Introduction
Cervical cancer is the second most common cancer
among women worldwide and a leading cause of mortality in women
(1). The most important risk
factor is infection with oncogenic high-risk human papillomaviruses
(HPVs), including HPV16 and HPV18 (2). Developments in the diagnosis and
treatment in cervical cancer have been observed; however, the
five-year survival rate is ~65% in patients with lymph node
metastases (3). Thus, it is
clinically important to investigate the molecular mechanism of
metastasis of cervical cancer.
MicroRNAs (miRNAs/miRs) are small non-coding RNA
sequences with ~18–25 nucleotides that modulate translational
efficiency or stability by targeting the 3′-untranslated region
(3′-UTR) of mRNA. Increasing evidence has demonstrated that miRNAs
serve a key role in the progression and oncogenesis of a variety of
cancers, including cervical cancer (4), gastric cancer (5), lung cancer (6) and breast cancer (7). In cervical cancer, numerous miRNAs
were demonstrated to be involved in cancer initiation, promotion
and progression. miR-506 acts as a tumor suppressor by inhibiting
cervical cancer growth (8).
miR-100 is downregulated in cervical cancer and may regulate cell
proliferation, cycle and apoptosis (9). miR-424 functions as a tumor
suppressor by inhibiting cell proliferation, migration and invasion
of cervical cancer (10). In
addition, Wang et al (4)
reported that miR-378 was aberrantly upregulated in
HPV16/18-positive cervical cancer and the increase of miR-378 may
also be induced by oncoprotein E6 and/or E7; however, the detailed
mechanism of miR-378 in cervical cancer requires further
investigation.
Autophagy-related protein 12 (ATG12) is a member of
the ATG family associated with autophagy. Numerous core ATGs in two
ubiquitin-like conjugation systems are essential for autophagosome
formation. In the first system, ATG12 is activated by ATG7,
transferred to ATG10 and ultimately attached to ATG5 (11). In the second system,
microtubule-associated protein 1 light chain 3 conjugates to lipid
phosphatidylethamine by ATG7 and ATG3. In addition to ATG5, a
recent study demonstrated that ATG12 also conjugates to ATG3
(12). The ATG12-ATG3 complex
promotes autolysosome formation under nutrient-rich conditions and
ATG12-ATG3 interacts with apoptosis-linked gene 2-interacting
protein X to promote basal autophagic flux and late endosome
function (12). Thus, these
previous studies indicated that ATG12 may serve a key role in
autophagy; however, the association between miR-378 and ATG12
remains to be investigated.
In the present study, miRNA-378 expression levels
were upregulated in cervical intraepithelial neoplasia (CIN) III
and cervical cancer tissues. Upregulation of the miRNA-378
expression levels significantly promoted cervical cancer migration
and invasion in vitro and in vivo. Furthermore, ATG12
was identified as a functional and direct target of miR-378. These
findings may provide novel insights into the molecular mechanism of
metastasis in cervical cancer.
Materials and methods
Tissues and cell lines
A total of 185 female patients (Table I) were involved in the present
study between January 2012 to January 2016 at the Department of
Gynecology, Qilu Hospital of Shandong University (Shandong, China).
In total, 60 normal cervix tissues were collected from patients who
underwent a hysterectomy due to benign gynecological diseases.
Additionally, 70 CIN III tissues were obtained from patients who
received cold-knife conization surgery and 55 cervical cancer
tissues were collected from patients who underwent radical
hysterectomy and were diagnosed with squamous cervical carcinoma.
Tissues were frozen in liquid nitrogen and stored at −80°C. Written
informed content was obtained from all patients and the present
study was approved by the Ethics Committee of Qilu Hospital of
Shandong University.
 | Table I.Main characteristics of patients
enrolled in the present study. |
Table I.
