Introduction
Atherosclerosis is an inflammatory disease about
creating an atheromatous plaque (1–3). The
pathobiology of atherosclerotic lesions is very complicated, but
generally, macrophages-derived foam cells contributes to rupture of
unstable plaques (4). Ruptures of
the fibrous cap expose thrombogenic material, eventually induce
thrombus formation in the lumen, resulting in ischemia (5). Research into the disease has led to
many compelling hypotheses about the pathophysiology of
atherosclerotic lesion formation and of complications such as
myocardial infarction and stroke (6).
Macrophages engulf modified lipoproteins and
transform themselves into lipid-loaded foam cells, which contribute
to the formation of the necrotic core in atheromatous plaques
(7). Various inflammatory factors
secreted by lipid-loaded macrophages also enlarge and expand the
local inflammatory reaction, which aggravates atherosclerosis
(6). The investigation of the
mechanism involved in oxidized low-density lipoprotein
(oxLDL)-induced macrophage inflammation and lipid uptake is
becoming increasingly important.
Long non-coding RNAs (lncRNAs) are non-coding RNAs
that are longer than 200 bp. The lncRNA nuclear paraspeckle
assembly transcript 1 (NEAT1) is widely expressed in various
tissues and participates in many biological activities, such as
adipogenesis and tumorigenesis, including breast cancer, leukemia,
ovarian cancer, hepatocellular carcinoma and laryngeal squamous
cancer (8–13). In a recent study, neat1_2, a longer
isoform of neat1, together with RNA-binding proteins, including
PSPC1, non-POU domain-containing octamer-binding (NONO) and SFPQ,
initiated the formation of subnuclear structures called
paraspeckles (14). Paraspeckles
can exert antiviral functions by stabilizing SFPQ, which suppresses
IL8 transcription (15).
In this study, we explored the role of
neat1-mediated paraspeckle formation in oxLDL-induced macrophage
inflammation and lipid uptake.
Materials and methods
Cell culture and transfection
The human monocyte cell line THP-1 was purchased
from American Type Culture Collection (ATCC; Manassas, VA, USA).
The cells were cultured in complete medium consisting of 10% fetal
bovine serum and RIPM-1640 (both from Gibco, Grand Island, NY, USA)
with penicillin and streptomycin (Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) at 37°C with 5% CO2. Before
treatment, the cells were supplied with 100 ng/ml PMA (79346;
Sigma-Aldrich; Merck KGaA) to differentiate them into macrophages.
Inhibitors including SB20358 (p-p38 inhibitor, S8307), SP600125
(p-JNK inhibitor, S5567), PD98059 (p-ERK inhibitor, P215) and
BAY11-7085 (p-p65 inhibitor, B5681) (all purchased from
Sigma-Aldrich; Merck KGaA), were added 30 min prior to oxLDL (40
µg/ml, YB-002; Yiyuan Biotechnology Co., Ltd., Guangzhou, China)
treatment for 24 h. Dimethyl sulphoxide (DMSO, D2650;
Sigma-Aldrich) was used as the control of inhibitors since these
chemical inhibitors were dissolved in DMSO. Finally, the cells were
harvested for later experiments.
siRNAs against NEAT1_1 (GGA ACA UUC UCA UUU AAU
Att), NEAT1_2 (GGG UAA AUC UCA AUC UUA Att), NONO (GGG GUG GUA UUA
AAC AAG UCA) and SFPQ (GGC AAA GGA UUC GGA UUU AUU), as well as a
negative control (nc) (GUACCUGACUAGUCGCAGAAG) were synthetized by
Ruibo Biotechnology Co., Ltd. (Guangzhou, China). Transfection was
performed using Lipo 3000 reagent (Life Technologies, Grand Island,
NY, USA) according to the manufacturer's instructions. After
transfection with siRNAs (2 µl, 50 nM) and culturing for 24 h, the
cells were supplied with human oxLDL (40 µg/ml, YB-002; Yiyuan
Biotechnology) for the indicated lengths of time.
RNA isolation and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted using a simple total RNA
extraction kit (Tiangen, Beijing, China). cDNA was synthesized
using random primers (Takara, Otsu, Japan). SYBR premix Ex Taq II
(Takara) master mix was used for RT-qPCR analysis, and the
amplification consisted of 95 for 30 sec and 40 cycles of 95 for 5
sec and 60 for 30 sec. RNA (18 sec) served as an endogenous
control. Primers were as follows: NEAT1 forward,
GAGAACCAAAGGGAGGGGTG and reverse, TGCTGCGTATGCAAGTCTGA; NEAT1_2
forward, ACATTGTACACAGCGAGGCA and reverse, CATTTGCCTTTGGGGTCAGC;
β-actin forward, TGACGTGGACATCCGCAAAG and reverse,
CTGGAAGGTGGACAGCGAGG.
