Introduction
Glioblastoma multiforme (GBM), also termed
glioblastoma, is a type of brain tumor derived from star-shaped
glial cells termed astrocytes, which support and protect neural
tissues in brain and spinal cord (1–3). It
is well-acknowledged that GBM is one of the most lethal and
difficult to treat types of human cancer due to its histological
heterogeneity, aggressive invasion and poor response to treatment
(4). Despite advances in research
into the molecular mechanisms underlying GBM occurrence and
development, treatment and prognosis remain poor and require
further investigation.
Previous studies have demonstrated that the
induction of epithelial to mesenchymal transition (EMT) is of
importance for carcinogenesis and cancer progression (5,6). EMT
is an important process at the cellular level, wherein epithelial
cells lose their polarity and are converted to more motile and
invasive mesenchymal phenotypes, thereby resulting in tumor
progression (7). Evidence has
suggested that the expression of microRNAs (miRNAs/miRs) may be
associated with EMT via the regulation of certain target genes
(8,9). miRNAs, a class of small non-coding
RNAs (20–22 nt in length) may bind to the 3′-untranslated region
(3′-UTR) of their target genes, thereby regulating gene expression.
Previous studies have demonstrated that miRNAs are involved in the
regulation of the expression of genes associated with development,
differentiation, proliferation and apoptosis (10,11).
Notably, miR-96, miR-182 and miR-183 are known to be associated
with EMT (12). In particular, it
has been reported decreased expression of miR-96-5p may be
associated with poor clinical outcomes for patients with colorectal
cancer (13), which provides a
basis for further investigation.
Astrocyte elevated gene-1 (AEG-1), also known termed
metadherin, was first identified to be a human immunodeficiency
virus-1- and tumor necrosis factor-α-inducible late response gene
in human fetal astrocytes (14).
Recent findings have suggested that AEG-1 may serve a dominant role
in the development and progression of multiple types of cancer,
including glioma (15), pancreatic
ductal adenocarcinoma (16),
cervical carcinoma (17),
colorectal cancer (18) and
hepatocellular carcinoma (19). In
addition, AEG-1 activates various signaling factors involved in
mediating EMT, including sonic hedgehog protein (20), transforming growth factor-β
(21), neurogenic locus notch
homolog protein 1 (22) and
protein Wnt (23). For GBM,
according to a previous bioinformatics study, AEG-1 is an
underlying target gene for miR-96 (Feng et al, unpublished
data). However, the functional role of miR-96 and its mechanism in
GBM remain poorly understood. Therefore, the identification of
novel miR-96 targets may provide novel insights into the molecular
mechanism underlying the miR-96-induced suppression of tumorigenic
properties in cancer cells.
The present study investigated the functional role
and mechanism of miR-96 in the migration and invasion,
proliferation, apoptosis and cell cycle progression of GBM. In
vitro approaches were used to explore whether AEG-1 may be a
direct target gene of miR-96, and whether miR-96 serves a role in
EMT by regulating AEG-1.
Materials and methods
Cell culture
The human glioma cell line U251 was purchased from
the American Type Culture Collection (Manassas, VA, USA). Cells
were cultured in Dulbecco's modified Eagle's medium (DMEM; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10%
heat-inactivated fetal bovine serum (FBS; Thermo Fisher Scientific,
Inc.), penicillin (100 U/ml) and streptomycin (100 mg/ml), in a
humidified atmosphere containing 5% (v/v) CO2 at 37°C,
for 18 h prior to transfection.
Transfection
The human glioma cells were seeded in different
culture plates (6, 24, 48 or 96-well) and cultured for 18 h prior
to transfection. Transfection was performed in U251 cells using
Lipofectamine RNAiMAX (Invitrogen; Thermo Fisher Scientific, Inc.),
according to the manufacturer's protocol. RNA with no homology to
any human genomic sequence was regarded as negative control
(miR-NC). The miRNA mimics and small interfering (si)RNA sequences
were designed and synthesized by View Solid Biotech Co., Ltd.
(Beijing, China). Three siRNAs (si-AEG1-744, si-AEG1-1432 and
si-AEG1-1883) were used in the present study. All forward and
reverse sequences are listed in Table
I.
 | Table I.Oligonucleotide sequences. |
Table I.
Oligonucleotide sequences.
| Name | Orientation | Sequence
(5′-3′) |
|---|
| A, Primers for
qPCR |
|
|
|---|
|
|---|
| AEG1-U1 | Forward |
CTAGTCTAGAGGCAGTATGTTTACATGTCA |
|
| Reverse |
CCGGAATTCGAATGGGGAGATACTAGGCTG |
| Mut-AEG1-U1 | Forward |
TTGTTTTTATACAATCACGGTTTTGGTCTGTGCTCAACAATAT |
|
| Reverse |
AAACCGTGATTGTATAAAAACAATCCCTATCAACTTCTCCTTT |
| GAPDH | Forward |
GAAGGTGAAGGTCGGAGTC |
|
| Reverse |
GAAGATGGTGATGGGATTTC |
|
| B, Primers for
mimic |
|
|
|
| miR-96 mimic | Forward |
GGCAGTATGTTTACATGTCA |
|
| Reverse |
CAGCCTAGTATCTCCCCATT |
| miR-NC mimic | Forward |
UCCUCCGAACGUGUCACGUTT |
|
| Reverse |
ACGUGACACGUUCGGAGAATT |
|
| C, Primers for
siRNA |
|
|
|
| si-AEG1-744 | Forward |
GCCAUCUGUAAUCUUAUCATT |
|
| Reverse |
ACGUGACACGUUCGGAGAATT |
| si-AEG1-1432 | Forward |
GCAACUUACAACCGCAUCATT |
|
| Reverse |
UGAUGCGGUUGUAAGUUGCTT |
| si-AEG1-1883 | Forward |
GCAAAGCAGCCACCAGAGATT |
|
| Reverse |
UCUCUGGUGGCUGCUUUGCTT |
| si-NC | Forward |
CGAAGGGAACACGGAUAACCU |
|
| Reverse |
CAGUACUUUUGUGUAGUACAA |
|
| D, Primers for
plasmid construction |
|
|
|
| AEG1-U1 | – |
CTAGTCTAGAGGCAGTATGTTTACATGTCACCGGAATTCGAATGGG |
|
|
| GAGATACTAGGCTG |
| Mut-AEG1-U1 | – |
AAACCGTGATTGTATAAAAACAATCCCTATCAACTTCTCCTTTTTGT |
|
|
|
TTTTATACAATCACGGTTTTGGTCTGTGCTCAACAATAT |
Cell proliferation assays
The proliferative abilities of transfected cells
were assessed using a Cell Counting Kit-8 (CCK-8) assay (Dojindo
Molecular Technologies, Inc., Kumamoto, Japan), following the
manufacturer's protocol. U251 cells were seeded at a density of
3,000 cells/well in 96-well culture plates and cultured at 37°C for
18 h prior to transfection. Subsequently, U251 cells were
transfected with miR-96 mimics and the corresponding miR-NC, in
addition to siRNA targeting AEG-1 (si-AEG1) and the corresponding
negative control siRNA (si-NC). Lipofectamine RNAiMAX (Invitrogen;
Thermo Fisher Scientific, Inc.) was added at a density of 0.3
µM/well and then miR-96 mimic or si-AEG1 was added (3 pM/well). The
culture medium was replaced with DMEM containing 10% CCK-8 solution
48 h post-transfection. Each group was repeated three times
independently. At 2–6 days post-transfection, the optical density
was measured at a wavelength of 450 nm using an ELISA reader
(PerkinElmer, Inc., Waltham, MA, USA). Based on the calculated
number of viable cell, the growth curve was produced.
