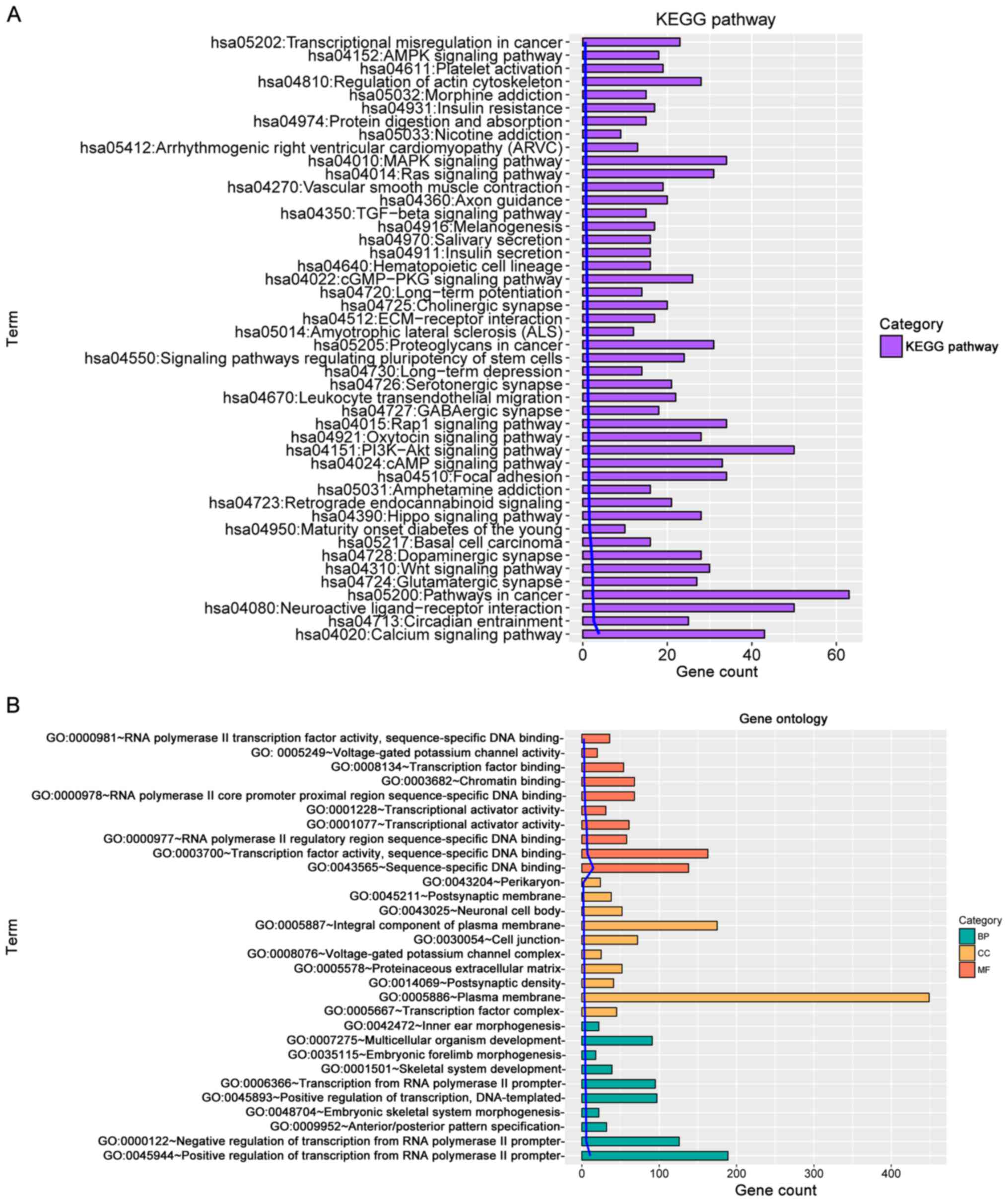
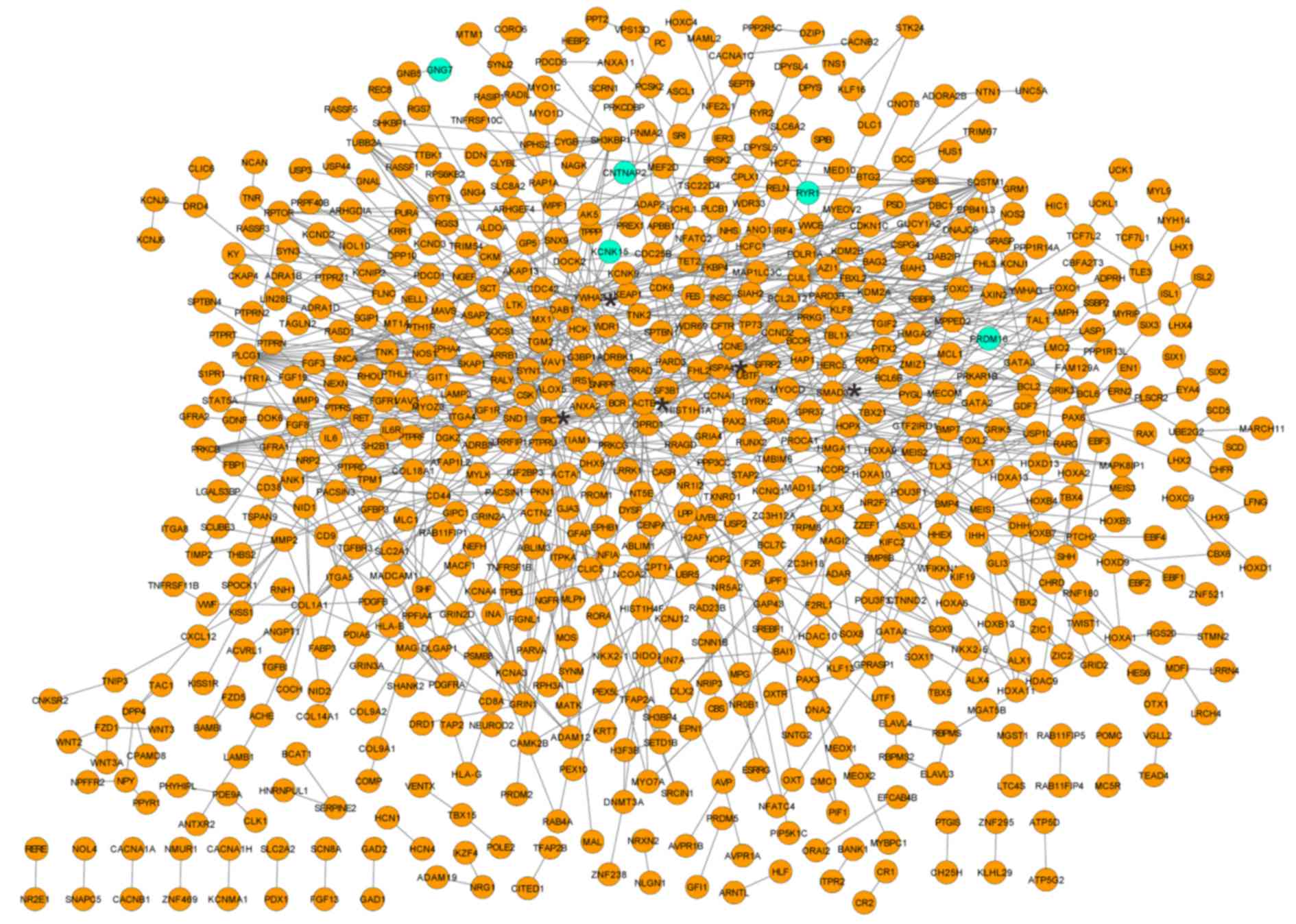
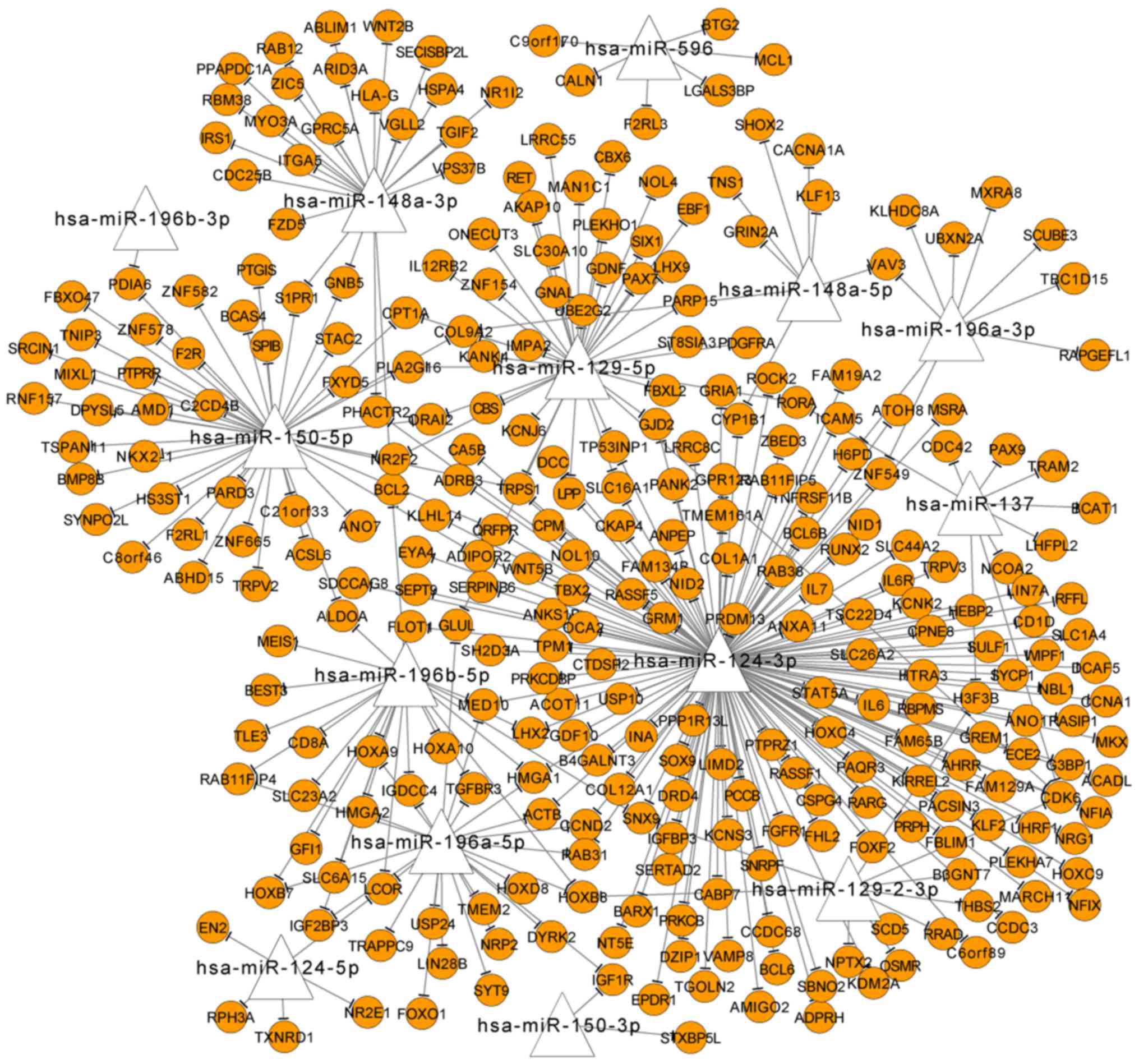
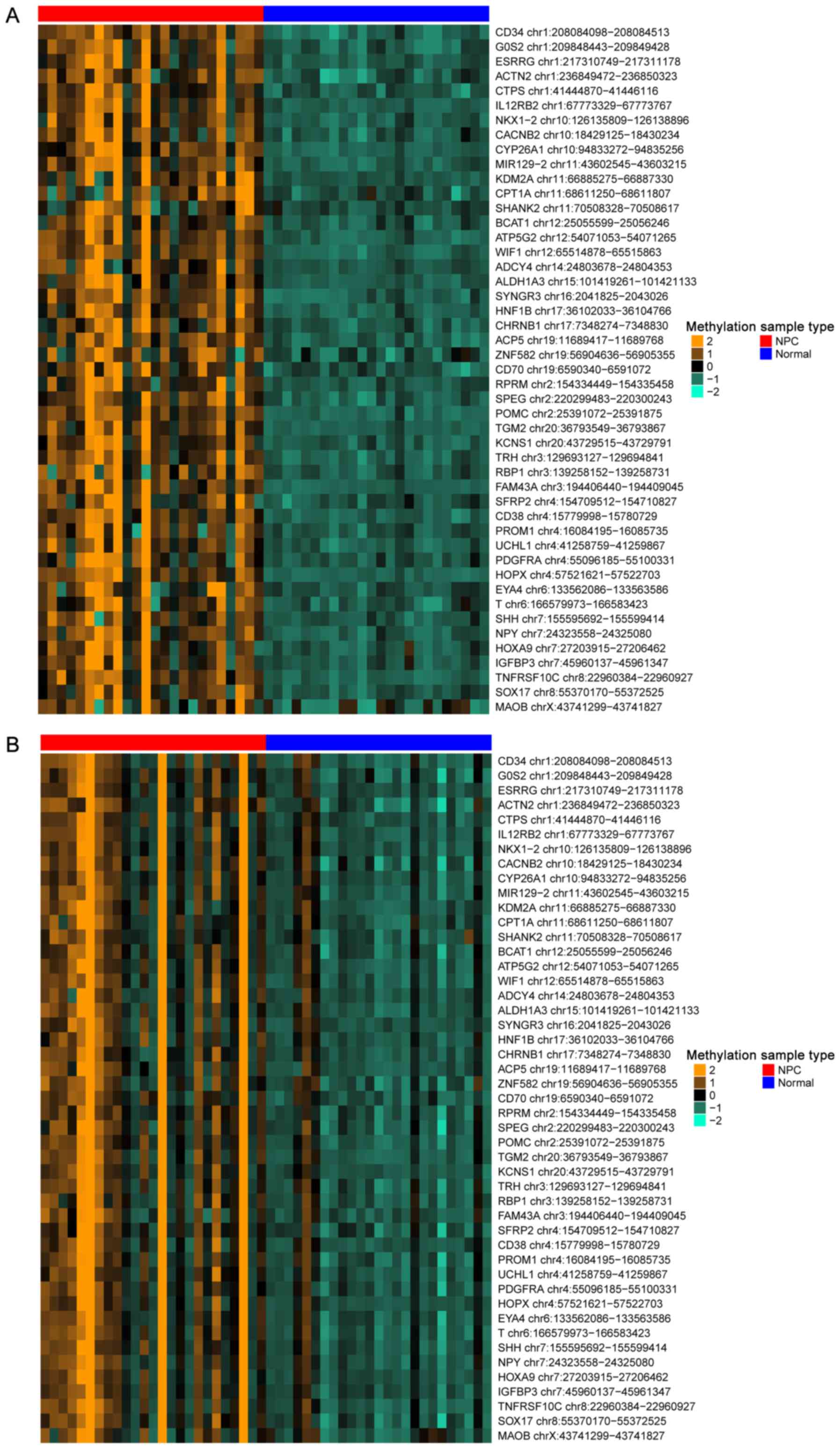
|
1
|
Tang LL, Chen WQ, Xue WQ, He YQ, Zheng RS,
Zeng YX and Jia WH: Global trends in incidence and mortality of
nasopharyngeal carcinoma. Cancer Lett. 374:22–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chang ET and Adami HO: The enigmatic
epidemiology of nasopharyngeal carcinoma. Cancer Epidemiol
Biomarkers Prev. 15:1765–1777. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tay JK, Chan SH, Lim CM, Siow CH, Goh HL
and Loh KS: The role of Epstein-Barr virus DNA load and serology as
screening tools for nasopharyngeal carcinoma. Otolaryngol Head Neck
Surg. 155:274–280. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Dai W, Zheng H, Cheung AK, Tang CS, Ko JM,
Wong BW, Leong MM, Sham PC, Cheung F, Kwong DL, et al: Whole-exome
sequencing identifies MST1R as a genetic susceptibility gene in
nasopharyngeal carcinoma. Proc Natl Acad Sci USA. 113:pp.
3317–3322. 2016; View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chan KC: Plasma Epstein-Barr virus DNA as
a biomarker for nasopharyngeal carcinoma. Chin J Cancer.
