|
1
|
Navarro-González JF, Mora-Fernández C,
Muros de Fuentes M and García-Pérez J: Inflammatory molecules and
pathways in the pathogenesis of diabetic nephropathy. Nat Rev
Nephrol. 7:327–340. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Reidy K, Kang HM, Hostetter T and Susztak
K: Molecular mechanisms of diabetic kidney disease. J Clin Invest.
124:2333–2340. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
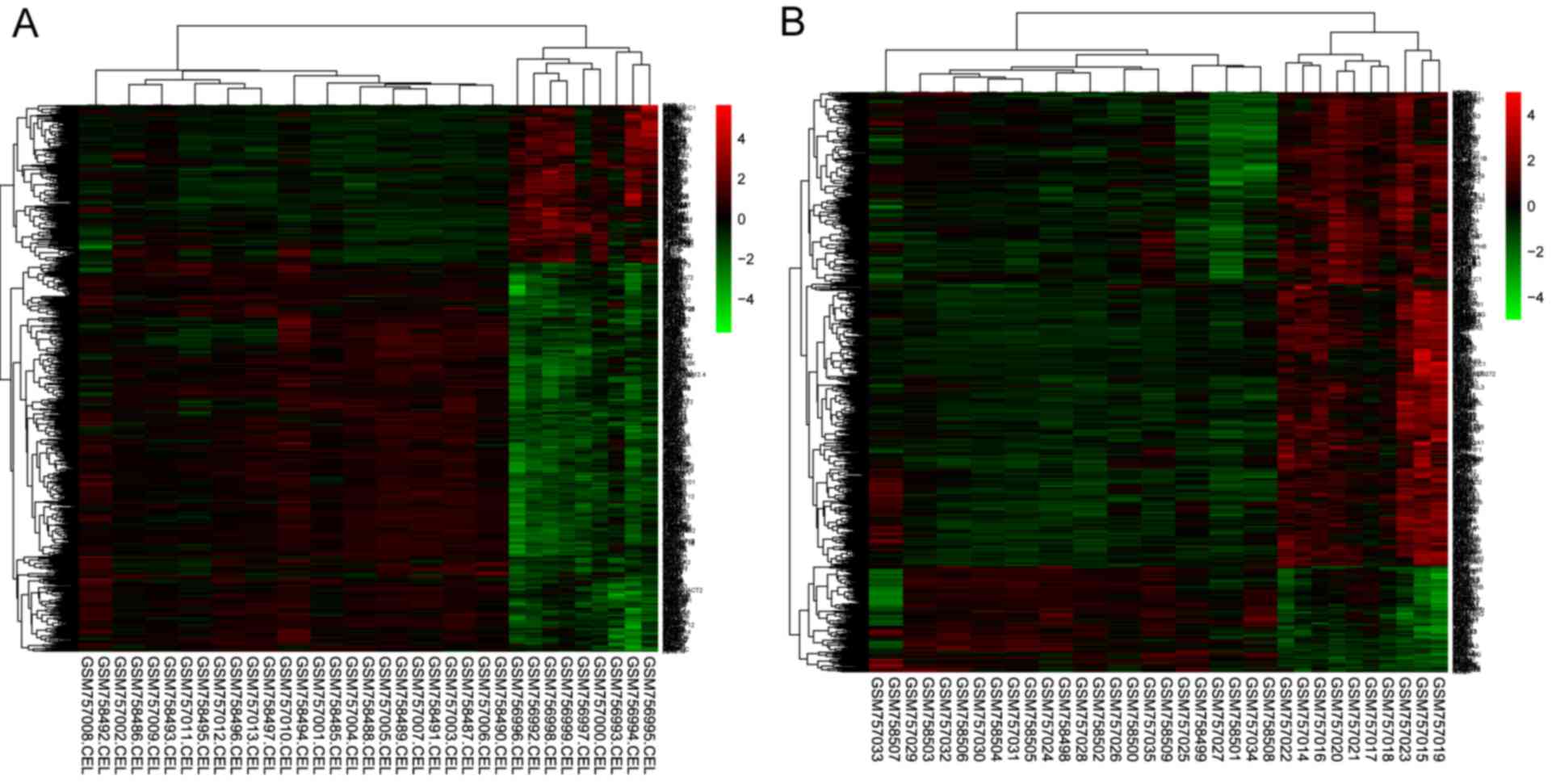
