc finger E-box-binding
homeobox 1 protein translation may be mediated by miR-223-3p to
inhibit the migration and invasion of human bladder cancer cells
(34,62). miR-223-3p may reflect changes in
inflammation, vascular calcification and pathophysiology of bones
(63). As a tumor suppressor,
enhanced levels of miR-223-3p inhibited tumorigenesis and
metastasis in HCC. miR-223-3p can promote HCC cell apoptosis
through the mammalian target of rapamycin pathway (64,65).
miR-223-3p may also be key in cell proliferation and exert
tumor-suppressive effects on hepatitis B virus-related HCC
(35). In addition, the
overexpression of miR-223-3p can increase HCC cell sensitivity to
anticancer drugs by repressing ATP-binding cassette sub-family B
member 1 at the mRNA and protein levels (66). These results demonstrate the
multiple functions of miR-223-3p in HCC.
The five hub genes of miR-223-3p identified in the
present stud were further analyzed by investigating potential
molecular mechanisms and the tumorigenesis of HCC. The hub genes
CHEK1, DNMT1, BIRC5, KIF23 and COL1A1 were all significantly
upregulated in HCC tissues, compared with non-cancerous controls.
By contrast, miR-223-3p was present at a decreased level in HCC
tissues. The AUC of SROC for miR-223-3p in diagnosing HCC was 0.89.
Therefore, the lower level of miR-223-3p may target these important
hub genes in certain pathways. For example, the CHEK1 gene was
involved in the cell cycle pathway, KIF23 and DNMT1 were enriched
at microRNA levels in cancer pathways, BIRC5 was identified in
cancer pathways and COL1A1 was identified in the collagen fibril
organization pathway. These five hub genes may be pivotal in
HCC.
As a novel tumor suppressor, CHEK1 is involved in
tumor prevention and therapeutics (67). However, the upregulation of CHEK1
in HCC may have a tumorigenic function, and the overexpression of
CHEK1 may be a reliable diagnostic indicator in patients with HCC.
Therefore, targeting the CHEK1/SYK(L) pathway may be a promising
therapeutic strategy for treating HCC (68,69).
DNMT1 is involved in tumorigenesis and cancer progression (70). A high expression level of DNMT1 has
been associated with HCC-related growth factors. This high level of
expression indirectly mediates the progression of HCC caused by
hepatitis B virus infection (71,72).
In addition, the upregulation of DNMT1 has been associated with
recurrence and poor outcome of HCC cases (73). The KIF23 gene acts an important
regulator in cytokinesis (74).
The upregulation of KIF23 leads to the earlier recurrence of HCC
(75). Therefore, DNMT1 and KIF23
may provide valuable information on the prognosis of patients with
HCC. BIRC5 is known as an apoptosis inhibitor protein in
malignancies; therefore BIRC5 may be a valuable diagnostic factor
for HCC. The co-suppression of OCT4/BIRC5 is beneficial for
treating patients with HCC (76,77),
and COL1A1 is an important the target in hepatic fibrosis, having
been confirmed as a potential prognostic biomarker in HCC (78–80).
Of note, in the present study, the upregulation of COL1A1 was
significantly correlated with the downregulation of miR-223
(r=−0.178; P=0.001), which indicated that COL1A1 may also be a
potential diagnostic biomarker in HCC.
In conclusion, increasing evidence has revealed that
the hub genes of miR-223-3p can directly or indirectly regulate the
occurrence, progression, diagnosis, prognosis and treatment of HCC.
However, the present study did not experimentally validate the
correlations between miR-223-3p and its hub genes. A number of
these hub genes had limited reports with miR-223-3p in HCC. In the
future, the predicted hub genes, which were identified from the PPI
analysis, require validation in in vivo and in vitro
experiments.
