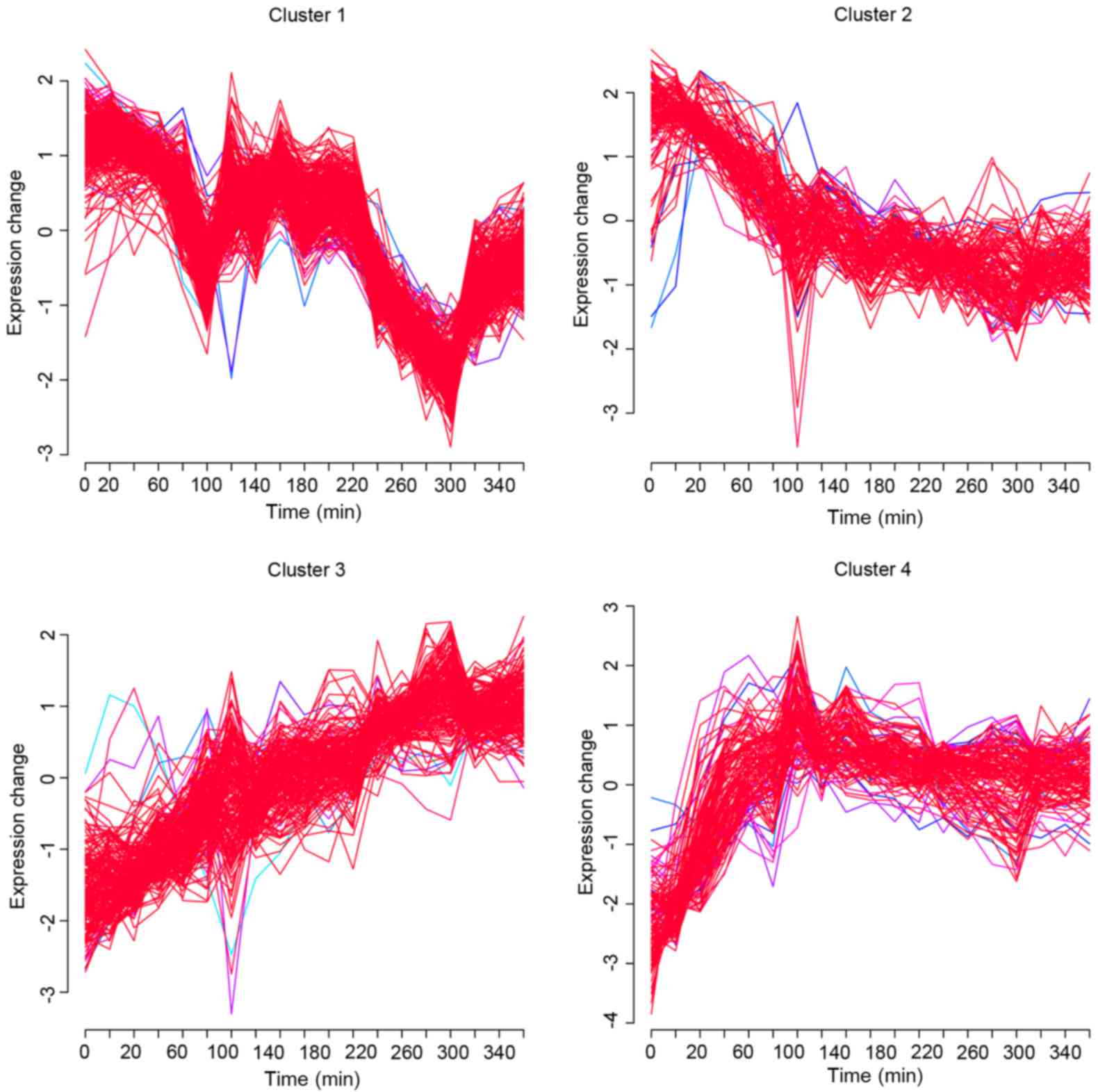
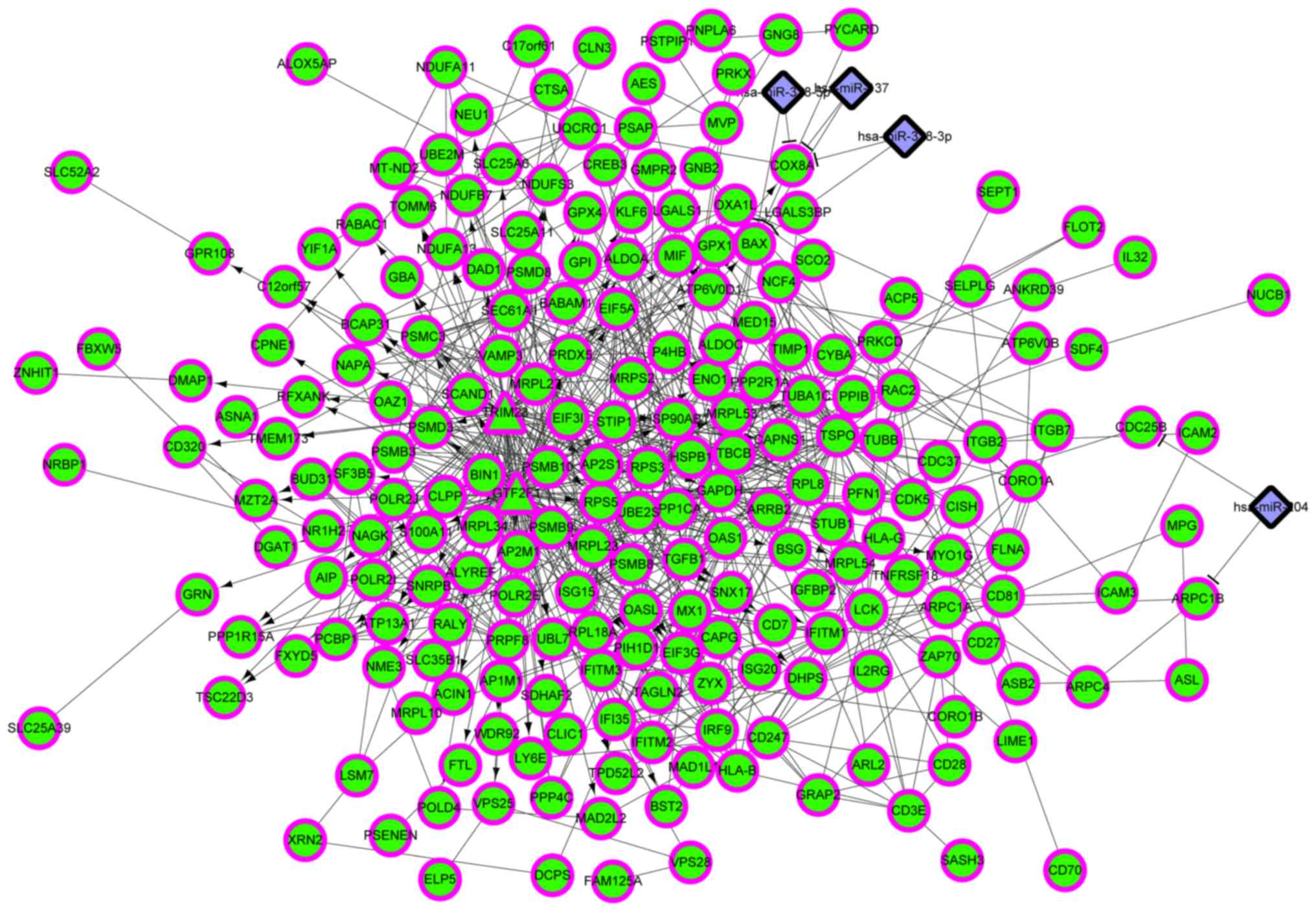
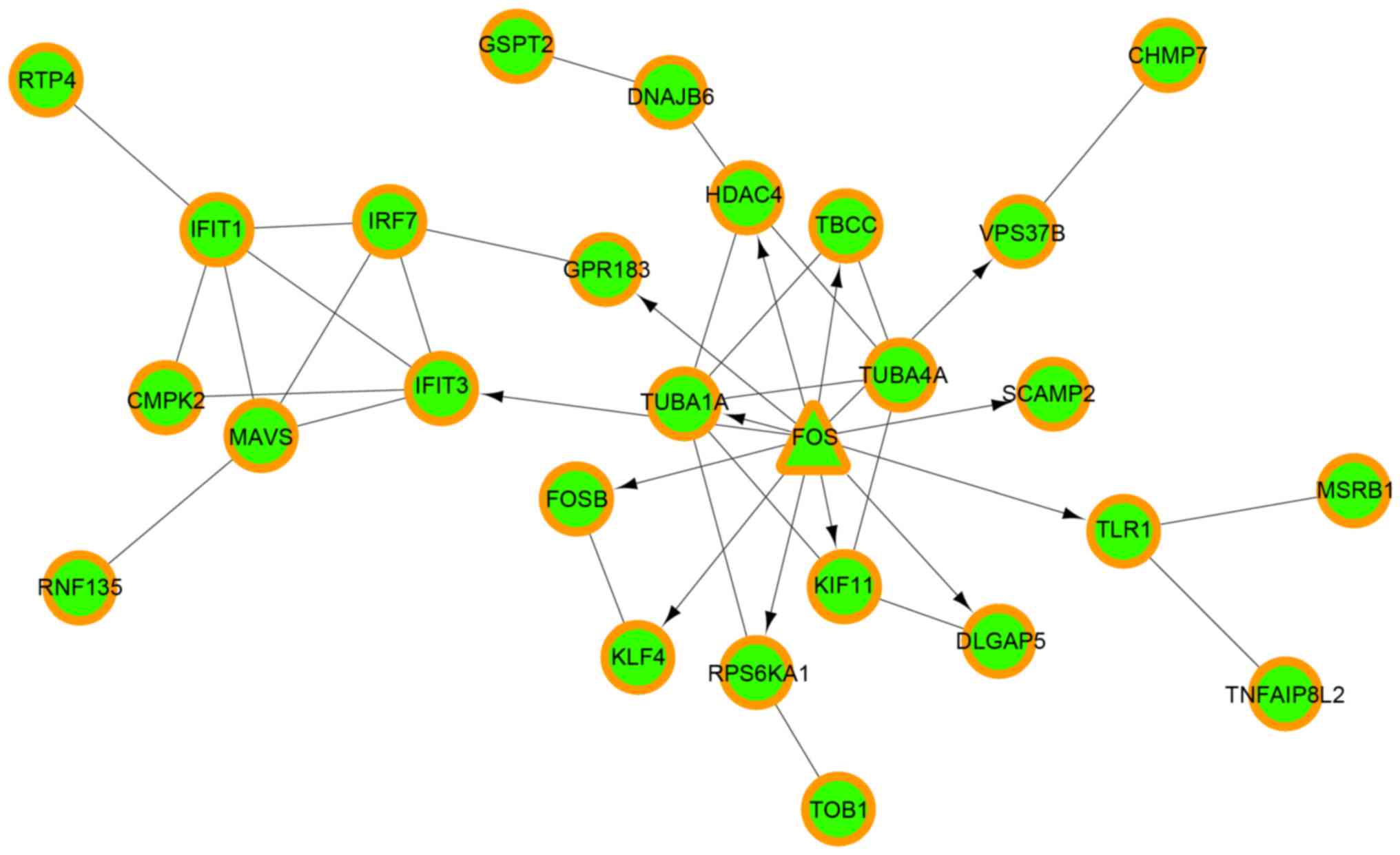
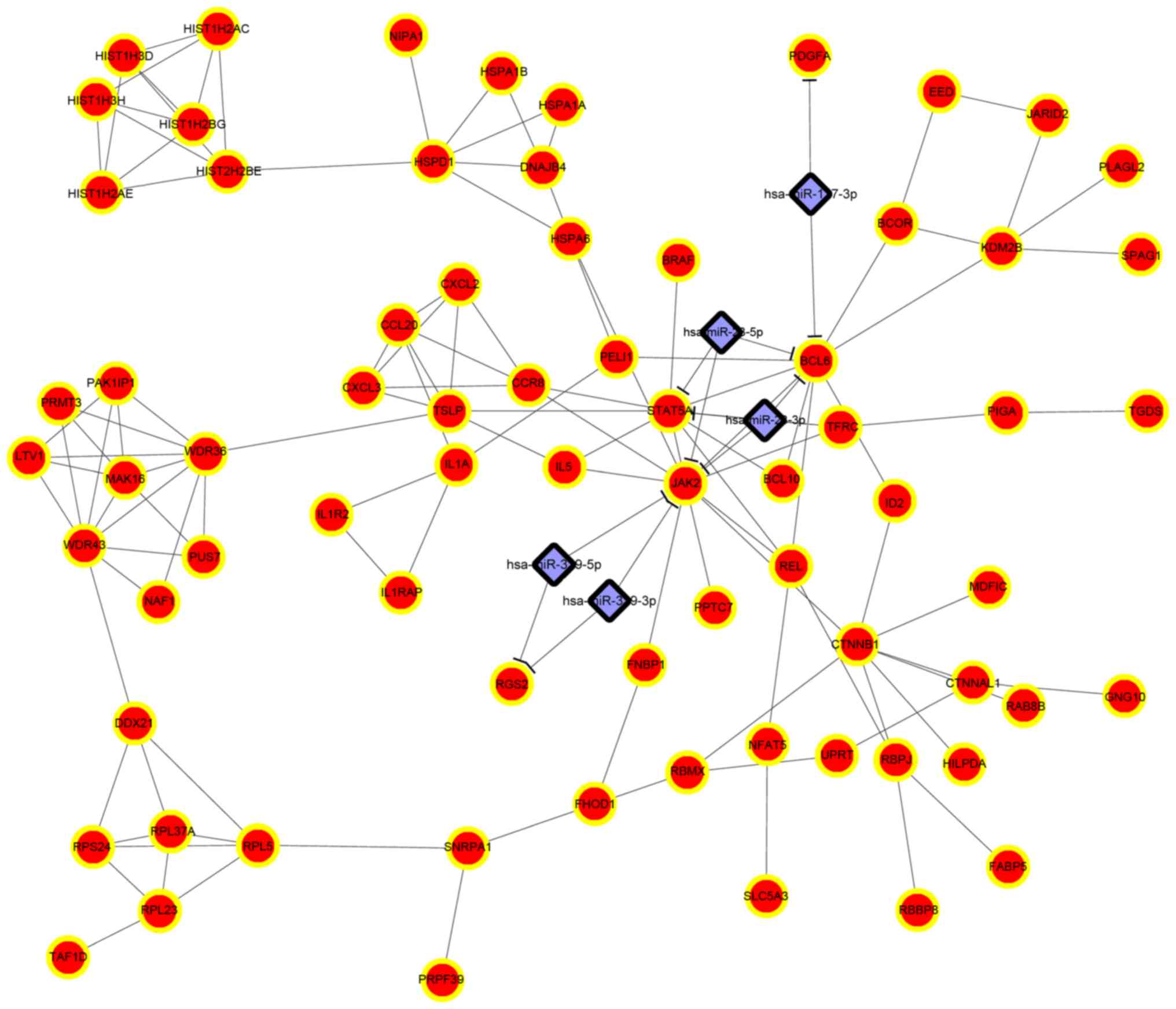
|
1
|
Thompson C and Powrie F: Regulatory T
cells. Curr Opin Pharmacol. 4:408–414. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Curiel TJ: Tregs and rethinking cancer
immunotherapy. J Clin Invest. 117:1167–1174. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Adeegbe DO and Nishikawa H: Natural and
induced T regulatory cells in cancer. Front immunol. 4:1902013.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Afzali B, Lombardi G, Lechler RI and Lord
GM: The role of T helper 17 (Th17) and regulatory T cells (Treg) in
human organ transplantation and autoimmune disease. Clin Exp
Immunol. 148:32–46. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wing K, Onishi Y, Prieto-Martin P,
Yamaguchi T, Miyara M, Fehervari Z, Nomura T and Sakaguchi S:
CTLA-4 control over Foxp3+ regulatory T cell function. Science.