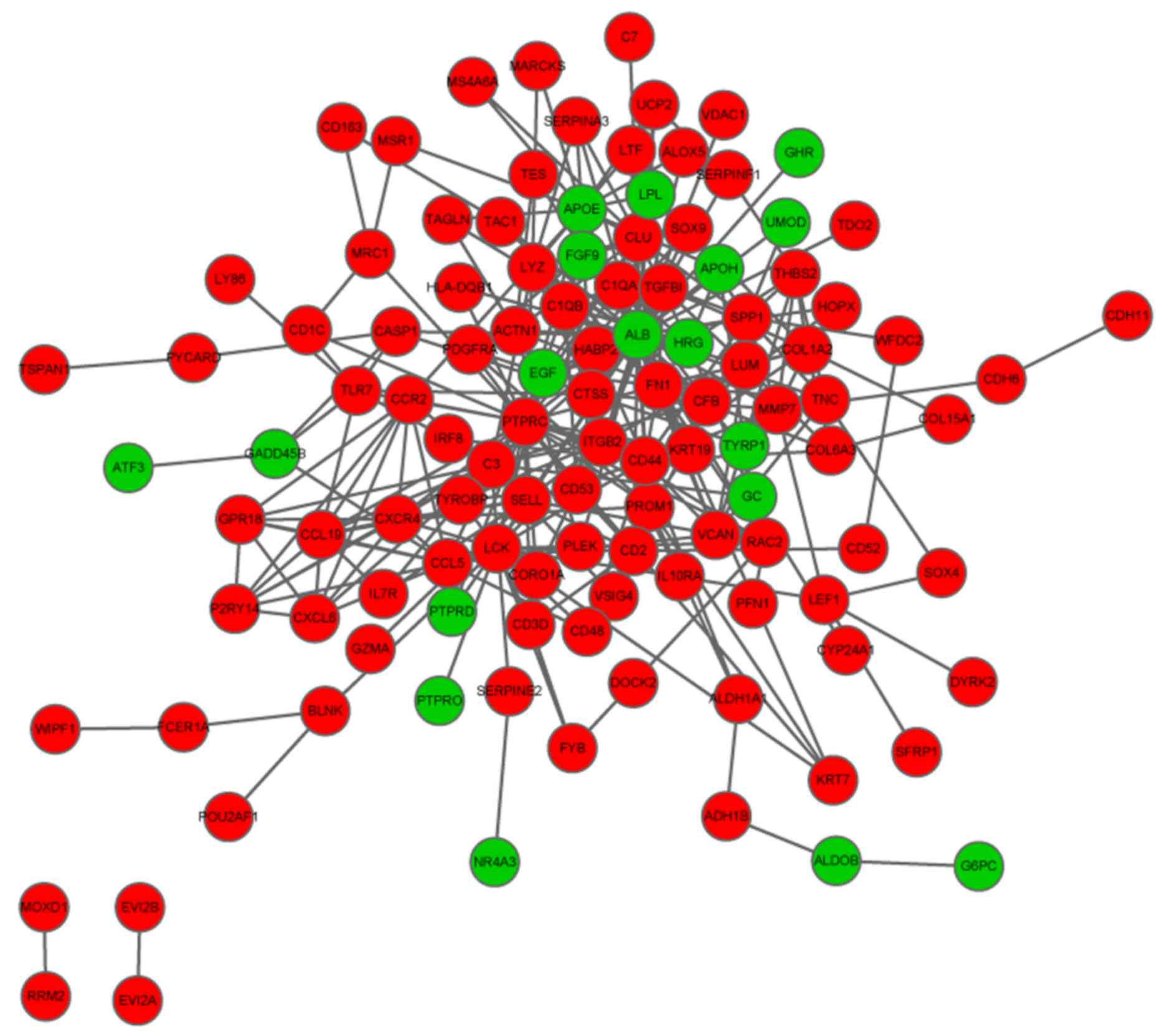
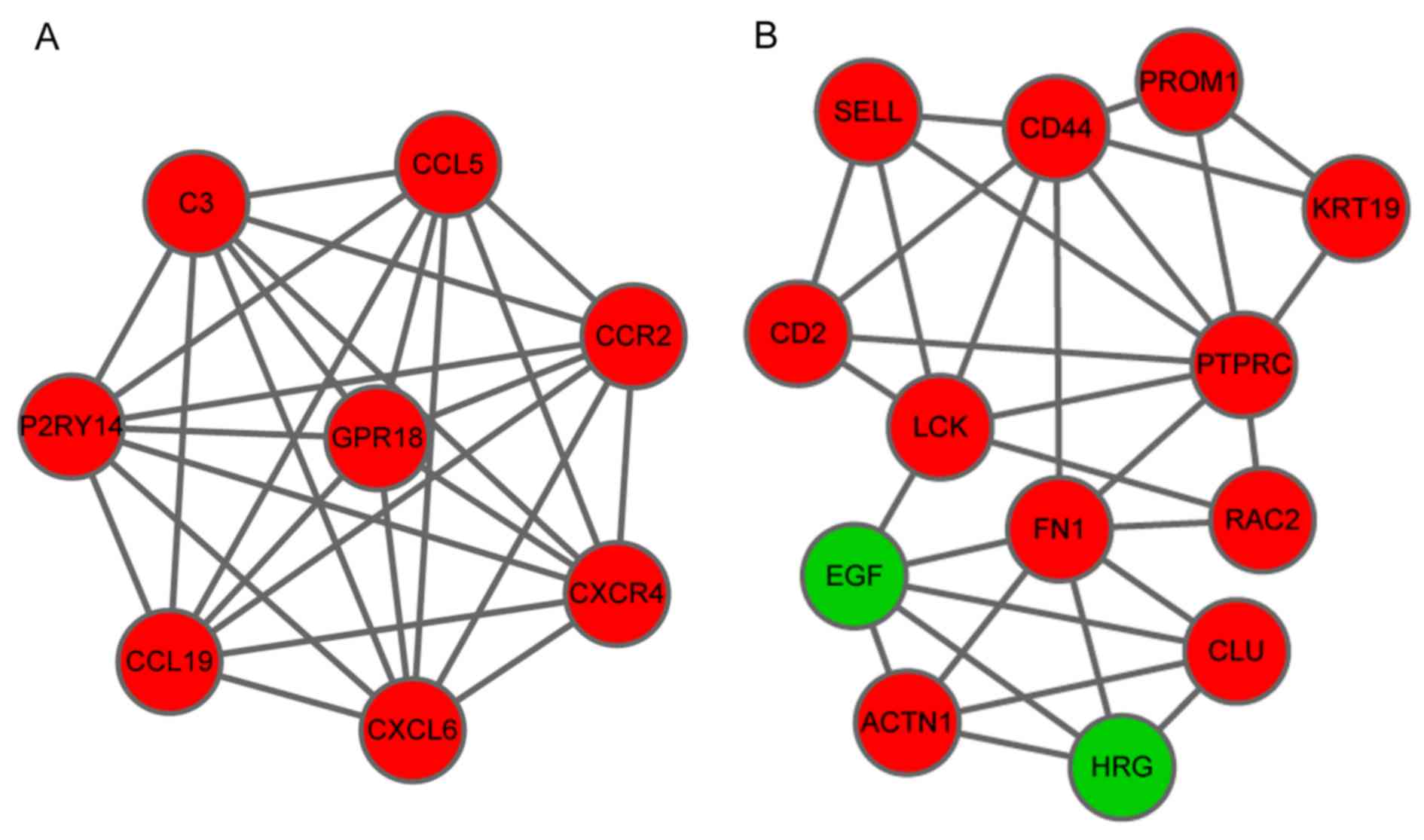
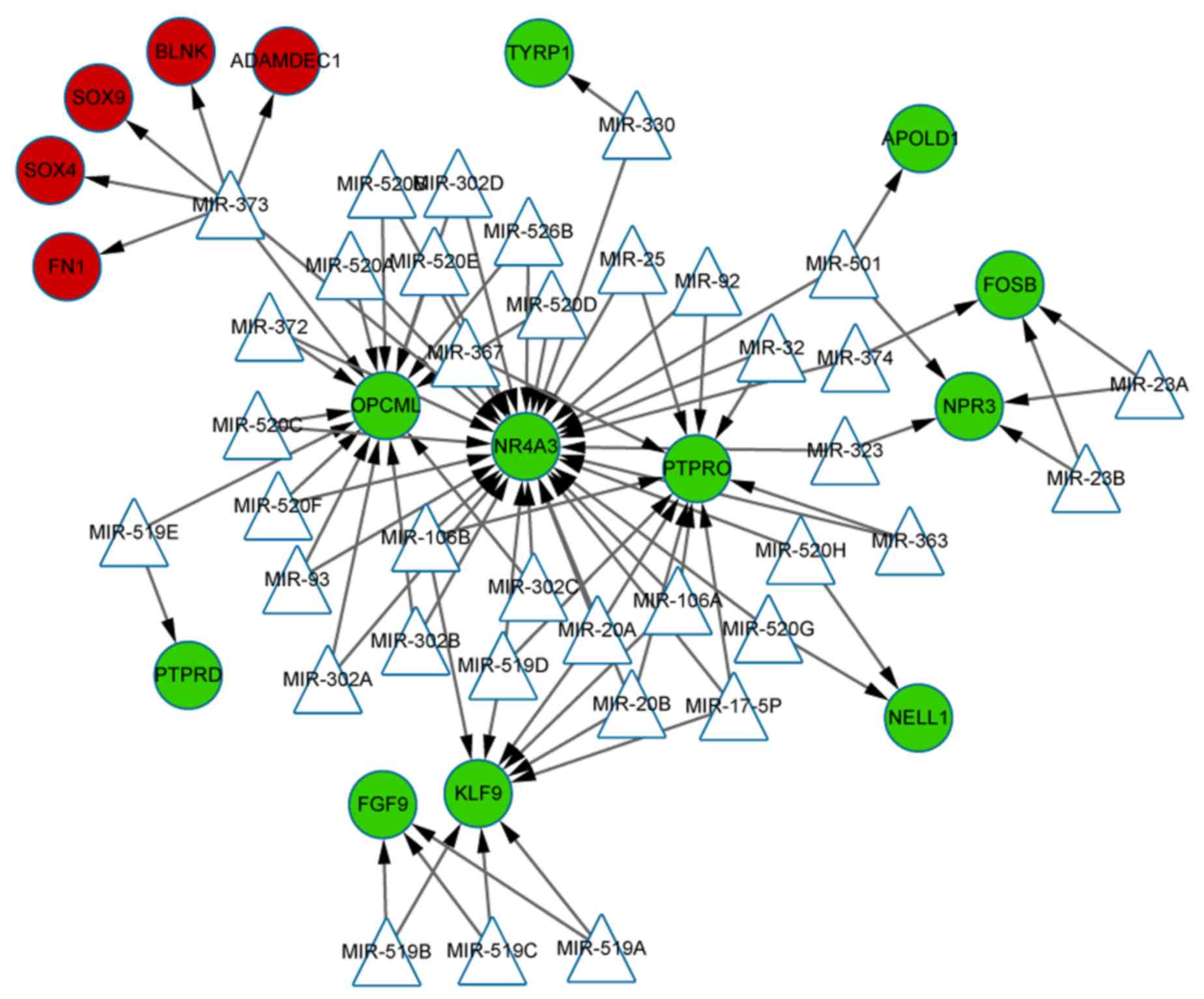
3
|
Nasri H: On the occasion of the world
diabetes day 2013; diabetes education and prevention; a nephrology
point of view. J Renal Inj Prev. 2:31–32. 2013.PubMed/NCBI
|
|
4
|
Xiao X, Ma B, Dong B, Zhao P, Tai N, Chen
L, Wong FS and Wen L: Cellular and humoral immune responses in the
early stages of diabetic nephropathy in NOD mice. J Autoimmun.