In the present study, a decreased level of
miR-223-3p in HCC was markedly associated with its diagnostic
value, possibly by targeting potential genes in different
biological pathways. Therefore, the present study revealed that
miR-223-3p merits attention as a significant biomarker in the
diagnostic setting and tumorigenesis of HCC. Additional experiments
and analyses are required to confirm the potential molecular
mechanisms and the prognostic role of miR-223-3p in HCC.
Acknowledgements
This study was supported by the fund of the National
Natural Science Foundation of China (grant no. NSFC81560386).
Glossary
Abbreviations
Abbreviations:
|
miR-223-3p
|
microRNA-223-3p
|
|
HCC
|
hepatocellular carcinoma
|
|
GEO
|
Gene Expression Omnibus
|
|
TCGA
|
The Cancer Genome Atlas
|
|
ROC
|
receiver operating curve
|
|
AUC
|
area under the curve
|
|
NLP
|
natural language processing
|
|
GO
|
Gene Ontology
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
PPI
|
protein-protein interaction
|
References
|
1
|
Mo Z, Zheng S, Lv Z, Zhuang Y, Lan X, Wang
F, Lu X, Zhao Y and Zhou S: Senescence marker protein 30 (SMP30)
serves as a potential prognostic indicator in hepatocellular
carcinoma. Sci Rep. 6:393762016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lafaro KJ, Demirjian AN and Pawlik TM:
Epidemiology of hepatocellular carcinoma. Surg Oncol Clin N Am.
24:1–17. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zheng W, Yao M, Qian Q, Sai W, Qiu L, Yang
J, Wu W, Dong Z and Yao D: Oncogenic secretory clusterin in
hepatocellular carcinoma: Expression at early staging and emerging
molecular target. Oncotarget. 8:52321–52332. 2016.PubMed/NCBI
|
|
4
|
Huang W, Cui X, Chen Y, Shao M, Shao X,
Shen Y, Liu Q, Wu M, Liu J, Ni W, et al: High VRK1 expression
contributes to cell proliferation and survival in hepatocellular
carcinoma. Pathol Res Pract. 212:171–178. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yu C, Cao Q, Chen P, Yang S, Gong X, Deng
M, Ruan B and Li L: Tissue transglutaminase 2 exerts a
tumor-promoting role in hepatitis B virus-related hepatocellular
carcinoma. Tumour Biol. 2016.(Epub ahead of print). View Article : Google Scholar
|
|
6
|
Mazzola A, Costantino A, Petta S,
Bartolotta TV, Raineri M, Sacco R, Brancatelli G, Cammà C and
Cabibbo G: Recurrence of hepatocellular carcinoma after liver
transplantation: An update. Future Oncol. 11:2923–2936. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jiang H, Zhang X, Tao Y, Shan L, Jiang Q,
Yu Y, Cai F and Ma L: Prognostic and clinicopathologic significance
of SIRT1 expression in hepatocellular carcinoma. Oncotarget.
8:52357–52365. 2016.PubMed/NCBI
|
|
8
|
Zhang T, Zhang X, Shi W, Xu J, Fan H,
Zhang S and Ni R: The DNA damage repair protein Ku70 regulates
tumor cell and hepatic carcinogenesis by interacting with FOXO4.
Pathol Res Pract. 212:153–161. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Forner A, Llovet JM and Bruix J:
Hepatocellular carcinoma. Lancet. 379:1245–1255. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ke M, Xu T, Li N, Ren Y, Shi A, Lv Y and
He H: Prognostic nutritional index predicts short-term outcomes
after liver resection for hepatocellular carcinoma within the Milan
criteria. Oncotarget. 7:81611–81620. 2016.PubMed/NCBI
|
|
11
|
Xu Y, Rong J, Duan S, Chen C, Li Y, Peng
B, Yi B, Zheng Z, Gao Y, Wang K, et al: High expression of GNA13 is
associated with poor prognosis in hepatocellular carcinoma. Sci
Rep. 6:359482016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li J, Gao JZ, Du JL and Wei LX: Prognostic
and clinicopathological significance of glypican-3 overexpression
in hepatocellular carcinoma: A meta-analysis. World J
Gastroenterol. 20:6336–6344. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang N, Gu J, Yin L, Wu J, Du MY, Ding K,
Huang T and He X: Incorporation of alpha-fetoprotein (AFP) into
subclassification of BCLC C stage hepatocellular carcinoma
according to a 5-year survival analysis based on the SEER database.