Cell migration and invasion
assays
U251 cells were seeded in 24-well culture plates at
a density of 1×105 cells/well and then cultured at 37°C
for 18 h prior to transfection. Lipofectamine RNAiMAX was added at
a density of 1.5 µl/well and either miR-96 mimic or si-AEG1 was
then added (15 pM/well). Cells were harvested 48 h
post-transfection. Following the manufacturer's protocol,
2×104 cells with 100 µl serum-free DMEM (Thermo Fisher
Scientific, Inc.) were seeded into the upper chamber of the
Transwell plates (Costar; Corning Incorporated, Corning, NY, USA)
for the migration assays, while cells with 100 µl serum-free DMEM
were plated into the upper chamber of an insert coated with
Matrigel (BD Biosciences, Franklin Lakes, NJ, USA) for the invasion
assays. The lower chambers were filled with 600 µl DMEM containing
10% FBS. Following 48 h of incubation, the cells remaining on the
upper membrane were removed with cotton swabs, whereas those that
had migrated or invaded through the membrane were fixed in 4%
polyformaldehyde and stained with 0.1% crystal violet for 20 min at
4°C. The number of cells was calculated to photograph five random
fields/filters using the fluorescence inversion microscope system
(Nikon Corporation, Tokyo, Japan) at a magnification of ×200. All
experiments were performed at least three times independently.
Cell cycle and apoptosis assays
U251 cells were seeded in six-well culture plates at
a density of 5×105 cells/well and cultured for 18 h
prior to transfection. Subsequently, U251 cells were transfected
with miR-96 mimics and the corresponding miR-NC, in addition to
si-AEG1 and si-NC. Cells were harvested 48 h post-transfection.
Lipofectamine RNAiMAX was added at a density of 7.5 µl/well, and
either miR-96 mimic or si-AEG1 was then added (75 pM/well). For the
cell cycle assay, cells were stained with propidium iodide (PI)
using a Cell Cycle kit (BD Biosciences), according to the
manufacturer's protocol. The cell cycle distribution was analyzed
using flow cytometry (BD Biosciences). For the cellular apoptosis
assay, cells were double-stained with Annexin V and PI using Roche
Annexin V/PI kits (Roche Diagnostics, Basel, Switzerland),
according to the manufacturer's protocol. The apoptotic cells were
detected using a flow cytometer (BD FACSCalibur; BD Biosciences)
and analyzed using BD CellQuest Pro software (version 5.1; BD
Biosciences). All experiments were performed three times,
independently.
Plasmid construction
A fragment of the AEG-1 3′-UTR (AEG1-U1) and a
mutated 3′-UTR of AEG-1 (AEG1-U1-Mut) that contained the putative
miR-96 binding sites were prepared for constructing the reporter
plasmids. In addition, DNA fragments were cloned downstream of the
luciferase gene in the pGL3-REPORT luciferase vector (Promega
Corporation, Madison, WI, USA). Primers used for the constructions
are listed in Table I. All the
constructions were confirmed via sequencing (BGI, Shenzhen,
China).
Luciferase assays
For the luciferase assays, cells were seeded into
48-well plates at a density of 5×104 cells/well for 24 h
and co-transfected with the experimental group (100 ng AEG1-U1, 100
ng hsa-mir-96-5p mimics and 5 ng Renilla) or the control
group (100 ng AEG1-U1, 100 ng hsa-mir-96-5p mimics empty vectors
and 5 ng Renilla). Transfection was performed using
Lipofectamine 2000® (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol. Cells
were harvested at 48 h and lysed using lysis buffer (Promega
Corporation). The luciferase reporter gene assay was implemented
using a dual-luciferase reporter assay system (Promega
Corporation), according to the manufacturer's instructions. Firefly
luciferase activity was normalized to Renilla luciferase
activity for each transfected well. All experiments were performed
at least three times.
