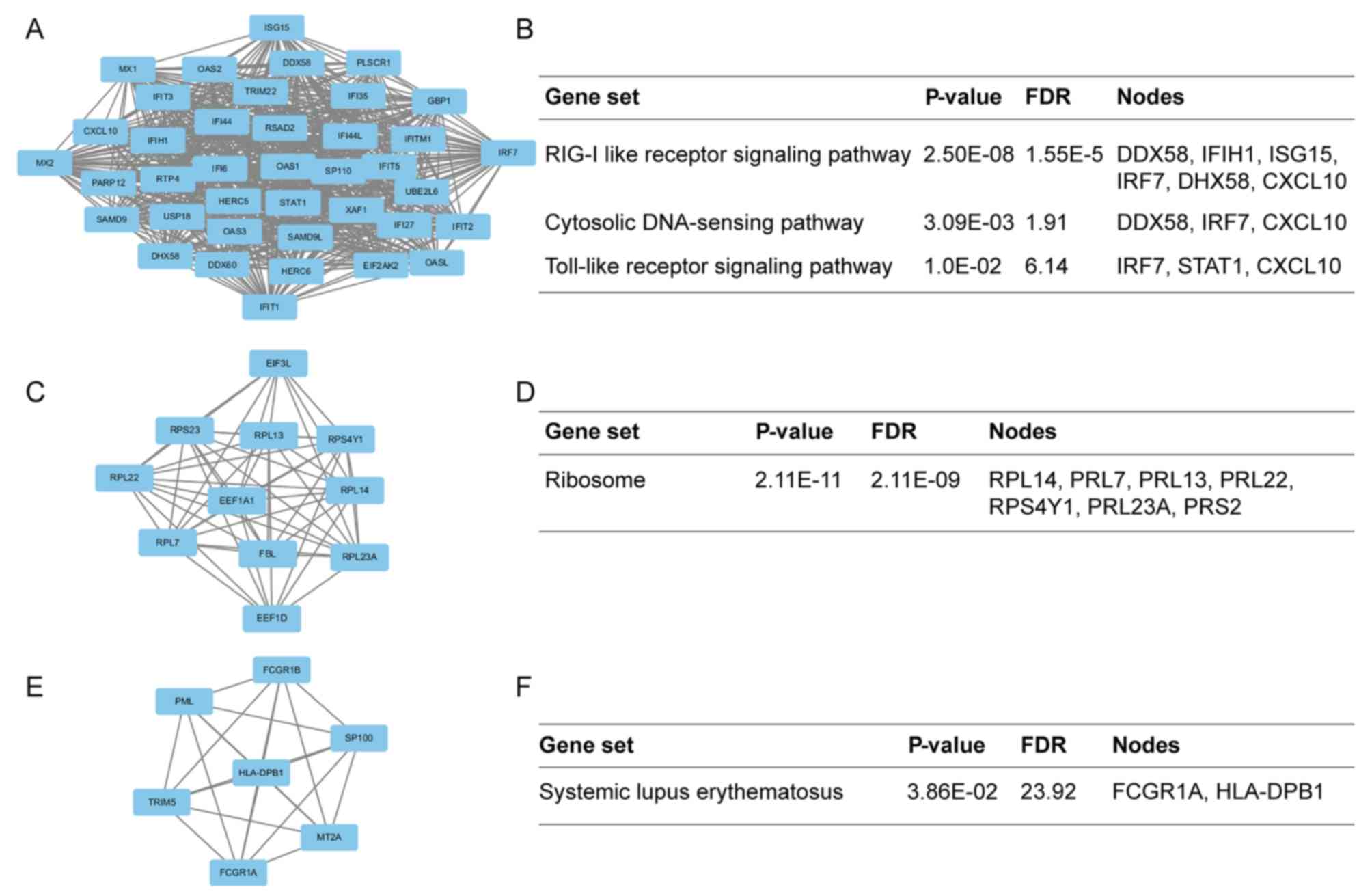
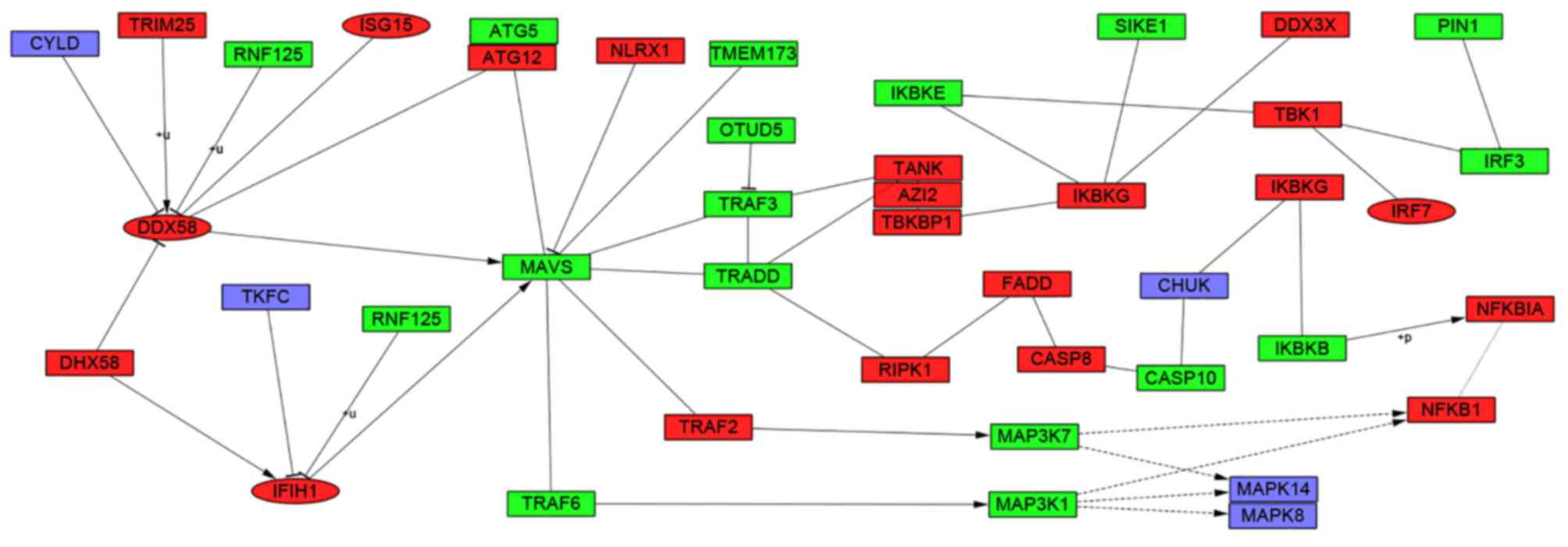
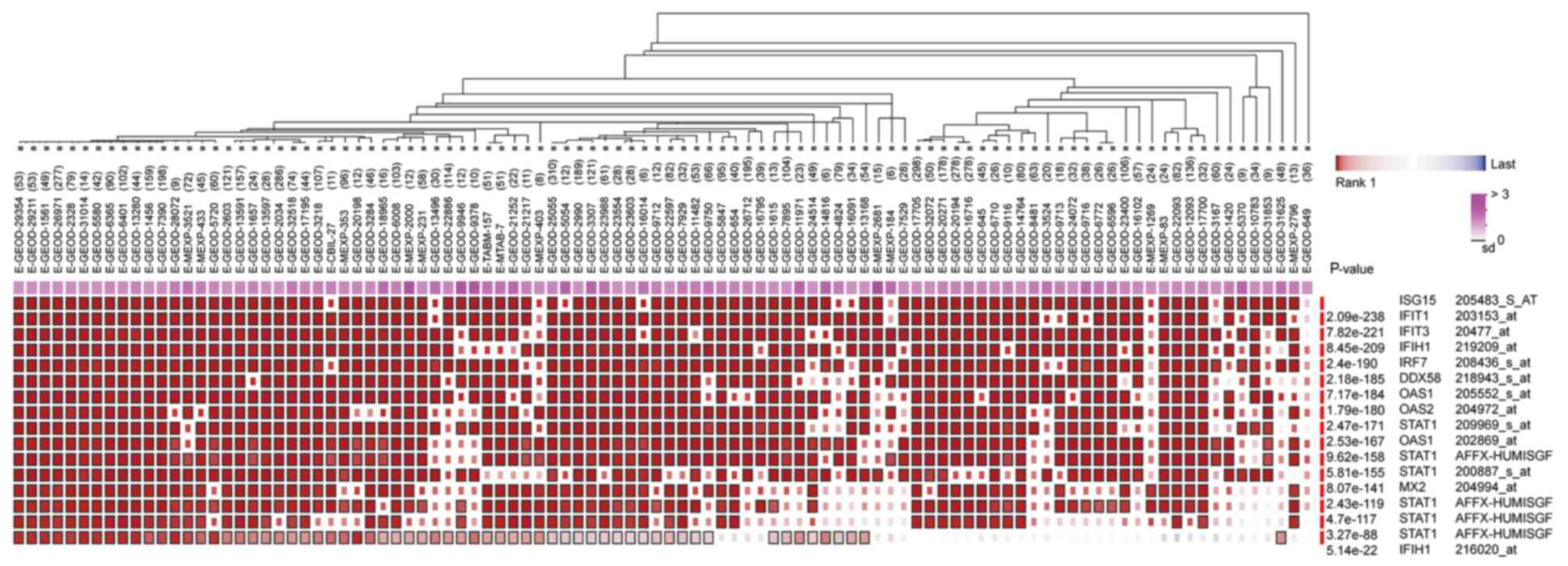
|
1
|
Weidenbusch M, Kulkarni OP and Anders HJ:
The innate immune system in human systemic lupus erythematosus.
Clin Sci (Lond). 131:625–634. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wu CJ, Guo J, Luo HC, Wei CD, Wang CF, Lan
Y and Wei YS: Association of CD40 polymorphisms and haplotype with
risk of systemic lupus erythematosus. Rheumatol Int. 36:45–52.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mok CC, Kwok RC and Yip PS: Effect of
renal disease on the standardized mortality ratio and life
expectancy of patients with systemic lupus erythematosus. Arthritis
Rheum. 65:2154–2160. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yang W and Lau YL: Solving the genetic
puzzle of systemic lupus erythematosus. Pediatr Nephrol.
30:1735–1748. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Dang J, Li J, Xin Q, Shan S, Bian X, Yuan
Q, Liu N, Ma X, Li Y and Liu Q: Gene-gene interaction of ATG5,
ATG7, BLK and BANK1 in systemic lupus erythematosus. Int J Rheum