Oncotarget. 7:81389–81401. 2016.PubMed/NCBI
|
|
14
|
Chen J, Wu FX, Luo HL, Liu JJ, Luo T, Bai
T, Li LQ and Fan XH: Berberine upregulates miR-22-3p to suppress
hepatocellular carcinoma cell proliferation by targeting Sp1. Am J
Transl Res. 8:4932–4941. 2016.PubMed/NCBI
|
|
15
|
Yates LA, Norbury CJ and Gilbert RJ: The
long and short of microRNA. Cell. 153:516–519. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu Z, Wang C, Jiao X, Zhao S, Liu X, Wang
Y and Zhang J: miR-221 promotes growth and invasion of
hepatocellular carcinoma cells by constitutive activation of NFκB.
Am J Transl Res. 8:4764–4777. 2016.PubMed/NCBI
|
|
17
|
Blanco-Calvo M, Calvo L, Figueroa A,
Haz-Conde M, Antón-Aparicio L and Valladares-Ayerbes M: Circulating
microRNAs: Molecular microsensors in gastrointestinal cancer.
Sensors. 12:9349–9362. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cao L, Xie B, Yang X, Liang H, Jiang X,
Zhang D, Xue P, Chen D and Shao Z: miR-324-5p suppresses
hepatocellular carcinoma cell invasion by counteracting ECM
degradation through post-transcriptionally downregulating ETS1 and
SP1. PLoS One. 10:e01330742015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zekri AN, Youssef AS, El-Desouky ED, Ahmed
OS, Lotfy MM, Nassar AA and Bahnassey AA: Serum microRNA panels as
potential biomarkers for early detection of hepatocellular
carcinoma on top of HCV infection. Tumour Biol. 37:12273–12286.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xue HY, Liu Y, Liao JZ, Lin JS, Li B, Yuan
WG, Lee RJ, Li L, Xu CR and He XX: Gold nanoparticles delivered
miR-375 for treatment of hepatocellular carcinoma. Oncotarget.
7:86675–86686. 2016.PubMed/NCBI
|
|
21
|
Shen S, Lin Y, Yuan X, Shen L, Chen J,
Chen L, Qin L and Shen B: Biomarker microRNAs for diagnosis,
prognosis and treatment of hepatocellular carcinoma: A functional
survey and comparison. Sci Rep. 6:383112016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhao L and Wang W: miR-125b suppresses the
proliferation of hepatocellular carcinoma cells by targeting
Sirtuin7. Int J Clin Exp Med. 8:18469–18475. 2015.PubMed/NCBI
|
|
23
|
Liu B, Sun T, Wu G, Shang-Guan H, Jiang
ZJ, Zhang JR and Zheng YF: miR-15a suppresses hepatocarcinoma cell
migration and invasion by directly targeting cMyb. Am J Transl Res.
9:520–532. 2017.PubMed/NCBI
|
|
24
|
He R, Yang L, Lin X, Chen X, Lin X, Wei F,
Liang X, Luo Y, Wu Y, Gan T, et al: miR-30a-5p suppresses cell
growth and enhances apoptosis of hepatocellular carcinoma cells via
targeting AEG-1. Int J Clin Exp Pathol. 8:15632–15641.
2015.PubMed/NCBI
|
|
25
|
Liu S, Liu K, Zhang W, Wang Y, Jin Z, Jia
B and Liu Y: miR-449a inhibits proliferation and invasion by
regulating ADAM10 in hepatocellular carcinoma. Am J Transl Res.