32:85–93. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tang W, Gao Y, Li Y and Shi G: Gene
networks implicated in diabetic kidney disease. Eur Rev Med
Pharmacol Sci. 16:1967–1973. 2012.PubMed/NCBI
|
|
6
|
Harjutsalo V and Groop PH: Epidemiology
and risk factors for diabetic kidney disease. Adv Chronic Kidney
Dis. 21:260–266. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Palmer ND, Ng MC, Hicks PJ, Mudgal P,
Langefeld CD, Freedman BI and Bowden DW: Evaluation of candidate
nephropathy susceptibility genes in a genome-wide association study
of African American diabetic kidney disease. PLoS One.
9:e882732014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Himmelfarb J and Tuttle KR: New therapies
for diabetic kidney disease. N Engl J Med. 369:2549–2550. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Anderberg RJ, Meek RL, Hudkins KL, Cooney
SK, Alpers CE, Leboeuf RC and Tuttle KR: Serum amyloid a and
inflammation in diabetic kidney disease and podocytes. Lab Invest.
95:6972015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Perez-Gomez MV, Sanchez-Niño MD, Sanz AB,
Zheng B, Martín-Cleary C, Ruiz-Ortega M, Ortiz A and
Fernandez-Fernandez B: Targeting inflammation in diabetic kidney
disease: Early clinical trials. Expert Opin Investig Drugs.
25:1045–1058. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Nasri H and Rafieiankopaei M: Protective
effects of herbal antioxidants on diabetic kidney disease. J Res
Med Sci. 19:82–83. 2014.PubMed/NCBI
|
|
12
|
Giuliani F, Di Maio M, Colucci G and
Perrone F: Conventional chemotherapy of advanced pancreatic cancer.
Curr Drug Targets. 13:795–801. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Brosius FC, Tuttle KR and Kretzler M: JAK
inhibition in the treatment of diabetic kidney disease.
Diabetologia. 59:1624–1627. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kato M, Zhang J, Wang M, Lanting L, Yuan
H, Rossi JJ and Natarajan R: MicroRNA-192 in diabetic kidney
glomeruli and its function in TGF-beta-induced collagen expression
via inhibition of E-box repressors. Proc Natl Acad Sci U S A.
104:pp. 3432–3437. 2007; View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chung ACK: MicroRNAs in diabetic kidney
disease. Adv Exp Med Biol. 888:253–269. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Woroniecka KI, Park AS, Mohtat D, Thomas
DB, Pullman JM and Susztak K: Transcriptome analysis of human
diabetic kidney disease. Diabetes. 60:2354–2369. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kauffmann A, Gentleman R and Huber W:
arrayQualityMetrics - a bioconductor package for quality assessment
of microarray data. Bioinformatics. 25:415–416. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Smyth GK: Limma: Linear Models for
Microarray Data. Springer; New York, NY: 2005
|
|
19
|
Kolde R: Pheatmap: Pretty Heatmaps.
2015.
|
|
20
|
Szklarczyk D, Franceschini A, Kuhn M,
Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork
P, et al: The STRING database in 2011: Functional interaction
networks of proteins, globally integrated and scored. Nucleic Acids
Res. 39:D561–D568. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Smoot ME, Ono K, Ruscheinski J, Wang PL
and Ideker T: Cytoscape 2.8: New features for data integration and
network visualization. Bioinformatics. 27:431–432. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Latora V and Marchiori M: A measure of
centrality based on network efficiency. New J Phys. 9:2007.
View Article : Google Scholar
|
|
23
|
Dagan T, Artzy-Randrup Y and Martin W:
Modular networks and cumulative impact of lateral transfer in
prokaryote genome evolution. Proc Natl Acad Sci USA. 105:pp.
10039–10044. 2008; View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang J, Duncan D, Shi Z and Zhang B:
WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): Update 2013.
