Introduction
Colorectal cancer (CRC) is the second most commonly
diagnosed cancer in females and the third in males worldwide, with
~1.4 million new cases and ~700,000 mortalities in 2012 (1). It is estimated that its incidence
will increase by 60% by 2030 (2).
Although various drug treatments, including active chemotherapeutic
drugs (e.g. fluoropyrimidines and oxaliplatin) and targeted drugs
(e.g. bevacizumab and cetuximab), have been applied to reduce the
incidence and mortality of CRC, the efficacy of these drugs in CRC
remains limited (3,4). Phytochemicals are expected to become
a novel option for CRC prevention and treatment (5). However, cancer is a complex disorder
associated with defects in multiple signaling pathways that confer
resistance to apoptosis.
Increasing evidence indicates that microRNAs
(miRNAs/miRs) contribute to the initiation and progression of
various types of cancer, including liver, lung and colorectal
cancers (6,7). As one of the first miRNAs to be
described, let-7 has been demonstrated to participate in the
development of cancer (8).
Furthermore, the RNA-binding protein Lin28 is an emerging oncogenic
driver which acts by restraining the biogenesis of let-7 (9). King et al (10) found that Lin28B is increased in CRC
patients and promotes the progression and metastasis of CRC. In
addition, a recent study by Tu et al (11) demonstrated that the Lin28/let-7
axis promotes invasive intestinal adenocarcinoma in murine models
by cooperating with the Wnt pathway. However, the role of the
Lin28/let-7 loop in the apoptosis of CRC cells is not well
understood.
The primary objective of the present study was to
examine whether Lin28/let-7 is involved in the apoptosis of CRC
cells and elucidate the underlying molecular mechanisms of this. It
was revealed that forced expression of let-7c, or silencing of
Lin28, led to reduced cell viability and increased apoptosis in CRC
cells, indicating the potential for this loop as a novel
therapeutic target for CRC.
Materials and methods
Patient samples
Cancer tissue samples and adjacent normal tissues
were obtained from 10 patients (6 male and 4 female; mean age,
55.3±5.7 years) with CRC who underwent surgical resection of
primary tumors at the Second Affiliated Hospital of Harbin Medical
University (Harbin, China) between 2016 and 2016. The patients did
not receive radiotherapy or chemotherapy prior to resection. The
specimens were immediately snap-frozen in liquid nitrogen and
stored at −80°C until use. All procedures were approved by the
Ethics Committee for the Use of Human Samples of Harbin Medical
University. Written informed consent was obtained from all
patients.
Immunohistochemistry and quantification.
Formalin-fixed, paraffin-embedded tissue sections (thickness, 5 µm)
were treated with xylene followed by a graded alcohol series, and
antigen retrieval was performed using 0.01 M citrate buffer.
Hydrogen peroxide was used for blocking of endogenous peroxidase
activity. Tissue sections were then treated with goat serum for 20
min. Subsequently, antibodies against Lin28 (#sc-293120; Santa Cruz
Biotechnology, Inc., Santa Cruz, CA, USA; dilution, 1:100) were
incubated with each section at 4°C overnight. An EliVision™ Plus
kit and a DAB kit (both from Beyotime Institute of Biotechnology,
Haimen, China) were used to detect the bound primary antibodies.
Staining was performed according to manufacturer's protocol. All
tumor slides were examined under a light microscope by two
independent pathologists. Lin28 signals in the tissues were
visually quantified using a scoring system in which the score for
the intensity of signal (0, no signal; 1, weak signal; 2,
intermediate signal; and 3, strong signal) was multiplied by the
score for the percentage of positive cells (0, 0%; 1, <25%; 2,
25–50%; and 3, >50%) to produce an overall score ranging from 0
to 9.
Cell culture and treatments
NCM460 normal human colonic epithelial cells and the
human CRC cell lines HCT116 and HT29 were obtained from the
Shanghai Cell Bank of the Chinese Academy of Sciences (Shanghai,
China). Cells were cultured in RPMI-1640 medium (Hyclone; GE
Healthcare Life Sciences, Logan, UT, USA) supplemented with 10%
fetal bovine serum (Hyclone; GE Healthcare Life Sciences), at 37°C
in a humidified atmosphere of 95% air and 5% CO2. Cells
transfected with siRNAs, let-7c or AMO-let-7c (Shanghai GenePharma,
Co., Ltd, Shanghai, China) were collected at 48 h post-transfection
for further measurements.
Transfection
Prior to transfection, HCT116 or HT29 cells were
grown in 25-cm2 cell culture flasks with 4 ml medium.