8:2609–2619. 2016.PubMed/NCBI
|
|
26
|
Liu Y, Zhang W, Liu K, Liu S, Ji B and
Wang Y: miR-138 suppresses cell proliferation and invasion by
inhibiting SOX9 in hepatocellular carcinoma. Am J Transl Res.
8:2159–2168. 2016.PubMed/NCBI
|
|
27
|
Yao H, Liu X, Chen S, Xia W and Chen X:
Decreased expression of serum miR-424 correlates with poor
prognosis of patients with hepatocellular carcinoma. Int J Clin Exp
Pathol. 8:14830–14835. 2015.PubMed/NCBI
|
|
28
|
Sun J, Fang K, Shen H and Qian Y:
MicroRNA-9 is a ponderable index for the prognosis of human
hepatocellular carcinoma. Int J Clin Exp Med. 8:17748–17756.
2015.PubMed/NCBI
|
|
29
|
Huang CS, Yu W, Cui H, Wang YJ, Zhang L,
Han F and Huang T: Increased expression of miR-21 predicts poor
prognosis in patients with hepatocellular carcinoma. Int J Clin Exp
Pathol. 8:7234–7238. 2015.PubMed/NCBI
|
|
30
|
Gan TQ, Tang RX, He RQ, Dang YW, Xie Y and
Chen G: Upregulated miR-1269 in hepatocellular carcinoma and its
clinical significance. Int J Clin Exp Med. 8:714–721.
2015.PubMed/NCBI
|
|
31
|
Sun XF, Sun JP, Hou HT, Li K, Liu X and Ge
QX: MicroRNA-27b exerts an oncogenic function by targeting Fbxw7 in
human hepatocellular carcinoma. Tumour Biol. 37:15325–15332. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kuasne H, Barros-Filho MC, Busso-Lopes A,
Marchi FA, Pinheiro M, Muñoz JJ, Scapulatempo-Neto C, Faria EF,
Guimarães GC, Lopes A, et al: Integrative miRNA and mRNA analysis
in penile carcinomas reveals markers and pathways with potential
clinical impact. Oncotarget. 8:15294–15306. 2017.PubMed/NCBI
|
|
33
|
Zhou X, Wen W, Zhu J, Huang Z, Zhang L,
Zhang H, Qi LW, Shan X, Wang T, Cheng W, et al: A six-microRNA
signature in plasma was identified as a potential biomarker in
diagnosis of esophageal squamous cell carcinoma. Oncotarget.
8:34468–34480. 2017.PubMed/NCBI
|
|
34
|
Guo J, Cao R, Yu X, Xiao Z and Chen Z:
MicroRNA-223-3p inhibits human bladder cancer cell migration and
invasion. Tumour Biol. 39:10104283176916782017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yu G, Chen X, Chen S, Ye W, Hou K and
Liang M: miR-19a, miR-122 and miR-223 are differentially regulated
by hepatitis B virus X protein and involve in cell proliferation in
hepatoma cells. J Transl Med. 14:1222016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Giray BG, Emekdas G, Tezcan S, Ulger M,
Serin MS, Sezgin O, Altintas E and Tiftik EN: Profiles of serum
microRNAs; miR-125b-5p and miR223-3p serve as novel biomarkers for
HBV-positive hepatocellular carcinoma. Mol Biol Rep. 41:4513–4519.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Bhattacharya S, Steele R, Shrivastava S,
Chakraborty S, Di Bisceglie AM and Ray RB: Serum miR-30e and
miR-223 as novel noninvasive biomarkers for hepatocellular
carcinoma. Am J Pathol. 186:242–247. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhou J, Yu L, Gao X, Hu J, Wang J, Dai Z,
Wang JF, Zhang Z, Lu S, Huang X, et al: Plasma microRNA panel to
diagnose hepatitis B virus-related hepatocellular carcinoma. J Clin
Oncol. 29:4781–4788. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Xu J, Wu C, Che X, Wang L, Yu D, Zhang T,
Huang L, Li H, Tan W, Wang C and Lin D: Circulating microRNAs,
miR-21, miR-122, and miR-223, in patients with hepatocellular
carcinoma or chronic hepatitis. Mol Carcinog. 50:136–142. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ji J, Zheng X, Forgues M, Yamashita T,
Wauthier EL, Reid LM, Wen X, Song Y, Wei JS, Khan J, et al:
Identification of microRNAs specific for epithelial cell adhesion
molecule-positive tumor cells in hepatocellular carcinoma.