Western blot analysis
The human glioma U251 cells were lysed in M-PER
Mammalian Protein Extraction Reagent (Thermo Fisher Scientific,
Inc.) in the presence of protease inhibitors at 4°C for 1 h. A
total of 20 µg of protein was loaded per lane. The protein lysates
were separated by electrophoresing on 10% SDS-PAGE gels, and the
separated proteins were transferred onto polyvinylidene fluoride
(PVDF) membranes (EMD Millipore, Billerica, MA, USA). Prior to
incubation, the PVDF membranes were blocked with 5% non-fat dried
milk at room temperature for 1 h. Subsequently, the PVDF membranes
were washed three times with TBS-Tween 20 (25 mM Tris-HCl, pH 8.0,
0.2 M NaCl, 0.1% Tween 20) following incubation with the following
primary antibodies at 4°C overnight: AEG-1 (cat. no. ab32081;
1:500; Abcam, Cambridge, UK) tubulin (cat. no. ab6046; 1:5,000;
Abcam) β-actin (cat. no. ab8229; 1:1,000; Abcam). Finally, the PVDF
membranes were incubated with corresponding secondary horseradish
peroxidase-conjugated goat-anti-mouse IgG (cat. no. 115-035-003;
1:5,000; Jackson ImmunoResearch Laboratories, Inc., West Grove, PA,
USA) and goat-anti-rabbit IgG (cat. no. 111-035-003; 1:10,000;
Jackson ImmunoResearch Laboratories, Inc.) at room temperature for
2 h. Immunoreactivity was determined using the Pierce enhanced
chemiluminescence western blotting substrate (Thermo Fisher
Scientific, Inc.), and protein levels were determined using a
bicinchoninic acid assay (Pierce; Thermo Fisher Scientific, Inc.)
and subsequently normalized to either β-actin or tubulin. Gel-Pro
analyzer version 5.1 (Beijing Sage Creation Science Co., Ltd.,
Beijing, China) was then used for densitometric analysis.
Total RNA extraction
Total RNA was isolated from glioma cells using
TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.), according to
the manufacturer's protocol. RNA was reverse-transcribed into cDNA
using a reverse transcription (RT) system (Takara Biotechnology,
Inc., Dalian, China) following the manufacturer's instructions. A
total of 10 µl of the reaction mix [1 µg of RNA template, 1 µl of
oligo (dT) adaptor primer (commonly 5′-T(18)VN-3′; 50 pM), 1 µl of
deoxyribonucelotide triphosphates (10 mM) and a remainder of
RNA-free H2O] was maintained for 5 min at 65°C.
Following this, the reaction mix was placed on ice and a further
reaction mixture was immediately added [4 µl of 2X reverse
transcription buffer, 0.5 µl of reverse transcriptase (200 U/µl)
and 5.5 µl of RNA-free H2O]. The subsequent reaction
conditions were performed as follows: 42°C for 60 min, 75°C for 15
min and then stored at 4°C prior to further experimentation.
Quantitative polymerase chain reaction
(qPCR) analysis
The qPCR was performed using the TransStart Green
qPCR SuperMix kit (Beijing Transgen Biotech Co., Ltd., Beijing,
China) protocol on a Real-Time PCR System (Applied Biosystems;
Thermo Fisher Scientific, Inc.). All primers for AEG-1 and GAPDH
are presented in Table I. The
reaction volume was 20 µl and the mixture contained 10 µl qPCR kit
Premix Ex Taq, 1 µl cDNA, 0.4 µl (10 µM) mRNA forward primer and
mRNA reverse primer or 0.4 µl (10 µM) GAPDH forward primer and
GAPDH reverse primer, and 8.2 µl ddH2O. The reaction
conditions were designed as follows: 94°C for 2 min, followed by 40