The let-7c mimics or AMO-let-7c (let-7c antisense
oligonucleotides), or their respective negative controls (NCs), as
well as 10 µl Lipofectamine 2000 (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) were separately mixed with 500
µl of Opti-MEM® I Reduced Serum Medium (Gibco; Thermo
Fisher Scientific, Inc.) for 5 min. Subsequently, the two mixtures
were combined and incubated for 20 min at room temperature. The
final concentration of miRNAs was 100 µM. The Lipofectamine-miRNA
mixtures were then added to the cells, which were incubated at 37°C
for 36 h prior to further experiments. The control cells underwent
the same transfection procedures without nucleic acid. The NC for
let-7c overexpression was a disordered sequence of let-7c. The
transfection protocol for the siRNAs (20 nM) was the same as that
for miRNAs. The sequences of the RNAs and controls were as follows:
let-7c mimics, 5′-UGAGGUAGUAGGUUGUAUGGUU-3′; AMO-let-7c,
5′-AACCAUACAACCUACUACCUCA-3; let-7c NC, 5′-UUCUCCGAACGUGUCACGUTT-3′
(sense) and 5′-ACGUGACACGUUCGGAGAATT-3′ (anti-sense); AMO-NC (NC
for AMO-let-7c), 5′-CAGUACUUUUGUGUAGUACAA-3′; si-Lin28 #1,
5′-GGAGACAGGUGCUACAACUUU-3′; si-Lin28 #2:
5′-UGACGUAUCUUGUGCGUUUUU-3′; si-Lin28 #3:
5′-AAAUGUGUCUCACGGGUUUUU-3′; scrambled siRNA,
5′-UGCGGAUUCUAUCUGUAU-3′.
MTT cell viability assay
HCT116 or HT29 cells were seeded in 96-well culture
plates in 200 µl medium with 1×104 cells/well, and
incubated at 37°C with 5% CO2. Following the
transfection of siRNAs or miRNAs, an MTT assay (Amresco, LLC,
Solon, OH, USA) was performed. Briefly, 20 µl of MTT solution (5
mg/ml) was added to each well, and the cells were continuously
incubated for 4 h. Following removal of cell culture medium,
formazan crystals were dissolved in 150 µl DMSO. The optical
density (OD) of each wells was measured with a microplate reader
(BioTek Instruments, Inc., Winooski, VT, USA) at 490 nm.
RNA isolation and quantification of
Lin28 and let-7c
Total RNA was extracted from CRC tissue samples or
HCT116 or HT29 cells by TRIzol® reagent (Invitrogen;
Thermo Fisher Scientific, Inc.) following the manufacturer's
protocol. Total RNA (0.5 µg) was then reverse transcribed using a
High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems;
Thermo Fisher Scientific, Inc.) to obtain cDNA. The temperature
protocol was as follows: 25°C for 10 min, 37°C for 120 min, 85°C
for 5 min, and hold at 4°C. The RNA level of Lin28 was determined
using a SYBR Green I incorporation method on an ABI 7500 Fast
Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific,
Inc.), with GAPDH as an internal control. The let-7c level was
measured using a mirVana™ qRT PCR miRNA Detection Kit (Ambion;
Thermo Fisher Scientific, Inc.), following the method described by
Liang et al (12). The
protocol was as follows: 95°C for 10 min; followed by 40 cycles of
95°C for 15 sec, 60°C for 30 sec, and 72°C for 30 sec. The primers
for qPCR were as follows: let-7c forward, 5′-GGGAGAGGTAGTAGGTTG-3′;
and let-7c reverse, 5′-TGGAGTCGGCAATTGCAC-3′; U6 forward,
5′-GCTTCGGCAGCACATATACTAAAAT-3′; U6 reverse,
5′-CGCTTCACGAATTTGCGTGTCAT-3′; Lin28 forward,
5′-TCTACCTCCTCAGCCAAAGA-3′; Lin28 reverse,
5′-TGGGATTCTGCTTCCTGTCT-3′; GAPDH forward,
5′-GGGGCTCTCTGCTCCTCCCTG-3′; and GAPDH reverse,
5′-CGGCCAAATCCGTTCACACCG-3′. Variations in the expression of let-7c
between different RNA samples were calculated after normalization
to U6. The data were analyzed using the 2−ΔΔCq method
(13); for the cell lines, cells
treated with Lipofectamine 2000 only were used as a control group.
The experiment was repeated independently five times.