Hepatology. 62:829–840. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Li G, Shen Q, Li C, Li D, Chen J and He M:
Identification of circulating microRNAs as novel potential
biomarkers for hepatocellular carcinoma detection: A systematic
review and meta-analysis. Clin Transl Oncol. 17:684–693. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Fiorino S, Bacchi-Reggiani ML, Visani M,
Acquaviva G, Fornelli A, Masetti M, Tura A, Grizzi F, Zanello M,
Mastrangelo L, et al: MicroRNAs as possible biomarkers for
diagnosis and prognosis of hepatitis B- and
C-related-hepatocellular-carcinoma. World J Gastroenterol.
22:3907–3936. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhao FY, Han J, Chen XW, Wang J, Wang XD,
Sun JG and Chen ZT: miR-223 enhances the sensitivity of non-small
cell lung cancer cells to erlotinib by targeting the insulin-like
growth factor-1 receptor. Int J Mol Med. 38:183–191. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Juzėnas S, Saltenienė V, Kupcinskas J,
Link A, Kiudelis G, Jonaitis L, Jarmalaite S, Kupcinskas L,
Malfertheiner P and Skieceviciene J: Analysis of deregulated
microRNAs and their target genes in gastric cancer. PLoS One.
10:e01323272015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wong QW, Lung RW, Law PT, Lai PB, Chan KY,
To KF and Wong N: MicroRNA-223 is commonly repressed in
hepatocellular carcinoma and potentiates expression of stathmin1.
Gastroenterology. 135:257–269. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhao WY, Wang DD, Song MQ, Yang L, Ye J
and Chen LB: Role of microRNA-223 and its target gene oncogene
c-myc in hepatocellular carcinoma pathogenesis. Zhonghua Gan Zang
Bing Za Zhi. 19:114–117. 2011.(In Chinese). PubMed/NCBI
|
|
47
|
Whiting PF, Rutjes AW, Westwood ME,
Mallett S, Deeks JJ, Reitsma JB, Leeflang MM, Sterne JA and Bossuyt
PM; QUADAS-2 Group, : QUADAS-2: A revised tool for the quality
assessment of diagnostic accuracy studies. Ann Intern Med.
155:529–536. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zintzaras E and Ioannidis JP:
Heterogeneity testing in meta-analysis of genome searches. Genet
Epidemiol. 28:123–137. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Deeks JJ, Macaskill P and Irwig L: The
performance of tests of publication bias and other sample size
effects in systematic reviews of diagnostic test accuracy was
assessed. J Clin Epidemiol. 58:882–893. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Huang WT, Wang HL, Yang H, Ren FH, Luo YH,
Huang CQ, Liang YY, Liang HW, Chen G and Dang YW: Lower expressed
miR-198 and its potential targets in hepatocellular carcinoma: A
clinicopathological and in silico study. Onco Targets Ther.
9:5163–5180. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Settles B: ABNER: An open source tool for
automatically tagging genes, proteins and other entity names in
text. Bioinformatics. 21:3191–3192. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43(Database issue): D447–D452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Khairy A, Hamza I, Shaker O and Yosry A:
Serum miRNA panel in egyptian patients with chronic hepatitis C
related hepatocellular carcinoma. Asian Pac J Cancer Prev.