Western blot analysis
The cells were harvested and lysed using total
protein lysis buffer (Cell Signaling Technology, Inc., Danvers, MA,
USA). A total of 30 µg of protein was separated on a 10%
polyacrylamide-SDS gel, blotted onto a PVDF membrane (Millipore,
Billerica, MA, USA) and blocked with 5% non-fat milk (Sangon
Biotech Co., Ltd., Shanghai, China). After incubation with the
primary antibody overnight at 4°C and the corresponding secondary
antibody at 37°C for 1 h, the membrane was developed using an ECL
kit (Pierce, Rockford, IL, USA). The antibodies used were as
follows: Rabbit anti-GAPDH (1:1,000) (GP10353; Nuoyang, Hangzhou,
China); rabbit anti-p-p65 (1:1,000; no. 3033); rabbit anti-p-p38
(1:1,000; no. 4511); rabbit anti-p-JNK (1:1,000; no. 9255); rabbit
anti-p-ERK (1:1,000; no. 3510) (all from Cell Signaling Technology,
Inc.); rabbit anti-cluster of differentiation 36 (CD36; 1:1,000;
ab133625); rabbit anti-LOX-1 (1:1,000; ab60178); rabbit anti-NONO
(1:1,000; ab70335); rabbit anti-SFPQ (1:1,000; ab38148) (all from
Abcam, Shanghai, China); goat anti-rabbit (1:5,000; GP853); and
goat anti-mouse (1:5,000; GP843) (both from Nuoyang).
Nuclear protein isolation
Nuclear protein was extracted using a Nuclear and
Cytoplasmic Protein Extraction kit (Beyotime Institute of
Biotechnology (Shanghai, China) according to the manufacturer's
instructions. For nuclear RNA extraction, after centrifugation to
isolate the cytoplasmic proteins, the pellet was dissolved using
TRIzol reagent (Life Technologies), then extracted using chloroform
and precipitated using isopropanol.
RIP
RNA protein immunoprecipitation was performed using
a Magna RIP kit (Millipore). In brief, the cells were washed with
ice-cold PBS and lysed on ice with RIP lysis buffer. A/G magnetic
beads with antibodies against IgG (Millipore), NONO or SFPQ (Abcam,
Cambridge, MA, USA) were allowed to settle for 30 min at room
temperature. Then, the cell lysate was immunoprecipitated with the
antibody-coated magnetic beads. After incubating the
magnetic-bead-bound complexes with 10% SDS and proteinase K, the
supernatant was used for RNA extraction. The first-strand cDNA was
synthesized using a cDNA synthesis kit (Applied Biosystems, Foster
City, CA, USA). Finally, qRT-PCR was performed for further
analysis.
Oil red staining
After incubating with oxLDL (40 µg/ml) for 24 h, the
cells were washed with PBS and stained using oil red stain
solution, which comprised 30% alcohol and 70% oil red solution.
Then, the cells were further stained with hematoxylin solution for
10 min. After washing with PBS, the cells were observed using an
Olympus light microscope (Olympus, Tokyo, Japan). Oil red solution
and hematoxylin were purchased from Jiancheng Bioengineering
Institute (Nanjing, China).
Statistical analysis
Data are shown as the mean ± standard deviation
(SD). Non parametric t-tests were used to compare the differences
between two groups. One-way analysis of variance (ANOVA) was used
to compare the differences among three or more groups. P-values
<0.05 were considered to indicate a statistically significant
difference.