Western blot
For western blot analysis, total protein samples
were extracted from HCT116 or HT29 cells. Cells were lysed with
radioimmunoprecipitation assay (RIPA)lysis buffer (Beyotime
Institute of Biotechnology, Haimen, China). Total protein was
quantified using a BCA Assay kit (Beyotime Institute of
Biotechnology) according to the manufacturer's protocol. Total
protein samples (50 µg) were separated on a 15% SDS-polyacrylamide
gel. After electrophoretic transfer of the proteins to a pure
nitrocellulose blotting membrane, the blots were blocked with 5%
non-fat dry milk (Beyotime Institute of Biotechnology) for 2 h at
room temperature, then incubated overnight at 4°C with rabbit
polyclonal antibodies against B-cell lymphoma 2 (Bcl-2) (#3498;
Cell Signaling Technology, Inc., Danvers, MA, USA; dilution,
1:800), Bcl-2-associated X protein (Bax) (#14796; Cell Signaling
Technology, Inc.; dilution, 1:800), Bcl-2-like 1
(BCL2L1)(#10783-1-AP; ProteinTech, Wuhan, China; dilution, 1:200)
and Lin28 (#sc-293120; Santa Cruz Biotechnology, Inc., Santa Cruz,
CA, USA; dilution, 1:200), and an anti-GAPDH antibody (#KC-5G4;
Kangchen Biotech, Shanghai, China; dilution, 1:2,000), which was
used as an internal control. The blots were subsequently incubated
with a DyLight 800-conjugated secondary antibody (#5151; Cell
Signaling Technology, Inc.; dilution, 1:1,000) for 2 h at room
temperature, and bands were detected using an Odyssey Infrared
Imaging System and analyzed using Odyssey software v1.2 (Infrared
Imaging System LI-COR Biosciences). The bands were quantified by
measuring the band intensity for each group.
Caspase-3 activity assay
HCT116 and HT29 cells were lysed in 50 µl of
ice-cold RIPA lysis buffer for 30 min. The caspase-3 activity assay
kit was obtained from the Beyotime Institute of Biotechnology. The
lysates were centrifuged at 16,000 × g for 15 min at 4°C. The
fluorogenic substrates for caspase-3 were labeled with the
p-nitroaniline (pNA). The enzyme activity was
determined by monitoring the fluorescence produced by free pNA
using a spectrofluorophotometer (RF-5301 PC; Shimadzu Corporation,
Kyoto, Japan) at 405 nm. Caspase-3 activity was expressed as
micromoles of pNA liberated per minute per microgram of protein,
following determination of total protein concentration using a BCA
Assay kit (Beyotime Institute of Biotechnology) according to the
manufacturer's protocol.
Luciferase reporter assays
The TargetScan database (http://www.targetscan.org/vert_71/) was utilized to
determine the direct target of let-7c. The BCL2L1 3′-UTR containing
the conserved let-7c-binding sites was synthesized by Invitrogen
and subcloned using the SacI and HindIII sites
downstream of the luciferase gene in a pMIR-REPORT Luciferase
vector (Promega Corporation, Madison, WI, USA). The luciferase
vector (100 ng) containing the 3′-UTR was cotransfected with let-7c
mimics into HEK 293 cells using Lipofectamine 2000. As an internal
control, 10 ng of Renilla luciferase reporters were also
included. At 36 h after transfection, the cells were collected and
dual luciferase activities were measured by a luminometer (Promega
Corporation) according to the manufacturer's instructions.
Statistical analysis
All data are presented as the mean ± standard error
of the mean. A Student's t-test was used for two-group comparisons,
and a one-way ANOVA followed by a Bonferroni test for multiple
comparisons was used for comparisons between three or more groups.
Two-tailed P<0.05 was considered to indicate a statistically
significant difference.
Results
Dysregulation of the Lin28/let-7 axis
in patients with CRC and in CRC cells
To investigate whether the Lin28/let-7 axis
participates in the process of CRC, immunohistochemistry, RT-qPCR
and western blot analyses were used to detect the levels of
Lin28/let-7 in patient-derived CRC tissues and in CRC cells.