Dis. 19:1284–1293. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang J, Liu Y, Zhao J, Xu J, Li S and Qin
X: P-glycoprotein gene MDR1 polymorphisms and susceptibility to
systemic lupus erythematosus in Guangxi population: A case-control
study. Rheumatol Int. 37:537–545. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Borrebaeck CA, Sturfelt G and Wingren C:
Recombinant antibody microarray for profiling the serum proteome of
SLE. Methods Mol Biol. 1134:67–78. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhu H, Luo H, Yan M, Zuo X and Li QZ:
Autoantigen microarray for High-throughput autoantibody profiling
in systemic lupus erythematosus. Genomics Proteomics
Bioinformatics. 13:210–218. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ducreux J, Houssiau FA, Vandepapelière P,
Jorgensen C, Lazaro E, Spertini F, Colaone F, Roucairol C, Laborie
M, Croughs T, et al: Interferon α kinoid induces neutralizing
anti-interferon α antibodies that decrease the expression of
interferon-induced and B cell activation associated transcripts:
Analysis of extended follow-up data from the interferon α kinoid
phase I/II study. Rheumatology (Oxford). 55:1901–1905. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Smith S, Fernando T, Wu PW, Seo J, Ní
Gabhann J, Piskareva O, McCarthy E, Howard D, O'Connell P, Conway
R, et al: MicroRNA-302d targets IRF9 to regulate the IFN-induced
gene expression in SLE. J Autoimmun. 79:105–111. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Humrich JY and Riemekasten G: Clinical
trials: The rise of IL-2 therapy-a novel biologic treatment for
SLE. Nat Rev Rheumatol. 12:695–696. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Deng Y and Tsao BP: Genetic susceptibility
to systemic lupus erythematosus in the genomic era. Nat Rev
Rheumatol. 6:683–692. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bentham J, Morris DL, Graham DSC, Pinder
CL, Tombleson P, Behrens TW, Martín J, Fairfax BP, Knight JC, Chen
L, et al: Genetic association analyses implicate aberrant
regulation of innate and adaptive immunity genes in the
pathogenesis of systemic lupus erythematosus. Nat Genet.