Main characteristics of patients
enrolled in the present study.
| Characteristic | Squamous carcinoma of
cervix | CIN III | N |
|---|
| Cases (n) | 55 | 70 | 60 |
| Age (years) | 44.2±7.2 | 39.6±7.3 | 48.8±5.6 |
| Surgery received | Radical
hysterectomy | CKC | Hysterectomy |
| Stage | IB-IIA | NA | NA |
| Lymph node
metastasis (n) |
|
|
|
|
Positive | 17 | NA | NA |
|
Negative | 38 | NA | NA |
The 2 cervical cancer cell lines, HeLa and C-33A,
were employed in the present study. HeLa and C-33A cells were
cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) with 10% fetal bovine serum (FBS; HyClone;
GE Healthcare Life Sciences, Logan, UT, USA) and 1%
penicillin/streptomycin. All cells were incubated in a humidified
incubator with 5% CO2 at 37°C.
Oligonucleotide construction and
transfection
To investigate the biological function of miR-378 in
cell migration and invasion, GIPZ-miR-378 was transfected into
C-33A cells to overexpress miR-378, and the miR-378 inhibitor was
transfected into HeLa cells to inhibit miR-378. Oligonucleotides
including miR-378 mimics (forward, 5′-ACUGGACUUGGAGUCAGAAGG-3′ and
reverse, 5′-UUCUGACUCCAAGUCCAGUUU-3′), miR-378 inhibitor
(5′-CCUUCUGACUCCAAGUCCAGU-3′) and mimic negative controls (forward,
5′-UUCUCCGAACGUGUCACGUTT-3′ and reverse,
5′-ACGUGACACGUUCGGAGAATT-3′); inhibitor negative control
(5′-CAGUACUUUUGUGUAGUACAA-3′) were purchased from Dharmacon
(Lafayette, CO, USA). RNA oligonucleotides were transfected at a
final concentration of 50 nM using Lipofectamine 2000 (Invitrogen,
Carlsbad, California, USA) according to the manufacturer's
protocol. Small interfering (si)RNA-ATG12 and scrambled negative
control were supplied by Shanghai GenePharma Co., Ltd. (Shanghai,
China). The sequences of the siRNA-ATG12 and negative control are
as follows: siRNA-ATG12 (forward, 5′-GUUGCAGCUUCCUACUUCATT-3′;
reverse, 5′-UGAAGUAGGAAGCUGAACTT-3′), scrambled negative control
siRNA was 5′-ACTACCGTTGTTATAGGTG-3′. The miR-378 overexpression
construct (GIPZ-miR-378) and control (GIPZ-NC) were provided by
Guangzhou Ribobio Co., Ltd. (Guangzhou, China). The multiplicity of
infection (MOI) of HeLa and C-33A cell was tested at 10, 20, 40 and
80 using lentiviral transfection. The optimum MOI of HeLa was 10
and MOI of C-33A was 20. Subsequent experiments were performed 48 h
after infection.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from cells or tissues using
TRIzol reagents (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocols. Reverse-transcribed
complementary DNA was synthesized at 37°C for 15 min and 80°C for 5
sec using a PrimeScript RT reagent kit (Takara Biotechnology, Co.,
Ltd., Dalian, China). Quantitative real-time PCR was performed with
StepOne plus (Applied Biosystems, Foster City, CA, USA) using SYBR
Premix ExTaq (Takara Biotechnology, Dalian, China). For miR-378
detection, U6 was used as an internal reference of miRNA. GAPDH was
used as an internal reference of mRNA. The following primers were
used: ATG12 (forward, 5′-CTGCTGGCGACACCAAGAAA; reverse
5′-CGTGTTCGCTCTACTGCCC-3′), miR-378 (forward,
5′-GGGACTGGACTTGGAGTCA-3′; reverse, 5′-GTGCGTGTCGTGGAGTCG-3′), U6
(forward 5′-CTCGCTTCGGCAGCACA-3′; reverse,
5′-AACGCTTCACGAATTTGCGT-3′), GAPDH (forward,
5′-GGAGCGAGATCCCTCCAAAAT-3′; reverse
5′-GGCTGTTGTCATACTTCTCATGG-3′).