Results
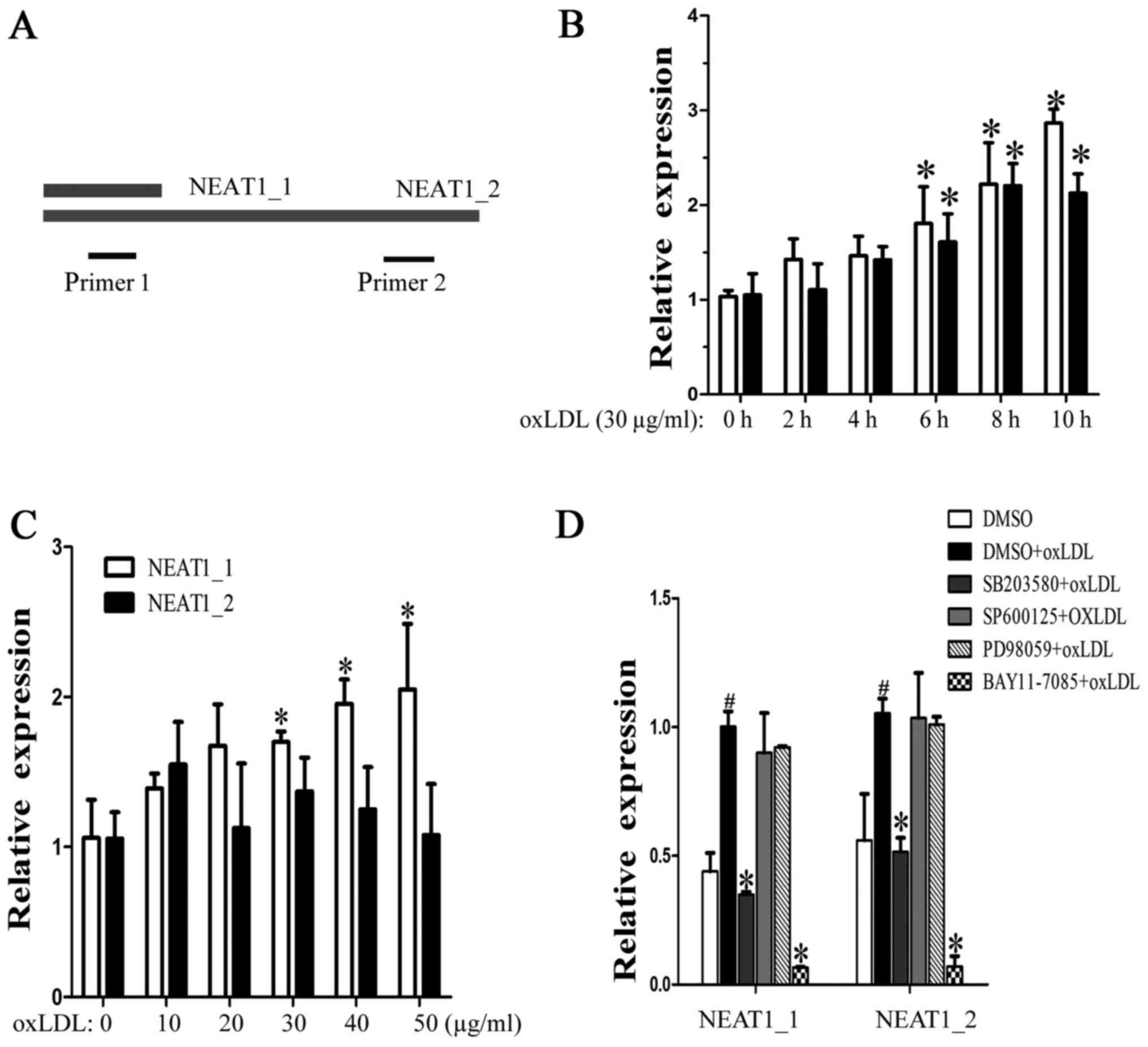
OxLDL induces NEAT1 and NEAT1_2
expression via p38 and NF-κB signaling
The lncRNA NEAT1 has two transcripts, NEAT1_1 and
NEAT1_2, which have the same transcription initiation site. Because
the entire NEAT1_1 sequence overlapped with the 5′ sequence of
NEAT1_2, we designed two primer pairs, NEAT1, to detect the
expression of both NEAT1_1 and NEAT1_2, and NEAT1_2, to detect only
NEAT1_2 (Fig. 1A). When the
macrophages were incubated with human oxLDL (30 µg/ml) for 2, 4, 6,
8 or 10 h, we found that both neat1 and NEAT1_2 increased over time
(Fig. 1B). Next, the macrophages
were stimulated with different concentrations of oxLDL (from 10 to
50 µg/ml) for 24 h. As shown in Fig.
1C, NEAT1 increased, but NEAT1_2 showed no significant changes,
which means that half-life period of NEAT1_2 is shorter than
NEAT1_1. To identify the regulatory pathway involved, MAPK and
NF-κB inhibitors were used. We found that p38 and NF-κB mediated
oxLDL-induced NEAT1 and NEAT1_2 expression (Fig. 1D). The above results show that
oxLDL can induce neat1 and NEAT1_2 transcription and that NEAT1_2
was less stable than neat1_1.