cycles of 94°C for 5 sec, 60°C for 15 sec and 72°C for 31 sec. The
values were normalized to the internal control products of GAPDH
and total protein was quantified using the 2−ΔΔCq method
(24). All reactions were
performed in triplicate.
Statistical analysis
All statistical calculations and analyses were
performed using Origin 9.0 software (OriginLab, Northampton, MA,
USA). Each experiment was repeated at least three times. The values
are expressed as the mean ± standard deviation. Either the
Student's t-test or one-way analysis of variance followed by the
Newman-Keuls method were performed to analyze the difference
between two groups and multiple groups, respectively. P<0.05 was
considered to indicate a statistically significant difference.
Results
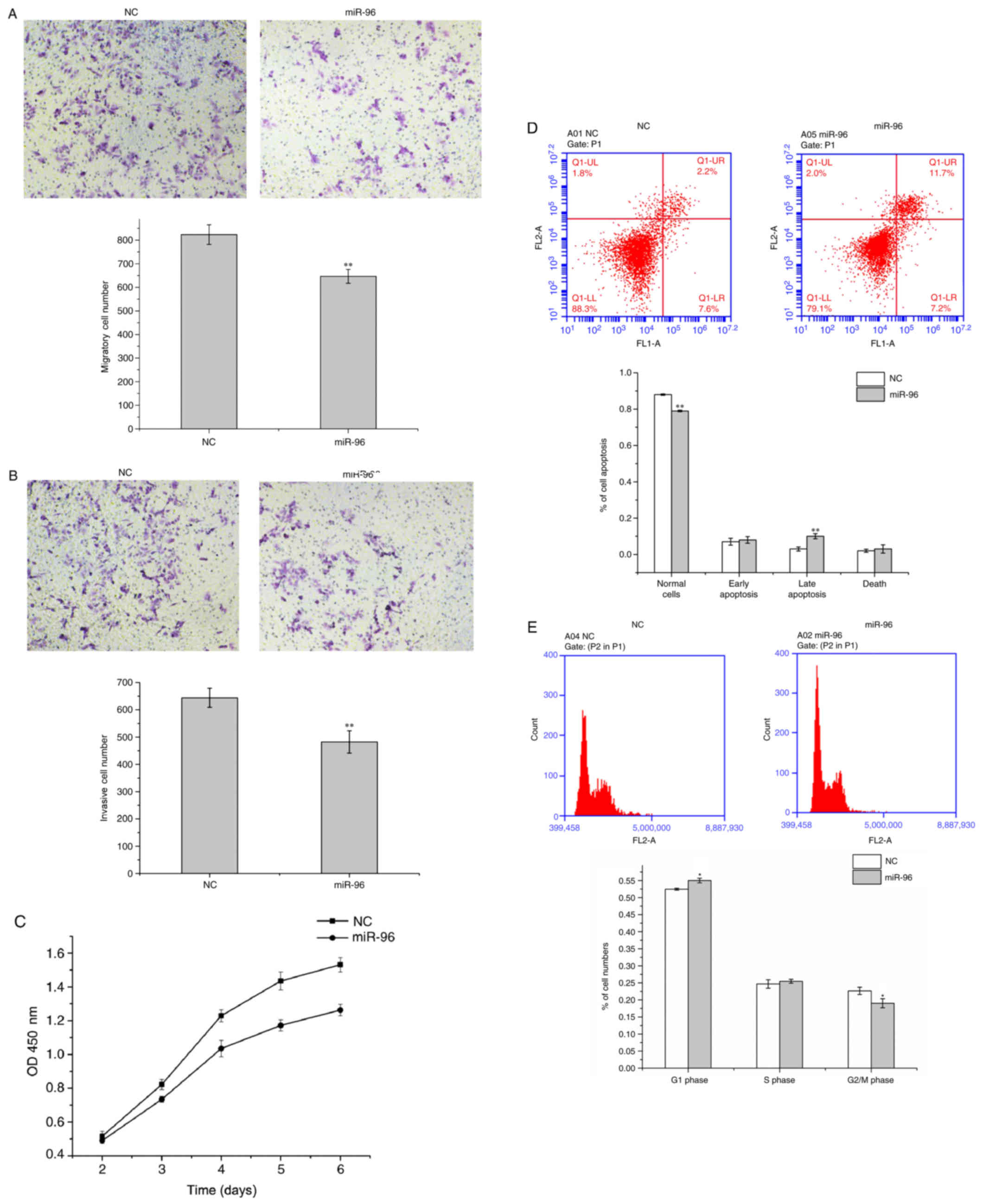
miR-96 inhibits migration, invasion,
proliferation and cell cycle progression in U251 cells, and
promotes their apoptosis
In order to evaluate the effect of miR-96 on the
migration, invasion, proliferation and cell cycle of U251 cells,
miR-96 mimics or miR-NC were transfected into U251 cells in
vitro. As presented in Fig. 1A and
B, it was observed that miR-96 notably decreased the migratory
and invasive abilities of U251 cells in vitro, demonstrated
by the Transwell assay (P<0.01). In addition, the CCK-8 assay
demonstrated that the proliferative ability of U251 cells
transfected with miR-96 mimic was decreased compared with U251
cells transfected with miR-NC. The results demonstrated that,
post-transfection, miR-96 markedly decreased the proliferation of
U251 cells, particularly at 4–6 days post-transfection (Fig. 1C). In addition, annexin V/PI
staining indicated that miR-96 was able to significantly promote
apoptosis in U251 cells (Fig. 1D).
It may be concluded from Fig. 1E
(P<0.05) that miR-96 markedly suppressed the cycle progression
of U251 cells and led to cell cycle arrest at G1 phase, as
demonstrated by the flow cytometry analysis.
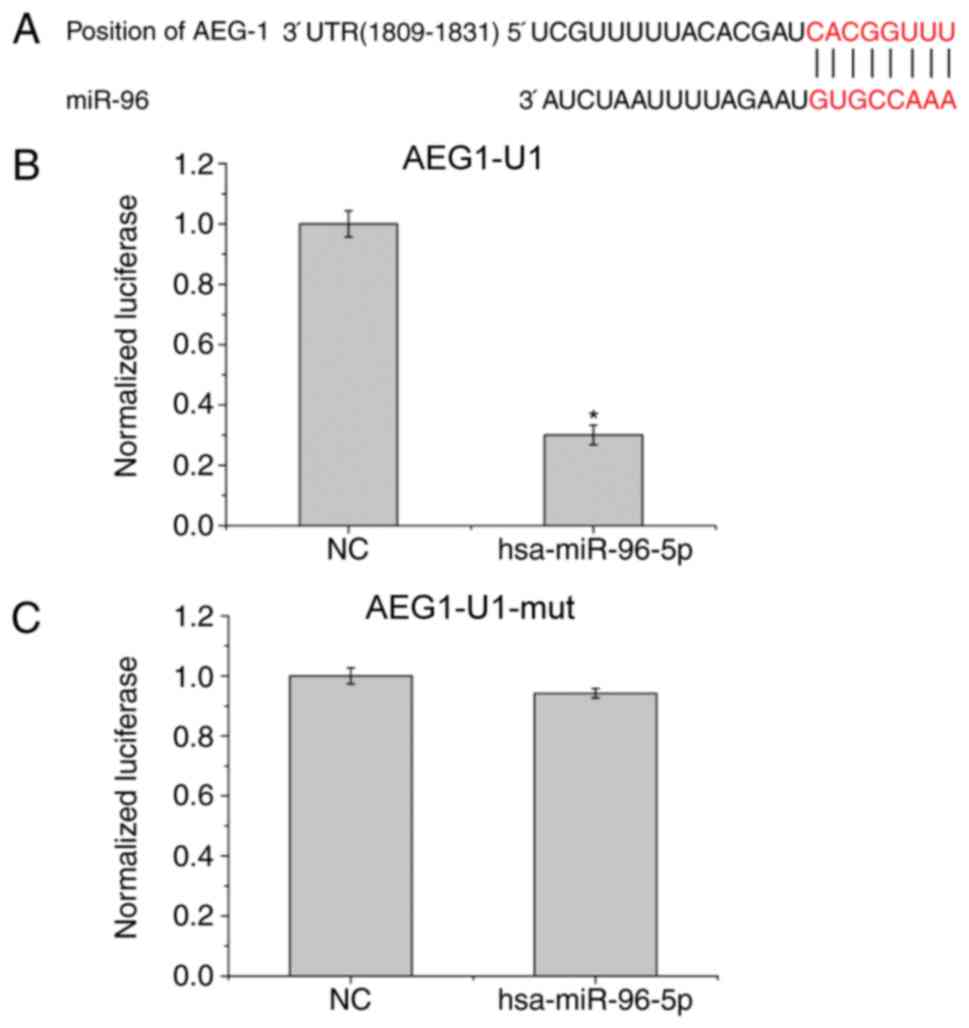
AEG-1 acts as a target for miR-96 in
GBM cells
Putative target genes of miR-96 in human cells were
screened using the tools miRNA.org,
TargetScan version 6.2 and RNA22, aiming to detect the molecular
inhibition mechanisms of miR-96 in the metastatic progression of
GBM cells (Feng et al, unpublished data). AEG-1 was
identified to be a target gene of miR-96; AEG-1 expression in GBM
was increased compared with the other predicted candidates. In
addition, AEG-1 expression was observed to increase as the tumor
grade of GBM increases. As presented in Fig. 2A, there exists one predicted
binding site in miR-96 which corresponds to the 3′-UTR of AEG-1,
which was inserted into the luciferase reporter gene plasmid
pGL3-3′-UTR following mutation to AEG1-U1. Subsequently, AEG1-U1
and AEG1-U1-mut were respectively co-transfected with the reporter
gene plasmid hsa-miR-96-5p into U251 cells. The luciferase reporter
assay demonstrated that miR-96 significantly decreased the
luciferase activity of the co-transfected U251 cells (P<0.05)
compared with NC (Fig. 2B),
indicating that the 1809–1831 gene sequence of AEG-1 is the target
of hsa-mir-96-5p. The luciferase activity of has-mir-96-5p
co-transfected with AEG1-U1-mut was almost stable (Fig. 2C).