17:2699–2703. 2016.PubMed/NCBI
|
|
54
|
Zuo D, Chen L, Liu X, et al: Combination
of miR-125b and miR-27a enhances sensitivity and specificity of
AFP-based diagnosis of hepatocellular carcinoma. Tumour Biol.
37:6539–6549. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Oksuz Z, Serin MS, Kaplan E, Dogen A,
Tezcan S, Aslan G, Emekdas G, Sezgin O, Altintas E and Tiftik EN:
Serum microRNAs; miR-30c-5p, miR-223-3p, miR-302c-3p and miR-17-5p
could be used as novel non-invasive biomarkers for HCV-positive
cirrhosis and hepatocellular carcinoma. Mol Biol Rep. 42:713–720.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Li LM, Hu ZB, Zhou ZX, Chen X, Liu FY,
Zhang JF, Shen HB, Zhang CY and Zen K: Serum microRNA profiles
serve as novel biomarkers for HBV infection and diagnosis of
HBV-positive hepatocarcinoma. Cancer Res. 70:9798–9807. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Laios A, O'Toole S, Flavin R, Martin C,
Kelly L, Ring M, Finn SP, Barrett C, Loda M, Gleeson N, et al:
Potential role of miR-9 and miR-223 in recurrent ovarian cancer.
Mol Cancer. 7:352008. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Tachibana H, Sho R, Takeda Y, Zhang X,
Yoshida Y, Narimatsu H, Otani K, Ishikawa S, Fukao A, Asao H and
Iino M: Circulating miR-223 in oral cancer: Its potential as a
novel diagnostic biomarker and therapeutic target. PLoS One.
11:e01596932016. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Swets JA: Measuring the accuracy of
diagnostic systems. Science. 240:1285–1293. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Han ZB, Zhong L, Teng MJ, Fan JW, Tang HM,
Wu JY, Chen HY, Wang ZW, Qiu GQ and Peng ZH: Identification of
recurrence-related microRNAs in hepatocellular carcinoma following
liver transplantation. Mol Oncol. 6:445–457. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Wei Y, Yang J, Yi L, Wang Y, Dong Z, Liu
Z, Ou-yang S, Wu H, Zhong Z, Yin Z, et al: miR-223-3p targeting
SEPT6 promotes the biological behavior of prostate cancer. Sci Rep.
4:75462014. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Zhi Y, Pan J, Shen W, He P, Zheng J, Zhou
X, Lu G, Chen Z and Zhou Z: Ginkgolide B inhibits human bladder
cancer cell migration and invasion through microRNA-223-3p. Cell
Physiol Biochem. 39:1787–1794. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Ulbing M, Kirsch AH, Leber B, Lemesch S,
Münzker J, Schweighofer N, Hofer D, Trummer O, Rosenkranz AR,
Müller H, et al: MicroRNAs 223–3p and 93-5p in patients with
chronic kidney disease before and after renal transplantation.