Nucleic Acids Res. 41:W77–W83. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
López-Hernandez FJ and López-Novoa JM:
Role of TGF-β in chronic kidney disease: An integration of tubular,
glomerular and vascular effects. Cell Tissue Res. 347:141–154.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhao J, Wang L, Cao AL, Jiang MQ, Chen X,
Wang Y, Wang YM, Wang H, Zhang XM and Peng W: HuangQi decoction
ameliorates renal fibrosis via tgf-βsmad signaling pathway in vivo
and in vitro. Cell Physiol Biochem. 38:1761–1774. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pezzolesi MG, Skupien J, Mychaleckyj JC,
Warram JH and Krolewski AS: Insights to the genetics of diabetic
nephropathy through a genome-wide association study of the GoKinD
collection. Semin Nephrol. 30:126–140. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Alldinger S, Gröters S, Miao Q, Fonfara S,
Kremmer E and Baumgörtner W: Roles of an extracellular matrix (ECM)
receptor and ECM processing enzymes in demyelinating canine
distemper encephalitis. Dtsch Tierarztl Wochenschr. 113:151–152.
2006.PubMed/NCBI
|
|
29
|
Mullen W, Delles C and Mischak H: EuroKUP
COST action: Urinary proteomics in the assessment of chronic kidney
disease. Curr Opin Nephrol Hypertens. 20:654–661. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Phillips AO and Steadman R: Diabetic
nephropathy: The central role of renal proximal tubular cells in
tubulointerstitial injury. Histol Histopathol. 17:247–252.
2002.PubMed/NCBI
|
|
31
|
Alvarez ML and Distefano JK: Functional
characterization of the plasmacytoma variant translocation 1 gene
(PVT1) in diabetic nephropathy. PLoS One. 6:e186712011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Alvarez ML and Distefano JK: The role of
non-coding RNAs in diabetic nephropathy: Potential applications as
biomarkers for disease development and progression. Diabetes Res
Clin Pract. 99:1–11. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Alvarez ML, Khosroheidari M, Eddy E and
Kiefer J: Role of microRNA 1207-5P and its host gene, the long
non-coding RNA Pvt1, as mediators of extracellular matrix
accumulation in the kidney: Implications for diabetic nephropathy.
PLoS One. 8:e774682013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sedger LM, Seddiki N and Ranasinghe C:
Cytokines and cytokine receptors as immunotherapeutics: Humble
beginnings and exciting futures. Cytokine Growth Factor Rev.
25:351–353. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ntemka A, Iliadis F, Papanikolaou N and
Grekas D: Network-centric analysis of genetic predisposition in
diabetic nephropathy. Hippokratia. 15:232–237. 2011.PubMed/NCBI
|
|
36
|
Skov V, Knudsen S, Olesen M, Hansen ML and
Rasmussen LM: Global gene expression profiling displays a network
of dysregulated genes in non-atherosclerotic arterial tissue from
patients with type 2 diabetes. Cardiovasc Diabetol. 11:1475–2840.
2012. View Article : Google Scholar
|
|
37
|
Heinzel A, Perco P, Mayer G, Oberbauer R,
Lukas A and Mayer B: From molecular signatures to predictive
biomarkers: Modeling disease pathophysiology and drug mechanism of
action. Front Cell Dev Biol. 2:372014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Darisipudi MN, Kulkarni OP, Sayyed SG, Ryu
M, Migliorini A, Sagrinati C, Parente E, Vater A, Eulberg D,
Klussmann S, et al: Dual blockade of the homeostatic chemokine
CXCL12 and the proinflammatory chemokine CCL2 has additive
protective effects on diabetic kidney disease. Am J Pathol.
179:116–124. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Long DA, Norman JT and Fine LG: Restoring
the renal microvasculature to treat chronic kidney disease. Nat Rev
Nephrol. 8:244–250. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wu J, Long Z, Cai H, Du C, Liu X, Yu S and
Wang Y: High expression of WISP1 in colon cancer is associated with
apoptosis, invasion and poor prognosis. Oncotarget. 7:49834–49847.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Valsecchi R, Coltella N, Belloni D,
Ponente M, Ten Hacken E, Scielzo C, Scarfò L, Bertilaccio MT,
Brambilla P, Lenti E, et al: HIF-1α regulates the interaction of
chronic lymphocytic leukemia cells with the tumor microenvironment.