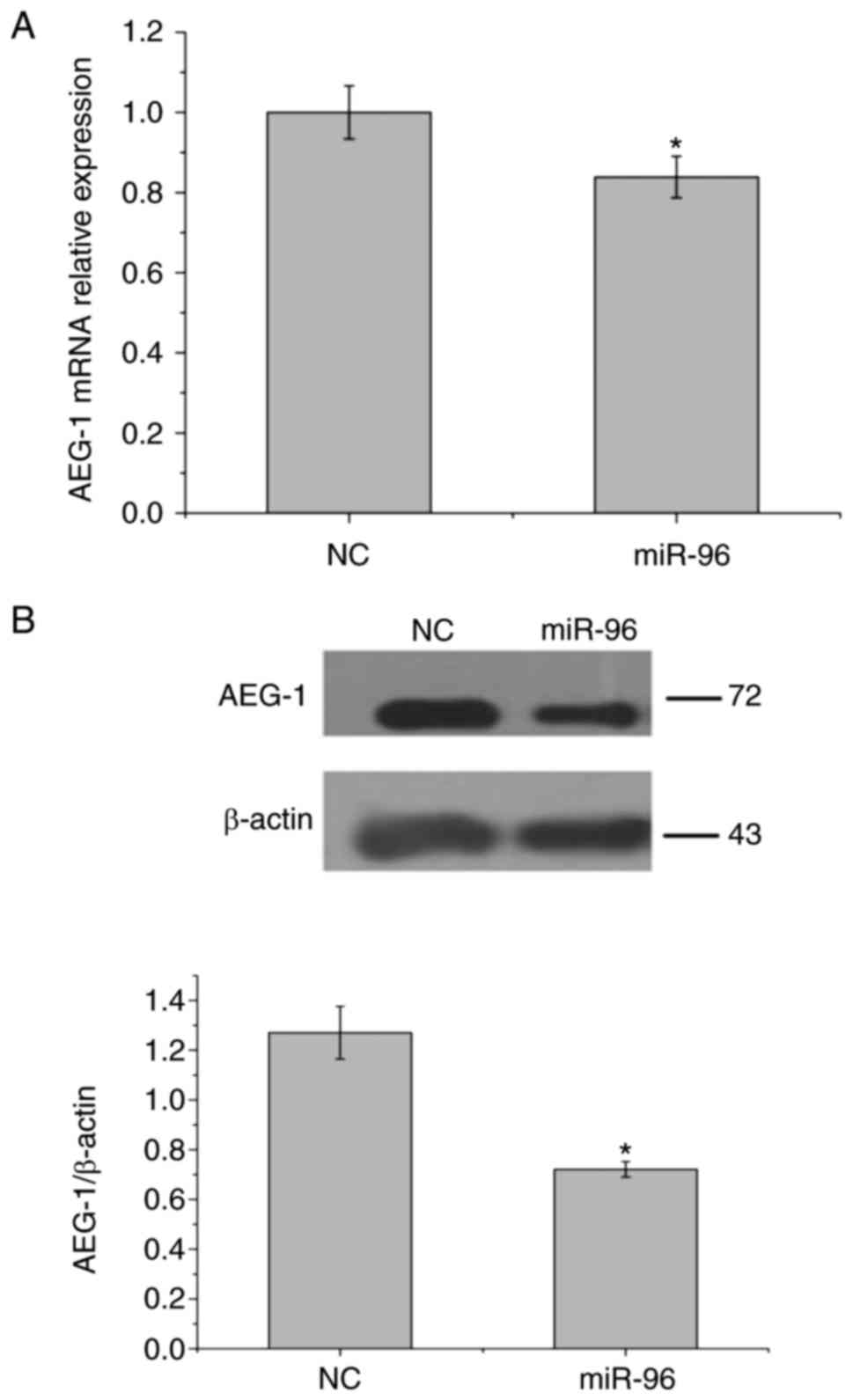
miR-96 downregulates the expression of
AEG-1 to inhibit EMT in GBM
In order to analyze the association between miR-96
and AEG-1, U251 cells were transfected with miR-96 mimics or
miR-NC, and analyzed using western blotting and RT-qPCR analysis.
Following transfection, the amount of AEG-1 mRNA was significantly
decreased compared with control mRNA as demonstrated by RT-qPCR
analysis (Fig. 3A). Western blot
analysis revealed that the expression of AEG-1 protein was
significantly downregulated in U251 cells transfected with miR-96,
compared with miR-NC (Fig. 3B;
P<0.05). These results suggested that miR-96 may negatively
regulate the expression of AEG-1 at the mRNA and protein levels.
Notably, previous research has demonstrated that miR-96 can enhance
EMT in tumor cells (25).