33:598–603. 2014.PubMed/NCBI
|
|
6
|
Jia W, Ren C, Wang L, Zhu B, Jia W, Gao M,
Zeng F, Zeng L, Xia X, Zhang X, et al: CD109 is identified as a
potential nasopharyngeal carcinoma biomarker using aptamer selected
by cell-SELEX. Oncotarget. 7P:1–55342. 2016.
|
|
7
|
Liu ZH, Hu JL, Liang JZ, Zhou AJ, Li MZ,
Yan SM, Zhang X, Gao S, Chen L, Zhong Q and Zeng MS: Far upstream
element-binding protein 1 is a prognostic biomarker and promotes
nasopharyngeal carcinoma progression. Cell Death Dis. 6:e19202015.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lu J, Xu X, Liu X, Peng Y, Zhang B, Wang
L, Luo H, Peng X, Li G, Tian W, et al: Predictive value of miR-9 as
a potential biomarker for nasopharyngeal carcinoma metastasis. Br J
Cancer. 110:392–398. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tian F, Yip SP, Kwong DL, Lin Z, Yang Z
and Wu VW: Promoter hypermethylation of tumor suppressor genes in
serum as potential biomarker for the diagnosis of nasopharyngeal
carcinoma. Cancer Epidemiol. 37:708–713. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Feng X, Ren C, Zhou W, Liu W, Zeng L, Li
G, Wang L, Li M, Zhu B, Yao K and Jiang X: Promoter
hypermethylation along with LOH, but not mutation, contributes to
inactivation of DLC-1 in nasopharyngeal carcinoma. Mol Carcinog.
53:858–870. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xiao X, Zhao W, Tian F, Zhou X, Zhang J,
Huang T, Hou B, Du C, Wang S, Mo Y, et al: Cytochrome b5 reductase
2 is a novel candidate tumor suppressor gene frequently inactivated
by promoter hypermethylation in human nasopharyngeal carcinoma.
Tumor Biol. 35:3755–3763. 2014. View Article : Google Scholar
|
|
12
|
Jiang W, Liu N, Chen XZ, Sun Y, Li B, Ren
XY, Qin WF, Jiang N, Xu YF, Li YQ, et al: Genome-wide
identification of a methylation gene panel as a prognostic
biomarker in nasopharyngeal carcinoma. Mol Cancer Ther.
14:2864–2873. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Dai W, Cheung AK, Ko JM, Cheng Y, Zheng H,
Ngan RK, Ng WT, Lee AW, Yau CC, Lee VH and Lung ML: Comparative
methylome analysis in solid tumors reveals aberrant methylation at
chromosome 6p in nasopharyngeal carcinoma. Cancer Med. 4:1079–1090.
2015. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Teschendorff AE, Marabita F, Lechner M,
Bartlett T, Tegner J, Gomez-Cabrero D and Beck S: A beta-mixture
quantile normalization method for correcting probe design bias in
Illumina Infinium 450 k DNA methylation data. Bioinformatics.
29:189–196. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Warden CD, Lee H, Tompkins JD, Li X, Wang
C, Riggs AD, Yu H, Jove R and Yuan YC: COHCAP: An integrative
genomic pipeline for single-nucleotide resolution DNA methylation
analysis. Nucleic Acids Res. 41:e1172013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sproul D, Nestor C, Culley J, Dickson JH,
Dixon JM, Harrison DJ, Meehan RR, Sims AH and Ramsahoye BH:
Transcriptionally repressed genes become aberrantly methylated and
distinguish tumors of different lineages in breast cancer. Proc
Natl Acad Sci USA. 108:pp. 4364–4369. 2011; View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Huang DW, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43(Database issue): D447–D452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Köhler S, Bauer S, Horn D and Robinson PN:
Walking the interactome for prioritization of candidate disease
genes. Am J Hum Genet. 82:949–958. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Dweep H and Gretz N: miRWalk2.0: A
comprehensive atlas of microRNA-target interactions. Nat Methods.
12:6972015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Davis AP, Murphy CG, Johnson R, Lay JM,
Lennon-Hopkins K, Saraceni-Richards C, Sciaky D, King BL,
Rosenstein MC, Wiegers TC and Mattingly CJ: The comparative
toxicogenomics database: Update 2013. Nucleic Acids Res.