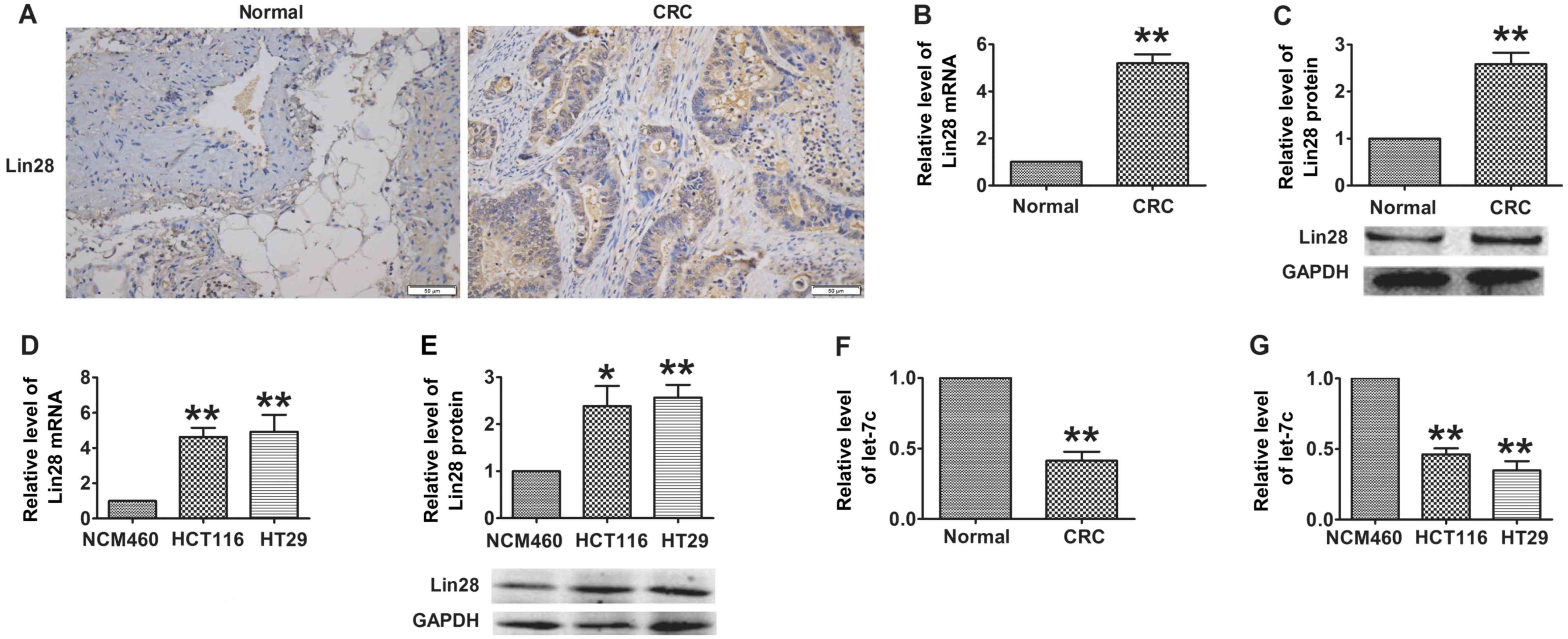
Immunohistochemistry revealed that Lin28, which was mainly located
in the cytoplasm of the cells, was overexpressed in CRC tissues
compared with adjacent normal colorectal tissues (7.22±0.94 vs
2.11±0.61, P<0.05; Fig. 1A). In
addition, as shown in Fig. 1B and
C, the expression of Lin28 was significantly increased at the
mRNA and protein levels in the CRC samples. In accord with the data
from patients, the mRNA and protein levels of Lin28 were revealed
to be markedly increased in CRC cells compared with NCM460 normal
colonic epithelial cells (Fig. 1D and
E). Furthermore, as a downstream factor of Lin28, the
expression of let-7c was markedly decreased in the CRC patient
tissues and in CRC cells compared with the respective normal
controls (Fig. 1F and G). These
results suggest that the dysregulation of the Lin28/let-7c axis may
contribute to the process of CRC.
Forced expression of let-7c induces
apoptosis in cultured CRC cells
To determine the role of let-7c in the process of
CRC, let-7c miRNA mimics were transfected into CRC cells (Fig. 2A). As shown in Fig. 2B, cell viability was decreased in
HCT116 and HT29 cells following let-7c mimic transfection. In
addition, overexpression of let-7c led to activation of caspase-3
activity in HCT116 and HT29 cells (Fig. 2C). Furthermore, western blot
analysis revealed altered Bcl-2 and Bax protein levels in let-7c
mimic-transfected CRC cells (Fig.
2D). These results suggest that enhanced expression of let-7c
could induce apoptosis in CRC cells.