322:271–275. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tai X, Van Laethem F, Pobezinsky L,
Guinter T, Sharrow SO, Adams A, Granger L, Kruhlak M, Lindsten T,
Thompson CB, et al: Basis of CTLA-4 function in regulatory and
conventional CD4+ T cells. Blood. 119:5155–5163. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sansom DM and Walker LS: The role of CD28
and cytotoxic T-lymphocyte antigen-4 (CTLA-4) in regulatory T-cell
biology. Immunol Rev. 212:131–148. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Huang CT, Workman CJ, Flies D, Pan X,
Marson AL, Zhou G, Hipkiss EL, Ravi S, Kowalski J, Levitsky HI, et
al: Role of LAG-3 in regulatory T cells. Immunity. 21:503–513.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Baratelli F, Lin Y, Zhu L, Yang SC,
Heuzé-Vourc'h N, Zeng G, Reckamp K, Dohadwala M, Sharma S and
Dubinett SM: Prostaglandin E2 induces FOXP3 gene expression and T
regulatory cell function in human CD4+ T cells. J Immunol.
175:1483–1490. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li J, Feng G, Liu J, Rong R, Luo F, Guo L,
Zhu T, Wang G and Chu Y: Renal cell carcinoma may evade the immune
system by converting CD4+ Foxp3-T cells into CD4+ CD25+ Foxp3+
regulatory T cells: Role of tumor COX-2-derived PGE2. Mol Med Rep.
3:959–963. 2010.PubMed/NCBI
|
|
11
|
Sutmuller RP, den Brok MH, Kramer M,
Bennink EJ, Toonen LW, Kullberg BJ, Joosten LA, Akira S, Netea MG
and Adema GJ: Toll-like receptor 2 controls expansion and function
of regulatory T cells. J Clin Invest. 116:485–494. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang Y, Luo F, Cai Y, Liu N, Wang L, Xu D
and Chu Y: TLR1/TLR2 agonist induces tumor regression by reciprocal
modulation of effector and regulatory T cells. J Immunol.
186:1963–1969. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Baban B, Chandler PR, Sharma MD, Pihkala
J, Koni PA, Munn DH and Mellor AL: IDO activates regulatory T cells
and blocks their conversion into Th17-like T cells. J Immunol.
183:2475–2483. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tran DQ, Andersson J, Wang R, Ramsey H,
Unutmaz D and Shevach EM: GARP (LRRC32) is essential for the
surface expression of latent TGF-beta on platelets and activated
FOXP3+ regulatory T cells. Proc Natl Acad Sci USA. 106:pp.
13445–13450. 2009; View Article : Google Scholar : PubMed/NCBI
|
|
15
|
He F, Chen H, Probst-Kepper M, Geffers R,
Eifes S, Del Sol A, Schughart K, Zeng AP and Balling R: PLAU
inferred from a correlation network is critical for suppressor
function of regulatory T cells. Mol Syst Biol. 8:6242012.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Probst-Kepper M, Geffers R, Kröger A,
Viegas N, Erck C, Hecht HJ, Lünsdorf H, Roubin R,
Moharregh-Khiabani D, Wagner K, et al: GARP: A key receptor
controlling FOXP3 in human regulatory T cells. J Cell Mol Med.