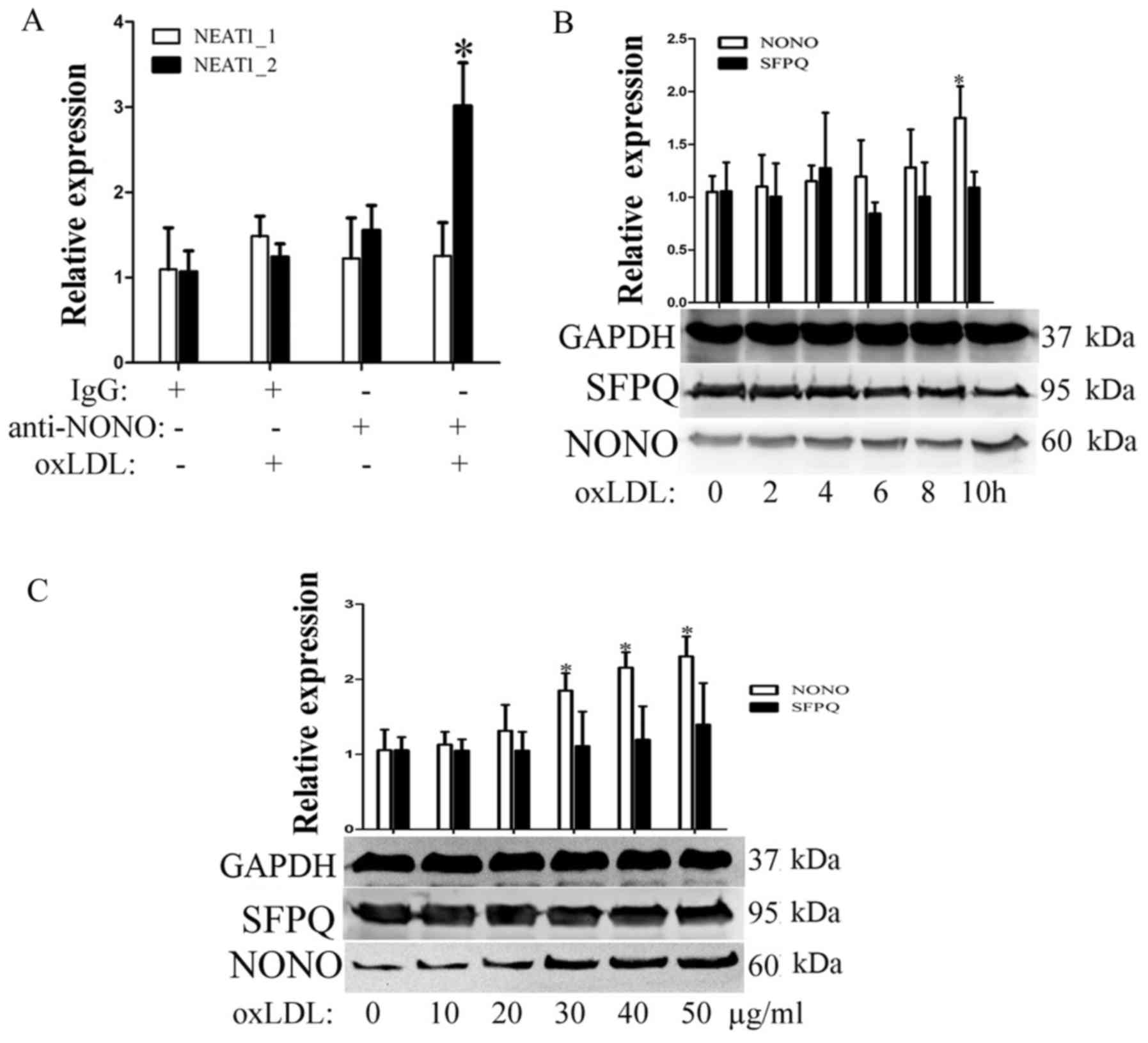
OxLDL induces NEAT1_2-mediated
paraspeckle formation
In previous study, the continued transcription of
the lncRNA NEAT1_2 promotes the formation of subnuclear structures
called paraspeckles. First, we detected the expression of two
paraspeckle-related proteins, NONO and SFPQ, during oxLDL
incubation. As shown in Fig. 2B and
C, SFPQ did not significantly change over the indicated times
or with the indicated concentrations of oxLDL. However, under the
same conditions, oxLDL can promote nono expression. To detect
whether oxLDL can induce paraspeckle formation, RIP experiments
were used to detect cross-linking between nono and NEAT1 or
NEAT1_2. As shown in Fig. 2A,
after 10 h of oxLDL stimulation, the complex that was
immunoprecipitated using a NONO antibody contained NEAT1_2 but not
neat1. Our results show that oxLDL stimulation can induce
NEAT1_2-mediated paraspeckle formation.
Neat1 promotes TNF-α secretion by
inducing p65 phosphorylation
To investigate whether paraspeckles participate in
the oxLDL-mediated secretion of proinflammatory factors,
macrophages were transfected with siRNAs against NEAT1_1, NEAT1_2,
NONO or SFPQ for 24 h prior to oxLDL stimulation. As shown in
Fig. 3A, TNF-α and iNOS
transcription were reduced by transfection with either siR-NEAT1_1
or siR-NEAT1_2. Then, we measured iNOS protein expression, but we
found no significant changes after siRNA transfection (Fig. 3B). TNF-α expression in the
supernatant of macrophages transfected with siRNAs under oxLDL
treatment was also analyzed, and we found that knocking down
NEAT1_1 or NEAT1_2 indeed decreased oxLDL-induced TNF-α secretion
(Fig. 3C). To determine whether
neat1 regulates TNF-α secretion by affecting proinflammatory
pathways, we analyzed the expression of MAPKs and NF-κB. As shown
in Fig. 3D and E, NEAT1 knockdown
had no significant effect on MAPKs or NF-κB at the early stage of
activation (2 h) by oxLDL, but it did decrease ERK and p65
phosphorylation at a later stage (8 h). The above results show that
NEAT1 promotes oxLDL-induced TNF-α secretion by regulating MAPKs
and NF-κB.