47:1457–1464. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Clough E and Barrett T: The gene
expression omnibus database. Methods Mol Biol. 1418:93–110. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Edgar R, Domrachev M and Lash AE: Gene
expression omnibus: NCBI gene expression and hybridization array
data repository. Nucleic Acids Res. 30:207–210. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The gene
ontology consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Huang da W, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Szklarczyk D, Morris JH, Cook H, Kuhn M,
Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al:
The STRING database in 2017: Quality-controlled protein-protein
association networks, made broadly accessible. Nucleic Acids Res.
45:D362–D368. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chin CH, Chen SH, Wu HH, Ho CW, Ko MT and
Lin CY: CytoHubba: Identifying hub objects and sub-networks from
complex interactome. BMC Syst Biol. 8 Suppl 4:S112014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bader GD and Hogue CW: An automated method
for finding molecular complexes in large protein interaction
networks. BMC Bioinformatics. 4:22003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Adler P, Kolde R, Kull M, Tkachenko A,
Peterson H, Reimand J and Vilo J: Mining for coexpression across
hundreds of datasets using novel rank aggregation and visualization
methods. Genome Biol. 10:R1392009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Rustici G, Kolesnikov N, Brandizi M,
Burdett T, Dylag M, Emam I, Farne A, Hastings E, Ison J, Keays M,
et al: ArrayExpress update-trends in database growth and links to
data analysis tools. Nucleic Acids Res. 41(Database Issue):
D987–D990. 2013.PubMed/NCBI
|
|
25
|
Yusuf JH, Kaliyaperumal D, Jayaraman M,
Ramanathan G and Devaraju P: Genetic selection pressure in TLR9
gene may enforce risk for SLE in Indian Tamils. Lupus. 26:307–310.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Armstrong DL, Zidovetzki R,
Alarcón-Riquelme ME, Tsao BP, Criswell LA, Kimberly RP, Harley JB,
Sivils KL, Vyse TJ, Gaffney PM, et al: GWAS identifies novel SLE
susceptibility genes and explains the association of the HLA
region. Genes Immun. 15:347–354. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Fattal I, Shental N, Mevorach D, Anaya JM,
Livneh A, Langevitz P, Zandman-Goddard G, Pauzner R, Lerner M,
Blank M, et al: An antibody profile of systemic lupus erythematosus
detected by antigen microarray. Immunology. 130:337–343. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Rios JJ, Perelygin AA, Long MT, Lear TL,
Zharkikh AA, Brinton MA and Adelson DL: Characterization of the
equine 2′-5′ oligoadenylate synthetase 1 (OAS1) and ribonuclease L
(RNASEL) innate immunity genes. BMC Genomics. 8:3132007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Getts DR, Chastain EM, Terry RL and Miller
SD: Virus infection, antiviral immunity, and autoimmunity. Immunol
Rev. 255:197–209. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Fernández-Trujillo MA, García-Rosado E,
Alonso MC, Castro D, Álvarez MC and Béjar J: Mx1, Mx2 and Mx3
proteins from the gilthead seabream (Sparus aurata) show in vitro
antiviral activity against RNA and DNA viruses. Mol Immunol.
56:630–636. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Relógio A, Schwager C, Richter A, Ansorge
W and Valcárcel J: Optimization of oligonucleotide-based DNA
microarrays. Nucleic Acids Res. 30:e512002. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ye S, Pang H, Gu YY, Shen N, Chen SL, Chen
XG, Hua J, Qian J and Rao ZH: Using GST-tag to capture protein
interaction of an interferon-inducible systemic lupus associated
gene, IFIT1. Zhonghua Yi Xue Za Zhi. 83:770–773. 2003.(In Chinese).