13:3343–3357. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gautier L, Cope L, Bolstad BM and Irizarry
RA: affy-analysis of Affymetrix GeneChip data at the probe level.
Bioinformatics. 20:307–315. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Futschik ME and Carlisle B: Noise-robust
soft clustering of gene expression time-course data. J Bioinform
Comput Biol. 3:965–988. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kumar L and Futschik E: Mfuzz: A software
package for soft clustering of microarray data. Bioinformation.
2:5–7. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xiao F, Zuo Z, Cai G, Kang S, Gao X and Li
T: miRecords: An integrated resource for microRNA-target
interactions. Nucleic Acids Res. 37:D105–D110. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Enright AJ, John B, Gaul U, Tuschl T,
Sander C and Marks DS: MicroRNA targets in Drosophila. Genome Biol.
5:R12003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang X: miRDB: A microRNA target
prediction and functional annotation database with a wiki
interface. RNA. 14:1012–1017. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Krek A, Grün D, Poy MN, Wolf R, Rosenberg
L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M
and Rajewsky N: Combinatorial microRNA target predictions. Nat
Genet. 37:495–500. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kertesz M, Iovino N, Unnerstall U, Gaul U
and Segal E: The role of site accessibility in microRNA target
recognition. Nat Genet. 39:1278–1284. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kohl M, Wiese S and Warscheid B:
Cytoscape: Software for visualization and analysis of biological
networks. Methods Mol Biol. 696:291–303. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Raney BJ, Cline MS, Rosenbloom KR, Dreszer
TR, Learned K, Barber GP, Meyer LR, Sloan CA, Malladi VS, Roskin
KM, et al: ENCODE whole-genome data in the UCSC genome browser
(2011 update). Nucleic Acids Res. 39:D871–D875. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Von Mering C, Jensen LJ, Kuhn M, Chaffron
S, Doerks T, Krüger B, Snel B and Bork P: STRING 7-recent
developments in the integration and prediction of protein
interactions. Nucleic Acids Res. 35:D358–D362. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Saito R, Smoot ME, Ono K, Ruscheinski J,
Wang PL, Lotia S, Pico AR, Bader GD and Ideker T: A travel guide to
Cytoscape plugins. Nat Methods. 9:1069–1076. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
He X and Zhang J: Why do hubs tend to be
essential in protein networks? PLoS Genet. 2:e882006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Binns D, Dimmer E, Huntley R, Barrell D,
O'Donovan C and Apweiler R: QuickGO: A web-based tool for gene
ontology searching. Bioinformatics. 25:3045–3046. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kanehisa M, Araki M, Goto S, Hattori M,
Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T
and Yamanishi Y: KEGG for linking genomes to life and the
environment. Nucleic Acids Res. 36:D480–D484. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Huang DW, Sherman BT, Tan Q, Collins JR,
Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC and Lempicki
RA: The DAVID gene functional classification tool: A novel
biological module-centric algorithm to functionally analyze large
gene lists. Genome Biol. 8:R1832007. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Fu S, Zhang M, Ou J, Liu H, Tan C, Liu J,
Chen H and Bei W: Construction and immune effect of Haemophilus
parasuis DNA vaccine encoding glyceraldehyde-3-phosphate
dehydrogenase (GAPDH) in mice. Vaccine. 30:6839–6844. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Madureira P, Baptista M, Vieira M, Liu H,
Tan C, Liu J, Chen H and Bei W: Streptococcus agalactiae GAPDH is a
virulence-associated immunomodulatory protein. J Immunol.