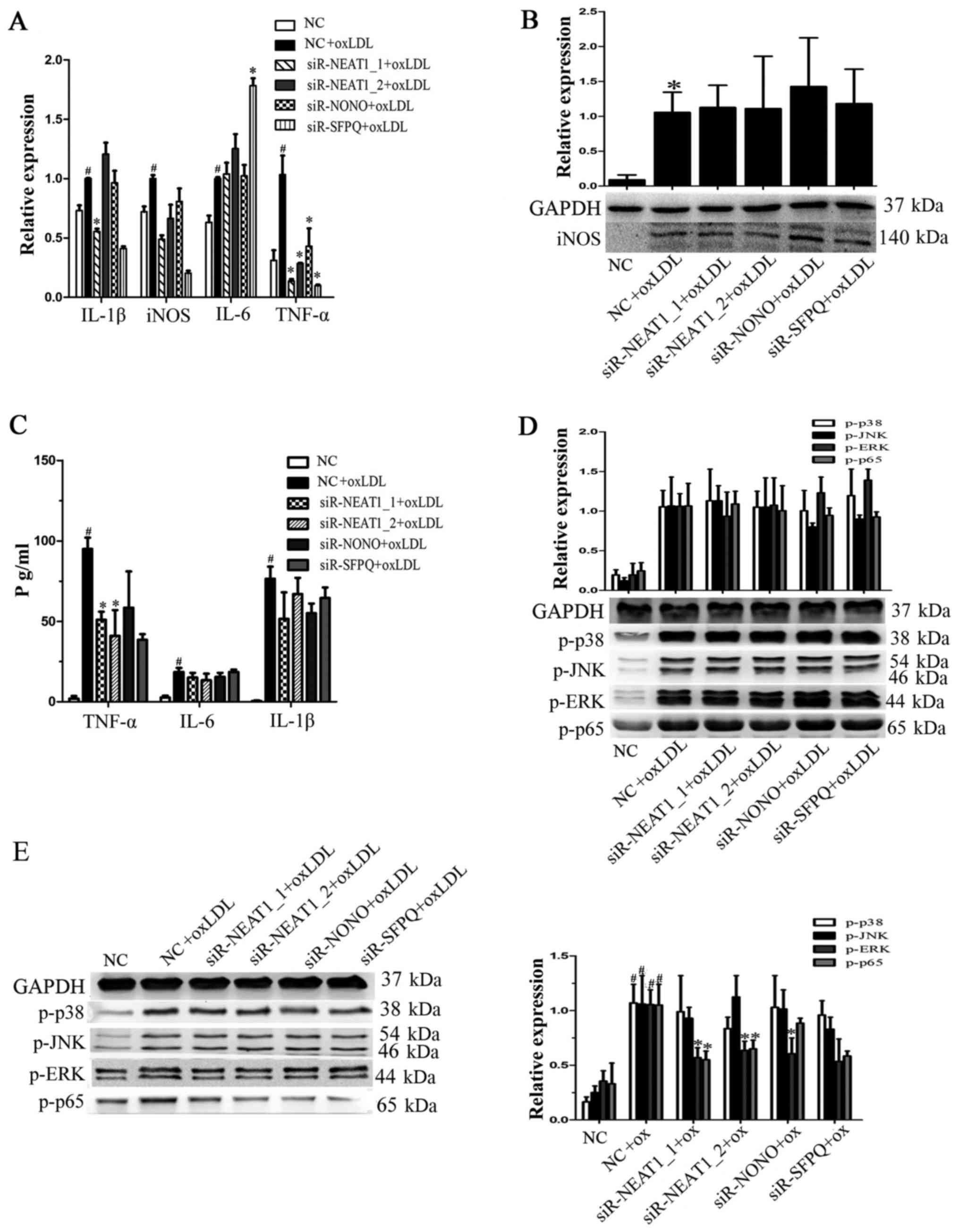 | Figure 3.NEAT1 promotes TNF-α secretion by
activating p65 phosphorylation. Prior to oxLDL treatment,
macrophages were transfected with a negative control, siR-NEAT1_1,
siR-NEAT1_2, siR-NONO or siR-SFPQ for 24 h. (A) Reverse
transcription-quantitative polymerase chain reaction was performed
to detect gene transcription. (B) Following transfection,
macrophages were incubated with oxLDL for a further 24 h. Western
blotting was performed to detect iNOS protein expression, and GAPDH
was an endogenous control. (C) Following transfection macrophages
were incubated with oxLDL for a further 10 h. The supernatant was
collected to analyze the levels of secreted TNF-α, IL-6 and IL-1β.