The following thermocycling conditions were used for
detection of mRNAs: Initial denaturation at 95°C for 5 sec; 40
cycles of 95°C for 30 sec, 62°C for 30 sec and 72°C for 30 sec. The
following thermocycling conditions were used for detection of miRs:
Initial denaturation at 95°C for 15 sec, 40 cycles of 94°C for 15
sec, 55°C for 30 sec and 70°C for 30 sec. All reactions were run in
triplicate. The 2−ΔΔCq method was used for
quantification (13).
Western blot analysis
A total of 5×106 HeLa cells and
8×106 C-33A cells were harvested and lysed using
radioimmunoprecipitation assay lysis buffer (Beyotime Institute of
Biotechnology, Shanghai, China). The concentration of cell protein
was measured with a Bicinchoninic Acid Protein Assay kit (Bio-Rad
Laboratories, Inc., Hercules CA, USA). A total of 30 µg/lane whole
cell protein was separated by 10% SDS-PAGE and blotted onto
nitrocellulose membranes (EMD Millipore, Billerica, MA, USA) and
blocked in Tris-buffered saline containing 0.1% Tween-20 with 5%
non-fat milk at room temperature for 1 h. The membranes were then
incubated with the following primary antibodies: CCND1 (1:2,000;
cat. no. ab21699; Abcam, Cambridge, MA, USA), phosphorylated-Rb
(Ser608; 1:5,000; cat. no. 8145S), ATG12 (1:2,000; cat. no. 4180T)
(both from Cell Signaling Technology, Inc., Danvers, MA, USA) and
β-actin (1:10,000, cat. no. ab49900; Abcam) at 4°C overnight,
followed by incubation with the horseradish peroxidase(HRP)
conjugated secondary antibodies (anti-mouse IgG; 1:10,000; cat. no.
7076; and anti-rabbit IgG; 1:10,000; cat. no. 7074; both from Cell
Signaling Technology, Inc.) at room temperature for 2 h. Protein
bands were visualized using the SuperSignal West Pico
chemiluminescent substrate (Pierce; Thermo Fisher Scientific,
Inc.). Protein band intensity was quantified by using the ImageJ
(version 1.6; National Institutes of Health, Bethesda, MD,
USA).
Luciferase reporter assay
To investigate the downstream targets of miR-378,
putative targets were analyzed using TargetScan (release 3.1;
www.targetscan.org/mamm_31/). For the
luciferase reporter assay, the pmirGLO dual-luciferase miRNA target
expression vector (Promega Corporation, Madison, WI, USA) was
employed in the present study to demonstrate the target of miR-378.
Using an Effectene transfection reagent according to the
manufacturer's protocol (Qiagen GmbH, Hilden, Germany), HeLa and
C-33A cells were co-transfected with pmirGLO-ATG12-3′-UTR-wild-type
(WT) or pmirGLO-ATG12-3′-UTR-mutant (MUT) plus miR-378 mimic or
negative control. MUT 3′-UTR was generated using QuikChange
Site-Directed Mutagenesis kit (Stratagene; Agilent Technologies,
Inc., Santa Clara, CA, USA). Luciferase activity was detected 48 h
post-cell transfection using the GloMax fluorescence reader
(Promega Corporation). Renilla luciferase was used as the
internal control.