Let-7c regulates BCL2L1 in a
post-transcriptional manner
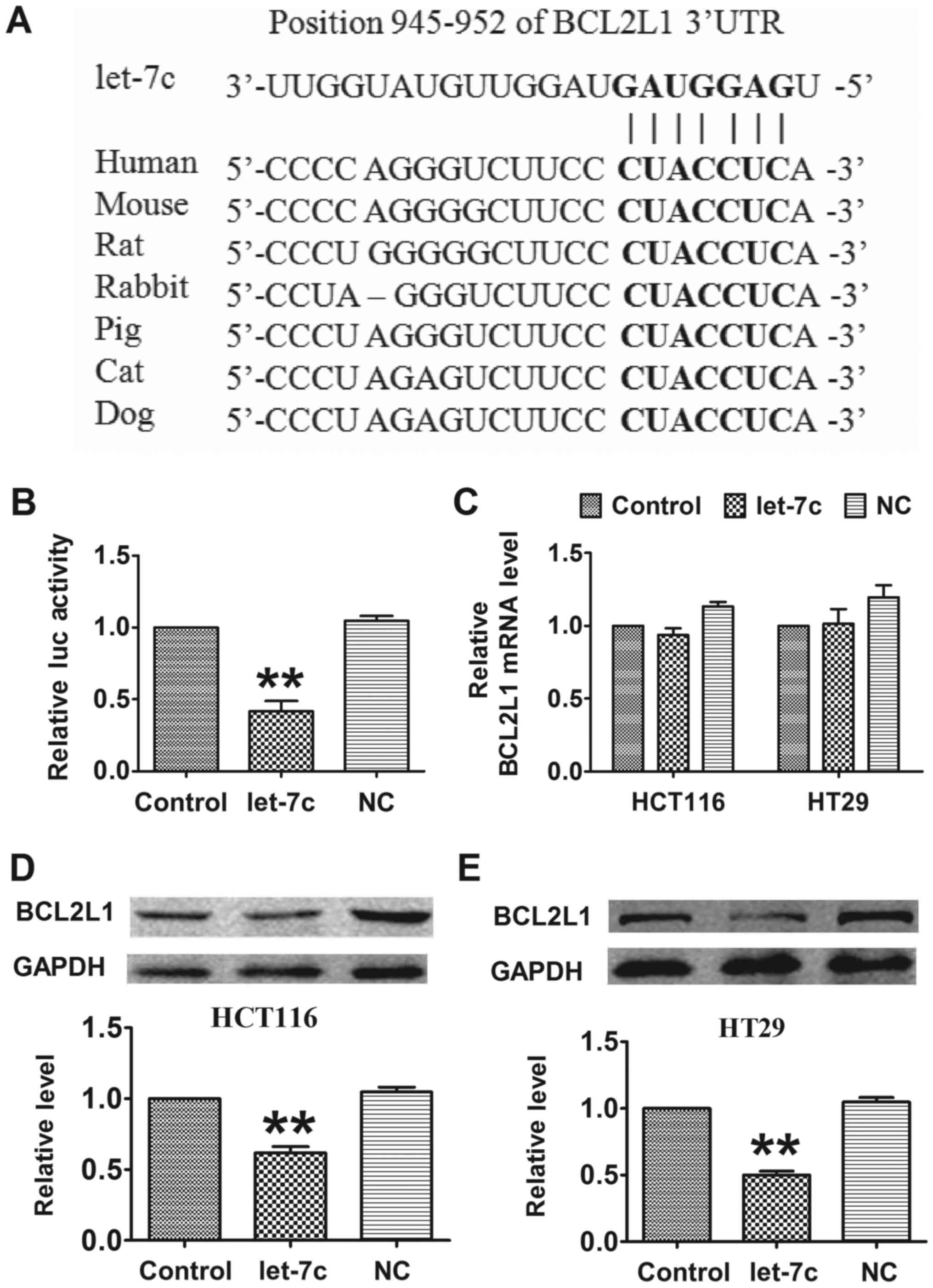
TargetScan, an online miRNA target prediction
database, was employed to determine the direct target of let-7c. As
depicted in Fig. 3A, let-7c has
seed sequence complementary with the binding site in the 3′
untranslated region (UTR) of BCL2L1, an antiapoptotic factor. A
luciferase assay showed that co-transfection of let-7c mimics with
a luciferase reporter vector carrying a portion of the wild-type
human BCL2L1 3′UTR led to a significant decrease in luciferase
activities compared with transfection with the luciferase vector
alone (control group), whereas transfection with the NC had no
significant effect (Fig. 3B).
Furthermore, overexpression of let-7c had no effect on the mRNA
level of BCL2L1 (Fig. 3C). By
contrast, forced expression of let-7c markedly decreased the
protein level of BCL2L1 in CRC cells compared with the
untransfected control group (Fig. 3D
and E). These results indicated that BCL2L1 is a direct target
of let-7c, and that let-7c regulates BCL2L1 in a
post-transcriptional manner.
Silencing of Lin28 promotes apoptosis
via upregulating let-7c in CRC cells
As a well-known RNA-binding protein, Lin28 has been
demonstrated to participate in various disease processes, including
cancer and lung fibrosis, by inhibiting the biogenesis of let-7
(13,14). In addition, Lin28 also contributes
to the initiation and invasion of CRC (10). Therefore, we hypothesized that
silencing of Lin28 may promote apoptosis in CRC by upregulating
let-7c. To address this, siRNAs were transfected into cells to
inhibit the expression of Lin28 (Fig.
4A), revealing that silencing of Lin28 increased the expression
of let-7c in CRC cells (Fig. 4B).