Blood. 127:1987–1997. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Patel V, Williams D, Hajarnis S, Hunter R,
Pontoqlio M, Somlo S and Iqarashi P: miR-17~92 miRNA cluster
promotes kidney cyst growth in polycystic kidney disease. Proc Natl
Acad Sci USA. 110:pp. 10765–10770. 2013; View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Phua YL and Ho J: MicroRNAs in the
pathogenesis of cystic kidney disease. Curr Opin Pediatr.
27:219–226. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Butz H, Rácz K, Hunyady L and Patócs A:
Crosstalk between TGF-β signaling and the microRNA machinery.
Trends Pharmacol Sci. 33:382–393. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Moura J, Børsheim E and Carvalho E: The
role of MicroRNAs in diabetic complications-special emphasis on
wound healing. Genes (Basel). 5:926–956. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Marques FZ, Campain AE, Tomaszewski M,
Zukowska-Szczechowska E, Yang YH, Charchar FJ and Morris BJ: Gene
expression profiling reveals renin mRNA overexpression in human
hypertensive kidneys and a role for microRNAs. Hypertension.
58:1093–1098. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Cloonan N, Brown MK, Steptoe AL, Wani S,
Chan WL, Forrest AR, Kolle G, Gabrielli B and Grimmond SM: The
miR-17-5p microRNA is a key regulator of the G1/S phase cell cycle
transition. Genome Biol. 9:R1272008. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Ozaltin F, Ibsirlioglu T, Taskiran EZ,
Baydar DE, Kaymaz F, Buyukcelik M, Kilic BD, Balat A, Iatropoulos
P, Asan E, et al: Disruption of PTPRO causes childhood-onset
nephrotic syndrome. Am J Hum Genet. 89:139–147. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Charba DS, Wiggins RC, Goyal M, Wharram
BL, Wiggins JE, McCarthy ET, Sharma R, Sharma M and Savin VJ:
Antibodies to protein tyrosine phosphatase receptor type O (PTPro)
increase glomerular albumin permeability (P(alb)). Ajp Renal
Physiology. 297:F138–F144. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Kato H, Gruenwald A, Suh JH, Miner JH,
Barisoni-Thomas L, Taketo MM, Faul C, Millar SE, Holzman LB and
Susztak K: Wnt/β-catenin pathway in podocytes integrates cell
adhesion, differentiation and survival. J Biol Chem.
286:26003–26015. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Xu X, Hong Y, Kong C, Xu L, Tan J, Liang
Q, Huang B and Lu J: Protein tyrosine phosphatase receptor-type O
(PTPRO) is co-regulated by E2F1 and miR-17-92. FEBS Lett.
582:2850–2856. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Conserva F, Pontrelli P, Accetturo M and
Gesualdo L: The pathogenesis of diabetic nephropathy: Focus on
microRNAs and proteomics. J Nephrol. 26:811–820. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Jiang ZS, Jia HX, Xing WJ, Han CD, Wang J,
Zhang ZJ and Qu W: Investigation of several biomarkers associated
with diabetic nephropathy. Exp Clin Endocrinol Diabetes. 123:1–6.
2015.PubMed/NCBI
|
|
54
|
Zucker SN, Fink EE, Bagati A, Mannava S,
Bianchi-Smiraglia A, Bogner PN, Wawrzyniak JA, Foley C, Leonova KI,
Grimm MJ, et al: Nrf2 amplifies oxidative stress via induction of
Klf9. Mol Cell. 53:916–928. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Dong H, You SH, Williams A, Wade MG, Yauk
CL and Thomas Zoeller R: Transient maternal hypothyroxinemia
potentiates the transcriptional response to exogenous thyroid
hormone in the fetal cerebral cortex before the onset of fetal
thyroid function: A messenger and microrna profiling study. Cereb
Cortex. 25:1735–1745. 2015. View Article : Google Scholar : PubMed/NCBI
|