PubMed/NCBI
|
|
33
|
Rönnblom L and Alm GV: An etiopathogenic
role for the type I IFN system in SLE. Trends Immunol. 22:427–431.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Rönnblom L: The importance of the type I
interferon system in autoimmunity. Clin Exp Rheumatol. 34 4 Suppl
98:S21–S24. 2016.
|
|
35
|
Lincez PJ, Shanina I and Horwitz MS:
Reduced expression of the MDA5 gene IFIH1 prevents autoimmune
diabetes. Diabetes. 64:2184–2193. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Robinson T, Kariuki SN, Franek BS, Kumabe
M, Kumar AA, Badaracco M, Mikolaitis RA, Guerrero G, Utset TO,
Drevlow BE, et al: Autoimmune disease risk variant of IFIH1 is
associated with increased sensitivity to IFN-α and serologic
autoimmunity in lupus patients. J Immunol. 187:1298–1303. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Goropevšek A, Gorenjak M, Gradišnik S, Dai
K, Holc I, Hojs R, Krajnc I, Pahor A and Avčin T: Increased levels
of STAT1 protein in blood CD4 T cells from systemic lupus
erythematosus patients are associated with perturbed homeostasis of
activated CD45RA-FOXP3hi regulatory subset and follow-up disease
severity. J Interferon Cytokine Res. 37:254–268. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zuo C, Sheng X, Ma M, Xia M and Ouyang L:
ISG15 in the tumorigenesis and treatment of cancer: An emerging
role in malignancies of the digestive system. Oncotarget.
7:74393–74409. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Recht M, Borden EC and Knight E Jr: A
human 15-kDa IFN-induced protein induces the secretion of
IFN-gamma. J Immunol. 147:2617–2623. 1991.PubMed/NCBI
|
|
40
|
Care MA, Stephenson SJ, Barnes NA, Fan I,
Zougman A, El-Sherbiny YM, Vital EM, Westhead DR, Tooze RM and
Doody GM: Network analysis identifies proinflammatory plasma cell
polarization for secretion of ISG15 in human autoimmunity. J
Immunol. 197:1447–1459. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Dong G, You M, Fan H, Ding L, Sun L and
Hou Y: STS-1 promotes IFN-α induced autophagy by activating the
JAK1-STAT1 signaling pathway in B cells. Eur J Immunol.
45:2377–2388. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Han C, Fu J, Liu Z, Huang H, Luo L and Yin
Z: Dipyrithione inhibits IFN-gamma-induced JAK/STAT1 signaling
pathway activation and IP-10/CXCL10 expression in RAW264.7 cells.
Inflamm Res. 59:809–816. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Li J, Zhao S, Yi M, Hu X, Li J, Xie H, Zhu
W and Chen M: Activation of JAK-STAT1 signal transduction pathway
in lesional skin and monocytes from patients with systemic lupus
erythematosus. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 36:109–115.
2011.PubMed/NCBI
|
|
44
|
Rönnblom L and Pascual V: The innate
immune system in SLE: Type I interferons and dendritic cells.
Lupus. 17:394–399. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Dixit E and Kagan JC: Intracellular
pathogen detection by RIG-I-like receptors. Adv Immunol.
117:99–125. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Chai HC, Chua KH, Lim SK and Phipps ME:
Insight into gene polymorphisms involved in toll-like
receptor/interferon signalling pathways for systemic lupus
erythematosus in South East Asia. J Immunol Res. 2014:5291672014.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Castiblanco J, Varela DC,
Castaño-Rodríguez N, Rojas-Villarraga A, Hincapié ME and Anaya JM:
TIRAP (MAL) S180L polymorphism is a common protective factor
against developing tuberculosis and systemic lupus erythematosus.
Infect Genet Evol. 8:541–544. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Kressler D, Hurt E and Bassler J: Driving
ribosome assembly. Biochim Biophys Acta. 1803:673–683. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Tan EM, Cohen AS, Fries JF, Masi AT,
McShane DJ, Rothfield NF, Schaller JG, Talal N and Winchester RJ:
The 1982 revised criteria for the classification of systemic lupus
erythematosus. Arthritis Rheum. 25:1271–1277. 1982. View Article : Google Scholar : PubMed/NCBI
|