Furthermore, silencing of Lin28 decreased CRC cell viability,
whereas inhibition of let-7c attenuated this effect (Fig. 4C and D). Further experiments showed
that silencing Lin28 promoted apoptosis in CRC cells, as determined
by measuring the activation of caspase-3 (Fig. 4E and F) and the dysregulation of
Bcl-2 and Bax protein (Fig. 4G and
H). Knockdown of let-7c mitigated the proapoptotic effect of
Lin28 inhibition (Fig. 4E-H).
These findings suggest that silencing of Lin28 induced apoptosis in
CRC cells by upregulating let-7c.
Discussion
In the present study, it was demonstrated that Lin28
was upregulated and let-7c was downregulated in CRC tissues and
cell lines. The forced expression of let-7c resulted in decreased
cell viability and promoted apoptosis in CRC cells. Furthermore, it
was demonstrated that BCL2L1 is a direct target of let-7c and may
act to mediate its proapoptotic effect. Additionally, silencing of
Lin28 decreased viability and promoted apoptosis in CRC cells,
whereas the knockdown of let-7c attenuated the proapoptotic action
of Lin28 inhibition. Taken together, the findings demonstrate the
dysregulation of the Lin28/let-7c axis in the progression of CRC,
and indicated that silencing of Lin28 can promote apoptosis in CRC,
which is mediated, at least in part, by increasing the expression
of let-7c. Therefore, targeting Lin28/let-7c could be a novel
strategy for the treatment of CRC.
Increasing numbers of studies have provided strong
evidence that miRNAs contribute to various diseases, including
cancer, cardiovascular diseases and diabetes (15–17).
As the earliest miRNA to be discovered, let-7 is crucial in the
progress of numerous diseases, particularly in various types of
cancer (8,18,19).
A study by Trang et al (20) demonstrated that loss of let-7
function enhanced lung tumor formation in vivo, whereas
forced expression of let-7 significantly decreased the tumor burden
in mice, indicating the inhibitory function of let-7 in cancer. Xia
et al (21) found that low
expression of let-7 may be a biomarker predicting poor prognosis in
patients with various cancers, particularly lung cancer. In
addition, Ghanbari et al (22) suggested that underexpressed
let-7a-5p and let-7f-5p in plasma and stool samples from patients
could serve as potential biomarkers for the early detection of CRC.
Han et al (23) reported
that let-7c inhibited the metastasis of CRC through the regulation
of matrix metalloproteinase-11 and PBX homeobox 3. However, the
underlying molecular mechanisms of let-7 in apoptosis, particularly
during cancer, are not well understood, despite several studies
reporting on the effect of let-7 on apoptosis (24,25).
In the present study, the results further confirmed the
proapoptotic action of let-7c by demonstrating the ability of
let-7c to induce apoptosis in CRC cells through repressing the
expression of BCL2L1; to the best of our knowledge, this is the
first report of such findings. The results suggest that let-7c may
be used for the treatment of CRC.
Bcl-2 family proteins serve critical roles in the
regulation of the intrinsic apoptosis pathway. The activation of
Bax, a key member of the Bcl-2 family, is able to promote
cytochrome c release and mitochondrial fission, which leads
to apoptosome formation and caspase-3 activation, in turn promoting
apoptosis (26). It has been
reported that BCL2L1 is able to stabilize the mitochondrial
localization of Bax while maintaining it in an inactive state
(27). We speculate that the
overexpression of let-7c results in the activation of Bax by
targeting BCL2L2, and which increases cytochrome c release
and promotes apoptosis in CRC.
A number of studies have demonstrated that the
biogenesis of let-7 is tightly controlled by Lin28, which,
conversely, is a direct target of let-7 (28,29).
Certain reports have demonstrated that the Lin28/let-7 loop
influences numerous biological processes, including proliferation,
differentiation, stem cell regeneration and cell aging (11,30).
In a recent study, Chien et al (31) revealed that Lin28/let-7 promoted
the transformation of oral squamous cell carcinoma cells into
cancer stem-like cells by regulating Oct4/Sox2 via modulation of
AT-rich interaction domain 3B and high mobility group AT-hook 2.
Other studies showed that Lin28/let-7 promoted cancer progression
and metastasis via regulating epithelial-mesenchymal transition
(EMT) (32,33): Liu et al (32) demonstrated that Lin28 may induce
EMT via downregulation of let-7a in breast cancer cells; and Fu
et al (33) reported that
miR-26a suppresses tumor growth and metastasis by targeting
Lin28B/let-7. Consistent with this result, a recent study by Liang
et al (12) demonstrated
that miR-26a inhibits lung fibrosis by repressing EMT via
disrupting the Lin28B/let-7d axis. However, the role of the
Lin28/let-7 axis in the process of apoptosis has not been well
established.