178:1379–1387. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chikuma S, Suita N, Okazaki IM, Shibayama
S and Honjo T: TRIM28 prevents autoinflammatory T cell development
in vivo. Nat Immunol. 13:596–603. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Betts BC, Abdel-Wahab O, Curran SA, St
Angelo ET, Koppikar P, Heller G, Levine RL and Young JW: Janus
kinase-2 inhibition induces durable tolerance to alloantigen by
human dendritic cell-stimulated T cells yet preserves immunity to
recall antigen. Blood. 118:5330–5339. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
O'Shea JJ and Plenge R: JAK and STAT
signaling molecules in immunoregulation and immune-mediated
disease. Immunity. 36:542–550. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Watts TH: TNF/TNFR family members in
costimulation of T cell responses. Annu Rev Immunol. 23:23–68.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Valencia X, Stephens G, Goldbach-Mansky R,
Wilson M, Shevach EM and Lipsky PE: TNF downmodulates the function
of human CD4+ CD25hi T-regulatory cells. Blood. 108:253–261. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Croft M: The role of TNF superfamily
members in T-cell function and diseases. Nature Reviews Immunology.
9:271–285. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ramirez-Montagut T, Chow A,
Hirschhorn-Cymerman D, Terwey TH, Kochman AA, Lu S, Miles RC,
Sakaguchi S, Houghton AN and van den Brink MR:
Glucocorticoid-induced TNF receptor family related gene activation
overcomes tolerance/ignorance to melanoma differentiation antigens
and enhances antitumor immunity. J Immunol. 176:6434–6442. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Kanamaru F, Youngnak P, Hashiguchi M,
Nishioka T, Takahashi T, Sakaguchi S, Ishikawa I and Azuma M:
Costimulation via glucocorticoid-induced TNF receptor in both
conventional and CD25+ regulatory CD4+ T cells. J Immunol.
172:7306–7314. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Rusca N and Monticelli S: MiR-146a in
immunity and disease. Mol Biol Int. 2011:4373012011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Lu LF, Boldin MP, Chaudhry A, Lin LL,
Taganov KD, Hanada T, Yoshimura A, Baltimore D and Rudensky AY:
Function of miR-146a in controlling Treg cell-mediated regulation
of Th1 responses. Cell. 142:914–929. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Huang B, Zhao J, Lei Z, Shen S, Li D, Shen
GX, Zhang GM and Feng ZH: miR-142-3p restricts cAMP production in
CD4+ CD25-T cells and CD4+ CD25+ TREG cells by targeting AC9 mRNA.
EMBO Rep. 10:180–185. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Lv M, Zhang X, Jia H, Li D, Zhang B, Zhang
H, Hong M, Jiang T, Jiang Q, Lu J, et al: An oncogenic role of
miR-142-3p in human T-cell acute lymphoblastic leukemia (T-ALL) by
targeting glucocorticoid receptor-α and cAMP/PKA pathways.
Leukemia. 26:769–777. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Burchill MA, Yang J, Vogtenhuber C, Blazar
BR and Farrar MA: IL-2 receptor β-dependent STAT5 activation is
required for the development of Foxp3+ regulatory T cells. J
Immunol. 178:280–290. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Yao Z, Kanno Y, Kerenyi M, Stephens G,
Durant L, Watford WT, Laurence A, Robinson GW, Shevach EM, Moriggl
R, et al: Nonredundant roles for Stat5a/b in directly regulating
Foxp3. Blood. 109:4368–4375. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Cohen AC, Nadeau KC, Tu W, Hwa V, Dionis
K, Bezrodnik L, Teper A, Gaillard M, Heinrich J, Krensky AM, et al:
Cutting edge: Decreased accumulation and regulatory function of
CD4+ CD25high T cells in human STAT5b deficiency. J Immunol.
177:2770–2774. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Tamiya T, Kashiwagi I, Takahashi R,
Yasukawa H and Yoshimura A: Suppressors of cytokine signaling
(SOCS) proteins and JAK/STAT pathways regulation of T-Cell
inflammation by SOCS1 and SOCS3. Arterioscler Thromb Vasc Biol.
31:980–985. 2011. View Article : Google Scholar : PubMed/NCBI
|