(D) Following transfection macrophages were incubated with oxLDL
for 2 h. Western blotting was performed to detect p-p38, p-JNK,
p-ERK and p-p65 protein expression, and GAPDH was an endogenous
control. (E) Following transfection macrophages were incubated with
oxLDL for a further 8 h. Western blotting was performed to detect
p-p38, p-JNK, p-ERK and p-p65 protein expression, and GAPDH was an
endogenous control. #P<0.05 vs. NC; *P<0.05 vs. NC
+ oxLDL. oxLDL, oxidized low-density lipoprotein; TNF-α, tumor
necrosis factor-α; NEAT, nuclear paraspeckle assembly transcript 1;
NONO, non-POU domain-containing octamer-binding; SFPQ, splicing
factor proline and glutamine rich; IL, interleukin; p,
phosphorylated; JNK, c-Jun N terminal kinase; ERK, extracellular
signal-regulated kinase; siR, small interfering RNA. |
Neat1 suppresses lipid uptake by
binding CD36 mRNA
Since lipid uptake by macrophages plays an important
role in the development of atherosclerosis, we also explored the
effect of paraspeckles on lipid uptake in macrophages. As shown in
Fig. 4A, transfection with either
siR-NEAT1_1 or siR-NEAT1_2 promotes Dil-labeled oxLDL (Dil-oxLDL)
uptake by macrophages. To determine the mechanism, we also analyzed
the transcription of scavenger receptors, including SRA, CD36 and
LOX-1, after transfection with siR-NEAT1_1 or siR-NEAT1_2 under
oxLDL treatment. As shown in Fig.
4B, only transfection with siR-NEAT1_2 promotes CD36 and LOX-1
transcription. However, we found that transfection with either
siR-NEAT1_1 or siR-NEAT1_2 promotes CD36 protein expression
(Fig. 4C). We suspect that NEAT1_2
inhibits CD36 expression by stabilizing CD36 mRNA in paraspeckles.
R-IPs were performed using the anti-NONO antibody. When macrophages
were treated with oxLDL, the immunoprecipitated complex contained
more CD36 mRNA (Fig. 4D). In
conclusion, NEAT1 suppressed lipid uptake in part by stabilizing
CD36 mRNA in paraspeckles.
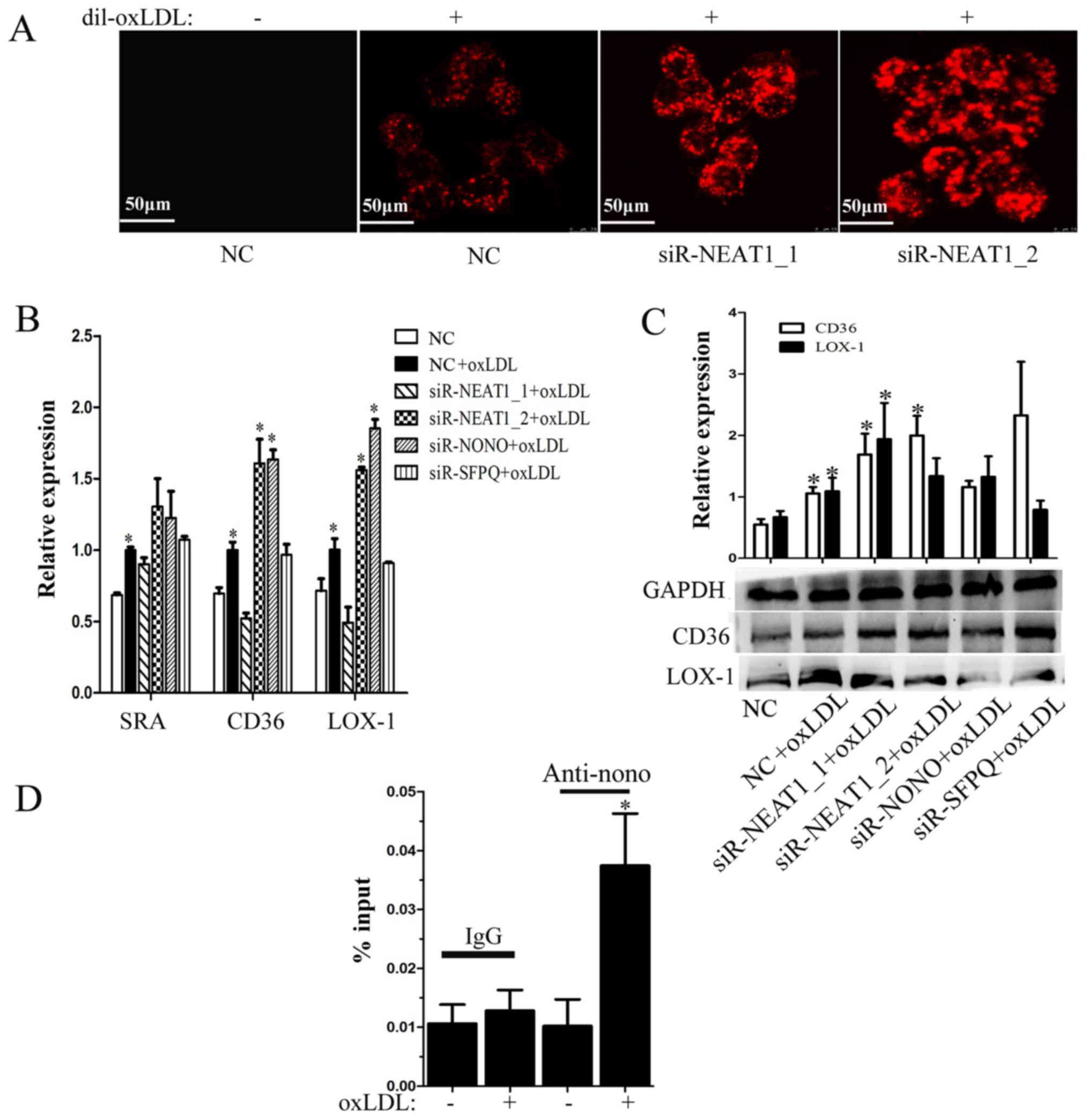 | Figure 4.NEAT1 inhibits lipid uptake in part by
suppressing CD36 expression. Macrophages were transfected with a
negative control, siR-NEAT1_1 or siR-NEAT1_2 for 24 h and (A) then
incubated with Dil-oxLDL for 30 min. Lipid uptake was evaluated
based on the fluorescence intensity of Dil-oxLDL. Scale bars, 50