41(Database issue): D1104–D1114. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Venza M, Visalli M, Catalano T, Beninati
C, Teti D and Venza I: DSS1 promoter hypomethylation and
overexpression predict poor prognosis in melanoma and squamous cell
carcinoma patients. Hum Pathol. 60:137–146. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ke L, Xiang Y, Guo X, Lu J, Xia W, Yu Y,
Peng Y, Wang L, Wang G, Ye Y, et al: c-Src activation promotes
nasopharyngeal carcinoma metastasis by inducing the
epithelial-mesenchymal transition via PI3K/Akt signaling pathway: A
new and promising target for NPC. Oncotarget. 7:28340–28355. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bo H, Gong Z, Zhang W, Li X, Zeng Y, Liao
Q, Chen P, Shi L, Lian Y, Jing Y, et al: Upregulated long
non-coding RNA AFAP1-AS1 expression is associated with progression
and poor prognosis of nasopharyngeal carcinoma. Oncotarget.
6:20404–20418. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Javelaud D and Mauviel A: Crosstalk
mechanisms between the mitogen-activated protein kinase pathways
and Smad signaling downstream of TGF-beta: implications for
carcinogenesis. Oncogene. 24:5742–5750. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xiao J, Xiang Q, Xiao YC, Su ZJ, Huang ZF,
Zhang QH, Tan Y, Li XK and Huang YD: The effect of transforming
growth factor-beta1 on nasopharyngeal carcinoma cells: Insensitive
to cell growth but functional to TGF-beta/Smad pathway. J Exp Clin
Cancer Res. 29:352010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Huang H, Sun P, Lei Z, Li M, Wang Y, Zhang
HT and Liu J: miR-145 inhibits invasion and metastasis by directly
targeting Smad3 in nasopharyngeal cancer. Tumour Biol.
36:4123–4131. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Busque L, Belisle C, Provost S, Giroux M
and Perreault C: Differential expression of SMAD3 transcripts is
not regulated by cis-acting genetic elements but has a gender
specificity. Genes Immun. 10:192–196. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Silva J, Teixeira AL, Lobo F, Maurício J
and Medeiros R: DNA repair system and prostate cancer progression:
The role of NBS1 polymorphism (rs1805794). DNA Cell Biol.
31:1182–1186. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ghorbani MJ, Salehi Z, Sabet EE and
Ejtehadi F: Anatlysis of HSPA1B A1267G gene polymorphism in peptic
ulcer. Mol Biol (Mosk). 48:728–732. 2014.(In Russian). View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chen HC, Chen GH, Chen YH, Liao WL, Liu
CY, Chang KP, Chang YS and Chen SJ: MicroRNA deregulation and
pathway alterations in nasopharyngeal carcinoma. Br J Cancer.
100:1002–1011. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang JP, Zeng C, Xu L, Gong J, Fang JH
and Zhuang SM: MicroRNA-148a suppresses the epithelial-mesenchymal
transition and metastasis of hepatoma cells by targeting Met/Snail
signaling. Oncogene. 33:4069–4076. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jarczak J, Kaba J and Bagnicka E: The
validation of housekeeping genes as a reference in quantitative
real time PCR analysis: Application in the milk somatic cells and
frozen whole blood of goats infected with caprine arthritis
encephalitis virus. Gene. 549:280–285. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yan Q, Zeng Z, Gong Z, Zhang W, Li X, He
B, Song Y, Li Q, Zeng Y, Liao Q, et al: EBV-miR-BART10-3p
facilitates epithelial-mesenchymal transition and promotes
metastasis of nasopharyngeal carcinoma by targeting BTRC.
Oncotarget. 6:41766–41782. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chen CH, Chuang SM, Yang MF, Liao JW, Yu
SL and Chen JJ: A novel function of YWHAZ/β-catenin axis in
promoting epithelial-mesenchymal transition and lung cancer
metastasis. Mol Cancer Res. 10:1319–1331. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Maia BM, Rocha RM and Calin GA: Clinical
significance of the interaction between non-coding RNAs and the
epigenetics machinery: Challenges and opportunities in oncology.
Epigenetics. 9:75–80. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Vymetalkova V, Vodicka P, Pardini B, Rosa
F, Levy M, Schneiderova M, Liska V, Vodickova L, Nilsson TK and
Farkas SA: Epigenome-wide analysis of DNA methylation reveals a
rectal cancer-specific epigenomic signature. Epigenomics.
8:1193–1207. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Bandres E, Agirre X, Bitarte N, Ramirez N,
Zarate R, Roman-Gomez J, Prosper F and Garcia-Foncillas J:
Epigenetic regulation of microRNA expression in colorectal cancer.
Int J Cancer. 125:2737–2743. 2009. View Article : Google Scholar : PubMed/NCBI
|