Bone. 95:115–123. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Dong Z, Qi R, Guo X, Zhao X, Li Y, Zeng Z,
Bai W, Chang X, Hao L, Chen Y, et al: miR-223 modulates
hepatocellular carcinoma cell proliferation through promoting
apoptosis via the Rab1-mediated mTOR activation. Biochem Biophys
Res Commun. 483:630–637. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Dong YW, Wang R, Cai QQ, Qi B, Wu W, Zhang
YH and Wu XZ: Sulfatide epigenetically regulates miR-223 and
promotes the migration of human hepatocellular carcinoma cells. J
Hepatol. 60:792–801. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Yang T, Zheng ZM, Li XN, Li ZF, Wang Y,
Geng YF, Bai L and Zhang XB: miR-223 modulates multidrug resistance
via downregulation of ABCB1 in hepatocellular carcinoma cells. Exp
Biol Med (Maywood). 238:1024–1032. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Wang XM, Li J, Feng XC, Wang Q, Guan DY
and Shen ZH: Involvement of the role of Chk1 in lithium-induced
G2/M phase cell cycle arrest in hepatocellular carcinoma cells. J
Cell Biochem. 104:1181–1191. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Xie Y, Wei RR, Huang GL, Zhang MY, Yuan YF
and Wang HY: Checkpoint kinase 1 is negatively regulated by miR-497
in hepatocellular carcinoma. Med Oncol. 31:8442014. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Hong J, Hu K, Yuan Y, Sang Y, Bu Q, Chen
G, Yang L, Li B, Huang P, Chen D, et al: CHK1 targets spleen
tyrosine kinase (L) for proteolysis in hepatocellular carcinoma. J
Clin Invest. 122:2165–2175. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Yan L, Yang X and Davidson NE: Role of DNA
methylation and histone acetylation in steroid receptor expression
in breast cancer. J Mammary Gland Biol Neoplasia. 6:183–192. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Fang QL, Yin YR, Xie CR, Zhang S, Zhao WX,
Pan C, Wang XM and Yin ZY: Mechanistic and biological significance
of DNA methyltransferase 1 upregulated by growth factors in human
hepatocellular carcinoma. Int J Oncol. 46:782–790. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Li H, Yang F, Gao B, Yu Z, Liu X, Xie F
and Zhang J: Hepatitis B virus infection in hepatocellular
carcinoma tissues upregulates expression of DNA methyltransferases.
Int J Clin Exp Med. 8:4175–4185. 2015.PubMed/NCBI
|
|
73
|
Saito Y, Kanai Y, Nakagawa T, Sakamoto M,
Saito H, Ishii H and Hirohashi S: Increased protein expression of
DNA methyltransferase (DNMT) 1 is significantly correlated with the
malignant potential and poor prognosis of human hepatocellular
carcinomas. Int J Cancer. 105:527–532. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Neef R, Klein UR, Kopajtich R and Barr FA:
Cooperation between mitotic kinesins controls the late stages of
cytokinesis. Curr Biol. 16:301–307. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Wang SM, Ooi LL and Hui KM: Upregulation
of Rac GTPase-activating protein 1 is significantly associated with
the early recurrence of human hepatocellular carcinoma. Clin Cancer
Res. 17:6040–6051. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Cao L, Li C, Shen S, Yan Y, Ji W, Wang J,
Qian H, Jiang X, Li Z and Wu M: OCT4 increases BIRC5 and CCND1
expression and promotes cancer progression in hepatocellular
carcinoma. BMC Cancer. 13:822013. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Jin B, Wang W, Du G, Huang GZ, Han LT,
Tang ZY, Fan DG, Li J and Zhang SZ: Identifying hub genes and
dysregulated pathways in hepatocellular carcinoma. Eur Rev Med
Pharmacol Sci. 19:592–601. 2015.PubMed/NCBI
|
|
78
|
Koilan S, Hamilton D, Baburyan N, Padala
MK, Weber KT and Guntaka RV: Prevention of liver fibrosis by triple
helix-forming oligodeoxyribonucleotides targeted to the promoter
region of type I collagen gene. Oligonucleotides. 20:231–237. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Yang MR, Zhang Y, Wu XX and Chen W:
Critical genes of hepatocellular carcinoma revealed by network and
module analysis of RNA-seq data. Eur Rev Med Pharmacol Sci.
20:4248–4256. 2016.PubMed/NCBI
|
|
80
|
Hayashi M, Nomoto S, Hishida M, Inokawa Y,
Kanda M, Okamura Y, Nishikawa Y, Tanaka C, Kobayashi D, Yamada S,
et al: Identification of the collagen type 1 α 1 gene (COL1A1) as a
candidate survival-related factor associated with hepatocellular
carcinoma. BMC Cancer. 14:1082014. View Article : Google Scholar : PubMed/NCBI
|