µm. (B) Macrophages were transfected and then incubated with oxLDL
for 12 h. RT-qPCR was performed to detect the transcription of
associated genes. (C) Macrophages were transfected and then
incubated with oxLDL for 24 h. Western blotting was performed to
detect CD36 and LOX-1 protein expression, and GAPDH was an
endogenous control. *P<0.05 vs. NC + oxLDL. (D) Macrophages were
treated with oxLDL for 8 h. RNA protein immunoprecipitation was
performed using anti-nono (with IgG as control), and RT-qPCRs were
used to detect the level of CD36 mRNA in the immunoprecipitated
complex. *P<0.05 vs. (−) oxLDL. NEAT, nuclear paraspeckle
assembly transcript 1; CD36, cluster of differentiation 36; siRNA,
small interfering RNA; oxLDL, oxidized low-density lipoprotein;
Dil-oxLDL, Dil-labeled oxLDL; RT-qPCR, reverse
transcription-quantitative polymerase chain reaction; LOX-1,
lectin-like oxidized low-density lipoprotein receptor 1; SRA, serum
resistance associated protein; NONO, non-POU domain-containing
octamer-binding; SFPQ, splicing factor proline and glutamine rich;
IgG, immunoglobulin G. |
Discussion
Inflammation and lipid uptake in macrophages by
oxLDL play pivotal roles in the formation of atherosclerotic
plaques during the development of atherosclerosis. A deeper
understanding of the mechanism involved in the oxLDL-induced
secretion of proinflammatory factors and uptake of lipids by
macrophages may yield therapeutic targets for atherosclerosis. In
this study, we explore the possibility of lncRNAs participating in
oxLDL-induced secretion of proinflammatory factors and lipid uptake
by macrophages. Because NEAT1_2 may contribute to the formation of
subnuclear structures called paraspeckles (16), we mainly investigate the role of
paraspeckles in inflammation and lipid uptake. By transfecting
macrophages with siRNAs against NEAT1, NEAT1_2, NONO or SFPQ, which
would stop paraspeckle formation (17), we found that paraspeckles have
different functions in inflammation and lipid uptake: Paraspeckles
promote TNF-α secretion indirectly and inhibit CD36 expression by
directly binding CD36 mRNA. Moreover, we also found that the
constitutive paraspeckle protein NONO may perform its function
independently of paraspeckles (Fig.