In summary, the present study demonstrated
dysregulation of the Lin28/let-7c axis in CRC patients, and further
confirmed that inhibiting this axis promoted apoptosis in CRC cells
by suppressing BCL2L1 through increasing the expression of let-7c.
Therefore, this axis may be a novel therapeutic target for the
treatment of patients with CRC. Notably, Roos et al
(34) identified that
N-methyl-N-[3-(3-methyl[1,2,4]triazolo[4,3-b]pyridazin-6-yl)phenyl]acetamide,
which blocks the Lin28/let-7 interaction, rescued let-7 processing
and function, and induced differentiation of mouse embryonic stem
cells, eventually reducing tumor-sphere formation in 22Rv1 human
prostate carcinoma and Huh7 human hepatocellular carcinoma cells.
These findings may represent a new direction for the treatment of
CRC. More studies must be performed to further explore
small-molecule inhibitors of Lin28 and evaluate their therapeutic
potential in CRC.
Acknowledgements
This study was supported by the Scientific Research
Fund of Heilongjiang Provincial Education Department (grant no.
12541368) and Scientific Research of the Health and Family Planning
Commission of Heilongjiang Province of China (grant no.
2013045).
References
|
1
|
Sforza V, Martinelli E, Ciardiello F,
Gambardella V, Napolitano S, Martini G, Della Corte C, Cardone C,
Ferrara ML, Reginelli A, et al: Mechanisms of resistance to
anti-epidermal growth factor receptor inhibitors in metastatic
colorectal cancer. World J Gastroenterol. 22:6345–6361. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ciasca G, Papi M, Minelli E, Palmieri V
and De Spirito M: Changes in cellular mechanical properties during
onset or progression of colorectal cancer. World J Gastroenterol.
22:7203–7214. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
De Greef K, Rolfo C, Russo A, Chapelle T,
Bronte G, Passiglia F, Coelho A, Papadimitriou K and Peeters M:
Multisciplinary management of patients with liver metastasis from
colorectal cancer. World J Gastroenterol. 22:7215–7225. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sanz-Garcia E, Grasselli J, Argiles G,
Elez ME and Tabernero J: Current and advancing treatments for
metastatic colorectal cancer. Expert Opin Biol Ther. 16:93–110.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yin TF, Wang M, Qing Y, Lin YM and Wu D:
Research progress on chemopreventive effects of phytochemicals on
colorectal cancer and their mechanisms. World J Gastroenterol.
22:7058–7068. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Arora H, Qureshi R, Rizvi MA, Shrivastava
S and Parihar MS: Study of apoptosis-related interactions in
colorectal cancer. Tumour Biol. 37:14415–14425. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Behbahani GD, Ghahhari NM, Javidi MA,
Molan AF, Feizi N and Babashah S: MicroRNA-mediated
post-transcriptional regulation of epithelial to mesenchymal
transition in cancer. Pathol Oncol Res. 23:1–12. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sun X, Liu J, Xu C, Tang SC and Ren H: The
insights of Let-7 miRNAs in oncogenesis and stem cell potency. J
Cell Mol Med. 20:1779–1788. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Nguyen LH and Zhu H: Lin28 and let-7 in
cell metabolism and cancer. Transl Pediatr. 4:4–11. 2015.PubMed/NCBI
|
|
10
|
King CE, Cuatrecasas M, Castells A,
Sepulveda AR, Lee JS and Rustgi AK: LIN28B promotes colon cancer
progression and metastasis. Cancer Res. 71:4260–4268. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tu HC, Schwitalla S, Qian Z, LaPier GS,
Yermalovich A, Ku YC, Chen SC, Viswanathan SR, Zhu H, Nishihara R,
et al: LIN28 cooperates with WNT signaling to drive invasive
intestinal and colorectal adenocarcinoma in mice and humans. Genes
Dev. 29:1074–1086. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liang H, Liu S, Chen Y, Bai X, Liu L, Dong
Y, Hu M, Su X, Chen Y, Huangfu L, et al: miR-26a suppresses EMT by
disrupting the Lin28B/let-7d axis: Potential cross-talks among
miRNAs in IPF. J Mol Med (Berl). 94:655–665. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang H, Zhao Q, Deng K, Guo X and Xia J:
Lin28: An emerging important oncogene connecting several aspects of
cancer. Tumour Biol. 37:2841–2848. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Piletič K and Kunej T: MicroRNA epigenetic
signatures in human disease. Arch Toxicol. 90:2405–2419. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sethupathy P: The promise and challenge of
therapeutic MicroRNA silencing in diabetes and metabolic diseases.
Curr Diab Rep. 16:522016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shen E, Diao X, Wei C, Wu Z, Zhang L and
Hu B: MicroRNAs target gene and signaling pathway by bioinformatics
analysis in the cardiac hypertrophy. Biochem Biophys Res Commun.