Therefore, U251 cells were transfected with si-AEG1 or si-NC to
detect the expression levels of E-cadherin and vimentin, in order
to further identify whether AEG-1 serves a role in EMT. As depicted
in Fig. 3C, when compared with
corresponding si-NC, the expression of vimentin decreased, while
that of E-cadherin markedly increased, in U251 cells transfected
with si-AEG1. The results suggested that AEG-1 may inhibit the EMT
process. In conclusion, miR-96 may downregulate the expression of
AEG-1 to inhibit EMT in GBM.
Inhibition of AEG-1 exerted a similar
impact to that of miR-96 overexpression
Aiming to investigate the impact of AEG-1 on U251
cell migration, invasion, proliferation, apoptosis and cell cycle
progression, we transfected si-AEG1 or si-NC into U251 cells,
respectively. As presented in Fig.
4A, RT-qPCR analysis and western blotting were performed to
confirm that si-AEG1 was able to significantly attenuate the mRNA
and protein expression levels of AEG-1 in U251 cells (P<0.01).
The results suggested that the siRNA efficiency of AEG-1 was most
significant at the 744 and 1883 sites in U251 cells. Therefore,
these two siRNAs were combined in the following experiments to
obtain the best interfering effect (Fig. 4A). In addition, the inhibition of
AEG-1 exerted a similar impact to that of miR-96 overexpression in
the GBM cells, markedly repressing the cell migration, invasion,
proliferation and cell cycle progression of U251 cells and
promoting their apoptosis in vitro (Fig. 4B-F).
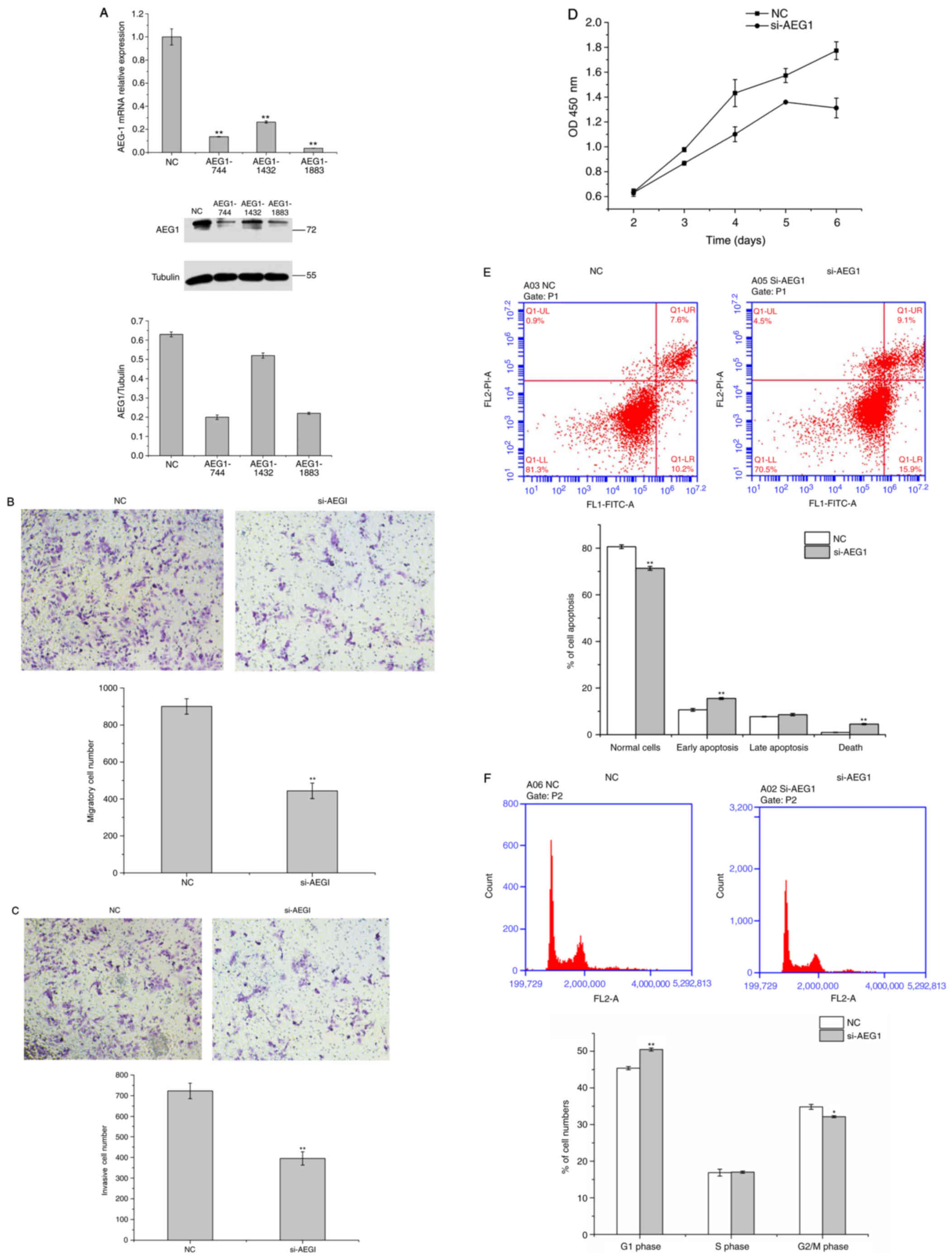 | Figure 4.Inhibition of AEG-1 exerts a similar
effect to that of miR-96 overexpression. (A) Reverse
transcription-quantitative polymerase chain reaction and western
blot analyses confirmed that si-AEG1 was able to significantly
inhibit AEG-1 expression at the mRNA and protein levels in U251
cells, and that the siRNA efficiency of AEG-1 was most marked at
the 744 and 1883 sites in U251 cells. AEG-1 repressed the (B) cell
migration, (C) invasion, (D) proliferation, (E) apoptosis and (F)
cell cycle progression of U251 cells in vitro. *P<0.05;
**P<0.01 vs. NC group. siRNA, small interfering RNA; NC,
negative control; AEG-1, astrocyte elevated gene-1; miR, microRNA;
OD, optical density; FITC, fluorescein isothiocyanate; PI,
propidium iodide. |
Discussion
miRNAs serve important roles in regulating gene
expression, inhibiting the translation of target genes by
recognizing binding sites in the mRNA 3′-UTR. It has previously
been reported that aberrantly-expressed miRNAs were associated with
cell cycle distribution, cellular migration and invasion, cellular
apoptosis and other processes in a variety of types of tumor
(10,11). miRNA may be regarded as a novel
prognostic marker in tumors. It was reported that miR-19a promoted
cell proliferation and invasion by targeting RhoB in human glioma
cells (26). An additional report
demonstrated that microRNA-198 expression was decreased and
exhibited prognostic value in human glioma (27). Notably, the effect of decreased
expression of miR-96-5p on poor survival in colorectal cancer
patients was investigated (13).