3E). But in our present study, the detailed mechanism by which
paraspeckles regulate p65 phosphorylation or paraspeckles pesist
CD36 mRNA is unknown. The role of paraspekcle in other biological
activities of macrophages such as migration, chemotaxis and
apoptosis is also needed to be explored. Collectively, these
results illustrate the complexity of paraspeckles: Paraspeckles may
affect biological activities by stabilizing target mRNAs or
proteins, and these stabilized proteins may function as
transcription factors to activate the transcription of other genes
(15). In our future research, we
aim to explore the detailed mechanism by which paraspeckles
regulate p65 phosphorylation. Additionally, we want to understand
whether paraspeckles influence TNF-α transcription by directly
changing the activity of the promoter.
In conclusion, we first explored the function of
NEAT1- and NEAT1_2-mediated paraspeckle formation in oxLDL-induced
secretion of proinflammatory factors and lipid uptake by
macrophages. Paraspeckles promote TNF-α secretion partially by
regulating p65 phosphorylation and suppressed lipid uptake
partially by stabilizing CD36 mRNA, which decreases CD36
expression.
References
|
1
|
Ross R: The pathogenesis of
atherosclerosis: A perspective for the 1990s. Nature. 362:801–809.
1993. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hansson GK and Hermansson A: The immune
system in atherosclerosis. Nat Immunol. 12:204–212. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ross R: Atherosclerosis-an inflammatory
disease. N Engl J Med. 340:115–126. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Finn AV, Nakano M, Narula J, Kolodgie FD
and Virmani R: Concept of vulnerable/unstable plaque. Arterioscler
Thromb Vasc Biol. 30:1282–1292. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Didangelos A, Simper D, Monaco C and Mayr
M: Proteomics of acute coronary syndromes. Curr Atheroscler Rep.
11:188–195. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Libby P, Ridker PM and Hansson GK:
Progress and challenges in translating the biology of
atherosclerosis. Nature. 473:317–325. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Moore KJ and Tabas I: Macrophages in the
pathogenesis of atherosclerosis. Cell. 145:341–355. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang P, Wu T, Zhou H, Jin Q, He G, Yu H,
Xuan L, Wang X, Tian L, Sun Y, et al: Long noncoding RNA NEAT1
promotes laryngeal squamous cell cancer through regulating
miR-107/CDK6 pathway. J Exp Clin Cancer Res. 35:222016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cooper DR, Carter G, Li P, Patel R, Watson
JE and Patel NA: Long non-coding RNA NEAT1 associates with SRp40 to
temporally regulate PPARγ2 splicing during adipogenesis in 3T3-L1
Cells. Genes (Basel). 5:1050–1063. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ke H, Zhao L, Feng X, Xu H, Zou L, Yang Q,
Su X, Peng L and Jiao B: NEAT1 is required for survival of breast
cancer cells through FUS and miR-548. Gene Regul Syst Bio. 10 Suppl
1:S11–S17. 2016.
|
|
11
|
Mang Y, Li L, Ran J, Zhang S, Liu J, Li L,
Chen Y, Liu J, Gao Y and Ren G: Long noncoding RNA NEAT1 promotes
cell proliferation and invasion by regulating hnRNP A2 expression
in hepatocellular carcinoma cells. Onco Targets Ther. 10:1003–1016.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gao C, Zhang J, Wang Q and Ren C:
Overexpression of lncRNA NEAT1 mitigates multidrug resistance by
inhibiting ABCG2 in leukemia. Oncol Lett. 12:1051–1057.
2016.PubMed/NCBI
|
|
13
|
Chai Y, Liu J, Zhang Z and Liu L:
HuR-regulated lncRNA NEAT1 stability in tumorigenesis and
progression of ovarian cancer. Cancer Med. 5:1588–1598. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Naganuma T, Nakagawa S, Tanigawa A, Sasaki
YF, Goshima N and Hirose T: Alternative 3′-end processing of long
noncoding RNA initiates construction of nuclear paraspeckles. EMBO
J. 31:4020–4034. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Imamura K, Imamachi N, Akizuki G, Kumakura
M, Kawaguchi A, Nagata K, Kato A, Kawaguchi Y, Sato H, Yoneda M, et
al: Long noncoding RNA NEAT1-dependent SFPQ relocation from
promoter region to paraspeckle mediates IL8 expression upon immune
stimuli. Mol Cell. 53:393–406. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fox AH, Lam YW, Leung AK, Lyon CE,
Andersen J, Mann M and Lamond AI: Paraspeckles: A novel nuclear
domain. Curr Biol. 12:13–25. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Anantharaman A, Jadaliha M, Tripathi V,
Nakagawa S, Hirose T, Jantsch MF, Prasanth SG and Prasanth KV:
Paraspeckles modulate the intranuclear distribution of
paraspeckle-associated Ctn RNA. Sci Rep. 6:340432016. View Article : Google Scholar : PubMed/NCBI
|