397:380–385. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bao MH, Feng X, Zhang YW, Lou XY, Cheng Y
and Zhou HH: Let-7 in cardiovascular diseases, heart development
and cardiovascular differentiation from stem cells. Int J Mol Sci.
14:23086–23102. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li X, Wang B, Cui H, Du Y, Song Y, Yang L,
Zhang Q, Sun F, Luo D, Xu C, et al: let-7e replacement yields
potent anti-arrhythmic efficacy via targeting beta 1-adrenergic
receptor in rat heart. J Cell Mol Med. 18:1334–1343. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Trang P, Medina PP, Wiggins JF, Ruffino L,
Kelnar K, Omotola M, Homer R, Brown D, Bader AG, Weidhaas JB and
Slack FJ: Regression of murine lung tumors by the let-7 microRNA.
Oncogene. 29:1580–1587. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xia Y, Zhu Y, Zhou X and Chen Y: Low
expression of let-7 predicts poor prognosis in patients with
multiple cancers: A meta-analysis. Tumour Biol. 35:5143–5148. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ghanbari R, Mosakhani N, Sarhadi VK,
Armengol G, Nouraee N, Mohammadkhani A, Khorrami S, Arefian E,
Paryan M, Malekzadeh R and Knuutila S: Simultaneous underexpression
of let-7a-5p and let-7f-5p microRNAs in plasma and stool samples
from early stage colorectal carcinoma. Biomark Cancer. 7 Suppl
1:S39–S48. 2016.
|
|
23
|
Han HB, Gu J, Zuo HJ, Chen ZG, Zhao W, Li
M, Ji DB, Lu YY and Zhang ZQ: Let-7c functions as a metastasis
suppressor by targeting MMP11 and PBX3 in colorectal cancer. J
Pathol. 226:544–555. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang T, Han P, He Y, Zhao C, Wang G, Yang
W, Shan M, Zhu Y, Yang C, Weng M, et al: Lin28A enhances
chemosensitivity of colon cancer cells to 5-FU by promoting
apoptosis in a let-7 independent manner. Tumour Biol. 37:7657–7665.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Geng L, Zhu B, Dai BH, Sui CJ, Xu F, Kan
T, Shen WF and Yang JM: A let-7/Fas double-negative feedback loop
regulates human colon carcinoma cells sensitivity to Fas-related
apoptosis. Biochem Biophys Res Commun. 408:494–499. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Youle RJ and Strasser A: The BCL-2 protein
family: Opposing activities that mediate cell death. Nat Rev Mol
Cell Biol. 9:47–59. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Renault TT, Teijido O, Antonsson B, Dejean
LM and Manon S: Regulation of Bax mitochondrial localization by
Bcl-2 and Bcl-× (L): Keep your friends close but your enemies
closer. Int J Biochem Cell Biol. 45:64–67. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
McDaniel K, Hall C, Sato K, Lairmore T,
Marzioni M, Glaser S, Meng F and Alpini G: Lin28 and let-7: Roles
and regulation in liver diseases. Am J Physiol Gastrointest Liver
Physiol. 310:G757–G765. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Huang Y: A mirror of two faces: Lin28 as a
master regulator of both miRNA and mRNA. Wiley Interdiscip Rev RNA.
3:483–494. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jun-Hao ET, Gupta RR and Shyh-Chang N:
Lin28 and let-7 in the metabolic physiology of aging. Trends
Endocrinol Metab. 27:132–141. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chien CS, Wang ML, Chu PY, Chang YL, Liu
WH, Yu CC, Lan YT, Huang PI, Lee YY, Chen YW, et al: Lin28B/Let-7
regulates expression of Oct4 and Sox2 and reprograms oral squamous
cell carcinoma cells to a stem-like state. Cancer Res.
75:2553–2565. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Liu Y, Li H, Feng J, Cui X, Huang W, Li Y,
Su F, Liu Q, Zhu J, Lv X, et al: Lin28 induces
epithelial-to-mesenchymal transition and stemness via
downregulation of let-7a in breast cancer cells. PLoS One.
8:e830832013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Fu X, Meng Z, Liang W, Tian Y, Wang X, Han
W, Lou G, Wang X, Lou F, Yen Y, et al: miR-26a enhances miRNA
biogenesis by targeting Lin28B and Zcchc11 to suppress tumor growth
and metastasis. Oncogene. 33:4296–4306. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Roos M, Pradère U, Ngondo RP, Behera A,
Allegrini S, Civenni G, Zagalak JA, Marchand JR, Menzi M, Towbin H,
et al: A small-molecule inhibitor of Lin28. ACS Chem Biol.
11:2773–2781. 2016. View Article : Google Scholar : PubMed/NCBI
|