Similarly, it has been reported that increased expression of miR-96
was may be associated with advanced stages of colorectal
adenocarcinoma, and it may predict an increased risk of disease
recurrence and poor overall survival (28). These previous results
notwithstanding, the functional role of miR-96 and its mechanism in
tumorigenesis remain largely unknown. In the present study, the
functions of miR-96 in the metabolism of GBM cells were
analyzed.
In the present study, it was observed that miR-96
expression was associated with various biological processes,
including cell migration, invasion, proliferation and the cell
cycle. Therefore, it was hypothesized that miR-96 may be a type of
miRNA that acts as a tumor suppressor in human malignancies and
serves a regulatory role in the occurrence and developmental
processes of human tumors. In order to investigate the functional
mechanisms of miR-96, its target genes were screened using target
gene prediction software, since the biological functions of miRNAs
depend on their downstream target genes. Out of all predicted
candidates, AEG-1 was highlighted due to the fact that it serves
important diagnostic and prognostic roles in multiple malignant
tumors. Subsequently, a luciferase assay was performed to confirm
that AEG-1 was a direct target gene of miR-96. AEG-1 has been
proven to exert functions in the development of various types of
cancer, including liver cancer (19,29),
breast cancer (30), colorectal
cancer (18,31), lung cancer (32) and ovarian cancer (33). It has been reported that C-C motif
chemokine 3/C-C chemokine receptor type 5-induced EMT may be
regulated by AEG-1 via extracellular signal-regulated kinase 1/2
and RAC-α serine/threonine protein kinase signaling in cardiac
myxoma (34). In addition, a study
analyzed the role of miR-302c-3p in suppressing the invasion and
proliferation of glioma cells by downregulating AEG-1 expression
(15). The present study suggested
that miR-96 is likely to repress the EMT process by down-regulating
the mRNA and protein expression levels of AEG-1. In addition, it
was observed that the inhibition of AEG-1 and overexpression of
miR-96 exerted similar effects on GBM cell migration, invasion,
proliferation and cell cycle progression. The results of the
present study suggested that miR-96 is likely to regulate the
expression of AEG-1 to suppress EMT, leading to an inhibition of
GBM cell migration, invasion, proliferation and cycle progression,
and a promotion of apoptosis.
In conclusion, miR-96 may be a novel
tumor-suppressing miRNA. miR-96 may suppress EMT by downregulating
AEG-1, resulting in an inhibition of the metabolism of GBM cells
in vitro. The results of the presents study provided a novel
insight into the occurrence and development of GBM. In addition,
miR-96 may be a potential therapeutic target for the treatment of
glioma.
Acknowledgements
The authors of the present study would like to
acknowledge Dr Xuying Qin (Department of Neurosurgery, Chinese
People's Liberation Army General Hospital, Beijing, China) for
technical help in the preparation of the present study, and Dr
Deming Shao (Beijing Taipu-Shunkang Institute for Laboratory
Medicine, Beijing, China) for assistance with data collection.
References
|
1
|
Rosenblum MK: The 2007 WHO classification
of nervous system tumors: Newly recognized members of the mixed
glioneuronal group. Brain Pathol. 17:308–313. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Furnari FB, Fenton T, Bachoo RM, Mukasa A,
Stommel JM, Stegh A, Hahn WC, Ligon KL, Louis DN, Brennan C, et al:
Malignant astrocytic glioma: Genetics, biology, and paths to
treatment. Genes Dev. 21:2683–2710. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Perry A, Aldape KD, George DH and Burger
PC: Small cell astrocytoma: An aggressive variant that is
clinicopathologically and genetically distinct from anaplastic
oligodendroglioma. Cancer. 101:2318–2326. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Stummer W, van den Bent MJ and Westphal M:
Cytoreductive surgery of glioblastoma as the key to successful
adjuvant therapies: New arguments in an old discussion. Acta
Neurochir (Wien). 153:1211–1218. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yang J and Weinberg RA:
Epithelial-mesenchymal transition: At the crossroads of development
and tumor metastasis. Dev Cell. 14:818–829. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wu Y and Zhou BP: New insights of
epithelial-mesenchymal transition in cancer metastasis. Acta
Biochim Biophys Sin (Shanghai). 40:643–650. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Thiery JP: Epithelial-mesenchymal
transitions in tumour progression. Nat Rev Cancer. 2:442–454. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ozcan S: MiR-30 family and EMT in human
fetal pancreatic islets. Islets. 1:283–285. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liu YN, Yin JJ, Abou-Kheir W, Hynes PG,
Casey OM, Fang L, Yi M, Stephens RM, Seng V, Sheppard-Tillman H, et
al: MiR-1 and miR-200 inhibit EMT via slug-dependent and
tumorigenesis via slug-independent mechanisms. Oncogene.
32:296–306. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tutar Y: miRNA and cancer; computational
and experimental approaches. Curr Pharm Biotechnol. 15:4292014.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mendell JT and Olson EN: MicroRNAs in
stress signaling and human disease. Cell. 148:1172–1187. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Weeraratne SD, Amani V, Teider N,
Pierre-Francois J, Winter D, Kye MJ, Sengupta S, Archer T, Remke M,
Bai AH, et al: Pleiotropic effects of miR-183~96~182 converge to
regulate cell survival, proliferation and migration in
medulloblastoma. Acta Neuropathol. 123:539–552. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ress AL, Stiegelbauer V, Winter E,
Schwarzenbacher D, Kiesslich T, Lax S, Jahn S, Deutsch A,
Bauernhofer T, Ling H, et al: MiR-96-5p influences cellular growth
and is associated with poor survival in colorectal cancer patients.
Mol Carcinog. 54:1442–1450. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Su ZZ, Kang DC, Chen Y, Pekarskaya O, Chao
W, Volsky DJ and Fisher PB: Identification and cloning of human
astrocyte genes displaying elevated expression after infection with
HIV-1 or exposure to HIV-1 envelope glycoprotein by rapid
subtraction hybridization, RaSH. Oncogene. 21:3592–3602. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang Y, Wei Y, Tong H, Chen L, Fan Y, Ji
Y, Jia W, Liu D and Wang G: MiR-302c-3p suppresses invasion and
proliferation of glioma cells via down-regulating metadherin (MTDH)
expression. Cancer Biol Ther. 16:1308–1315. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Huang Y, Ren GP, Xu C, Dong SF, Wang Y,
Gan Y, Zhu L and Feng TY: Expression of astrocyte elevated gene-1
(AEG-1) as a biomarker for aggressive pancreatic ductal
adenocarcinoma. BMC Cancer. 14:4792014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yu JQ, Zhou Q, Zhu H, Zheng FY and Chen
ZW: Overexpression of astrocyte elevated gene-1 (AEG-1) in cervical
cancer and its correlation with angiogenesis. Asian Pac J Cancer
Prev. 16:2277–2281. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Huang S, Wu B, Li D, Zhou W, Deng G, Zhang
K and Li Y: Knockdown of astrocyte elevated gene-1 inhibits tumor
growth and modifies microRNAs expression profiles in human
colorectal cancer cells. Biochem Biophys Res Commun. 444:338–345.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Srivastava J, Robertson CL, Gredler R,
Siddiq A, Rajasekaran D, Akiel MA, Emdad L, Mas V, Mukhopadhyay ND,
Fisher PB and Sarkar D: Astrocyte elevated gene-1 (AEG-1)
contributes to non-thyroidal illness syndrome (NTIS) associated
with hepatocellular carcinoma (HCC). J Biol Chem. 290:15549–15558.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Valcourt U, Kowanetz M, Niimi H, Heldin CH
and Moustakas A: TGF-beta and the Smad signaling pathway support
transcriptomic reprogramming during epithelial-mesenchymal cell
transition. Mol Biol Cell. 16:1987–2002. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shin SY, Rath O, Zebisch A, Choo SM, Kolch
W and Cho KH: Functional roles of multiple feedback loops in
extracellular signal-regulated kinase and Wnt signaling pathways
that regulate epithelial-mesenchymal transition. Cancer Res.
70:6715–6724. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Timmerman LA, Grego-Bessa J, Raya A,
Bertrán E, Pérez-Pomares JM, Díez J, Aranda S, Palomo S, McCormick
F, Izpisúa-Belmonte JC and de la Pompa JL: Notch promotes
epithelial-mesenchymal transition during cardiac development and
oncogenic transformation. Genes Dev. 18:99–115. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Karhadkar SS, Bova GS, Abdallah N, Dhara
S, Gardner D, Maitra A, Isaacs JT, Berman DM and Beachy PA:
Hedgehog signalling in prostate regeneration, neoplasia and
metastasis. Nature. 431:707–712. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang W, Qian P, Zhang X, Zhang M, Wang H,
Wu M, Kong X, Tan S, Ding K, Perry JK, et al: Autocrine/paracrine
human growth hormone-stimulated MicroRNA 96-182-183 cluster
promotes epithelial-mesenchymal transition and invasion in breast
cancer. J Biol Chem. 290:13812–13829. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen Q, Guo W, Zhang Y, Wu Y and Xiang J:
MiR-19a promotes cell proliferation and invasion by targeting RhoB
in human glioma cells. Neurosci Lett. 628:161–166. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Man HB, Bi WP and Man HH: Decreased
microRNA-198 expression and its prognostic significance in human
glioma. Genet Mol Res. 15:2016. View Article : Google Scholar
|
|
28
|
Rapti SM, Kontos CK, Papadopoulos IN and
Scorilas A: High miR-96 levels in colorectal adenocarcinoma predict
poor prognosis, particularly in patients without distant metastasis
at the time of initial diagnosis. Tumour Biol. 37:11815–11824.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li C, Wu X, Zhang W, Li J, Liu H, Hao M,
Wang J, Zhang H, Yang G, Hao M, et al: AEG-1 promotes metastasis
through downstream AKR1C2 and NF1 in liver cancer. Oncol Res.
22:203–211. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gollavilli PN, Kanugula AK, Koyyada R,
Karnewar S, Neeli PK and Kotamraju S: AMPK inhibits MTDH expression
via GSK3β and SIRT1 activation: Potential role in triple negative
breast cancer cell proliferation. FEBS J. 282:3971–3985. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang B, Shen ZL, Jiang KW, Zhao G, Wang
CY, Yan YC, Yang Y, Zhang JZ, Shen C, Gao ZD, et al: MicroRNA-217
functions as a prognosis predictor and inhibits colorectal cancer
cell proliferation and invasion via an AEG-1 dependent mechanism.
BMC Cancer. 15:4372015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
He W, He S, Wang Z, Shen H, Fang W, Zhang
Y, Qian W, Lin M, Yuan J, Wang J, et al: Astrocyte elevated
gene-1(AEG-1) induces epithelial-mesenchymal transition in lung
cancer through activating Wnt/β-catenin signaling. BMC Cancer.
15:1072015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhou B, Yang J, Shu B, Liu K, Xue L, Su N,
Liu J and Xi T: Overexpression of astrocyte-elevated gene-1 is
associated with ovarian cancer development and progression. Mol Med
Rep. 11:2981–2990. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Shi P, Fang C and Pang X: Astrocyte
elevated gene-1 regulates CCL3/CCR5-induced
epithelial-to-mesenchymal transition via Erk1/2 and Akt signaling
in cardiac myxoma. Oncol Rep. 34:1319–1326. 2015. View Article : Google Scholar : PubMed/NCBI
|