Immunohistochemistry (IHC) assay
Tissues were fixed in 4% paraformaldehyde for 24 h
at 4°C, paraffin embedded and cut into 4 µm sections. Slides were
deparaffinized and rehydrated using xylene and descending alcohol
series. Antigen retrieval was performed at 95°C for 5 min in sodium
citrate buffer (10 mM sodium citrate; 0.05% Tween-20; pH 6.0) and
washed 3 times for 2 min with PBS. Then slides were blocked with 3%
hydrogen peroxidase in methanol and 10% goat serum (Beijing
Solarbio Science & Technology Co., Ltd., Beijing, China) for 2
h at room temperature. Slides were incubated with ATG12 antibody
(1:500; cat. no. NBP2-15501, Novus Biologicals, LLC, Littleton, CO,
USA) overnight at 4°C and then with HRP-labeled goat anti-rabbit
IgG (1:50; cat. no. A0208; Beyotime Institute of Biotechnology)
antibody for 2 h at room temperature. Streptavidin-horseradish
peroxidase was added, and sections were stained with
3,3′-diaminobenzidine(DAB) substrate for 3 min and counterstained
with hematoxylin 30 sec at room temperature. For blank controls,
the primary antibodies were replaced with PBS solution (100 mM; pH
7.4). All slides were evaluated by two independent investigators
without prior knowledge of the clinical data of the specimens.
According to staining intensity, specimens were scored as absent
(0), weak (1), moderate (2) or intense (3). The tissues with score
0 and 1 were defined as negatively stained, while 2 and 3 defined
positive staining of ATG12. The slides were analyzed and imaged
using a bright-field microscope (Olympus BX50; Olympus Corporation,
Tokyo, Japan).
Cell migration and invasion
Cell migration and invasion potential was evaluated
using Transwell inserts (8 µm pores; BD Biosciences, San Jose, CA,
USA). All the experiments were repeated 3 times. C-33A/HeLa cells
transfected with si-ATG12, GIPZ-miR-378 and miR-378 inhibitor were
pre-cultured in RPMI-1640 medium without FBS at 37°C for 24 h in
12-well plates. For the migration assay, 1×105 cells
were placed in the top chamber of Transwell plates and incubated at
37°C for 24 h with serum-free RPMI-1640 medium. For the invasion
assay, 1×105 cells were placed in the top chamber of
Transwell plates, which were pre-coated with extracellular matrix
gel (Matrigel) (BD Biosciences) and incubated at 37°C for 24 h with
serum-free medium; RPMI-1640 medium with 15% FBS (HyClone; GE
Healthcare Life Sciences) was added to the lower chamber as a
chemoattractant. Following 48 h incubation, cells remaining on the
upper surface were wiped off using cotton swabs and cells on the
lower surface were fixed in 4% paraformaldehyde for 30 min at room
temperature and stained with 0.1% crystal violet for 30 min at room
temperature, and counted under a light microscope (magnification,
×100) in five random visual fields.
Metastasis assay in vivo
Immunodeficient, 5-week old female BALB/c nu/nu mice
(Vital River Laboratory Animal Technology Co., Ltd, Beijing, China)
were used for the metastasis assay in vivo. A total of 20
mice (10 in GIPZ-miR-378 group and 10 GIPZ-NC group) were kept
under specific pathogen free conditions, 12-h light/dark cycle at
20–30°C and all of mice had free access to food and water. A total
of 2×106 C-33A cells transfected with GIPZ-miR-378 or
GIPZ-NC were injected into mice via a tail vein injection.
Following 8 weeks, mice were sacrificed and the lungs were
dissected and fixed using 10% formalin for 24 h at 4°C and paraffin
embedded. The paraffin blocks were cut into 4 µm thick slices and
hematoxylin and eosin staining was performed. Hematoxylin staining
was performed for 45 sec and eosin staining for 30 sec, both at
room temperature. Histological examination was confirmed under
bright-field microscope. All animal experiments were approved by
the Ethics Committee of Qilu Hospital of Shandong University.
Statistical analysis
Statistical analysis was performed using SPSS
software (version 19.0; IBM Corp., Armonk, NY, USA). Differences in
miR-378 expression levels between groups were examined using
one-way analysis of variance, followed by a Tukey's post hoc test.
Comparisons in cell density, the relative levels of protein
expression and the number of tumor foci between the two different
transfection groups were presented as the mean ± standard deviation
from at least three independent experiments and were analyzed using
an unpaired Student's t-test. P<0.05 was considered to indicate
a statistically significant difference.
Results
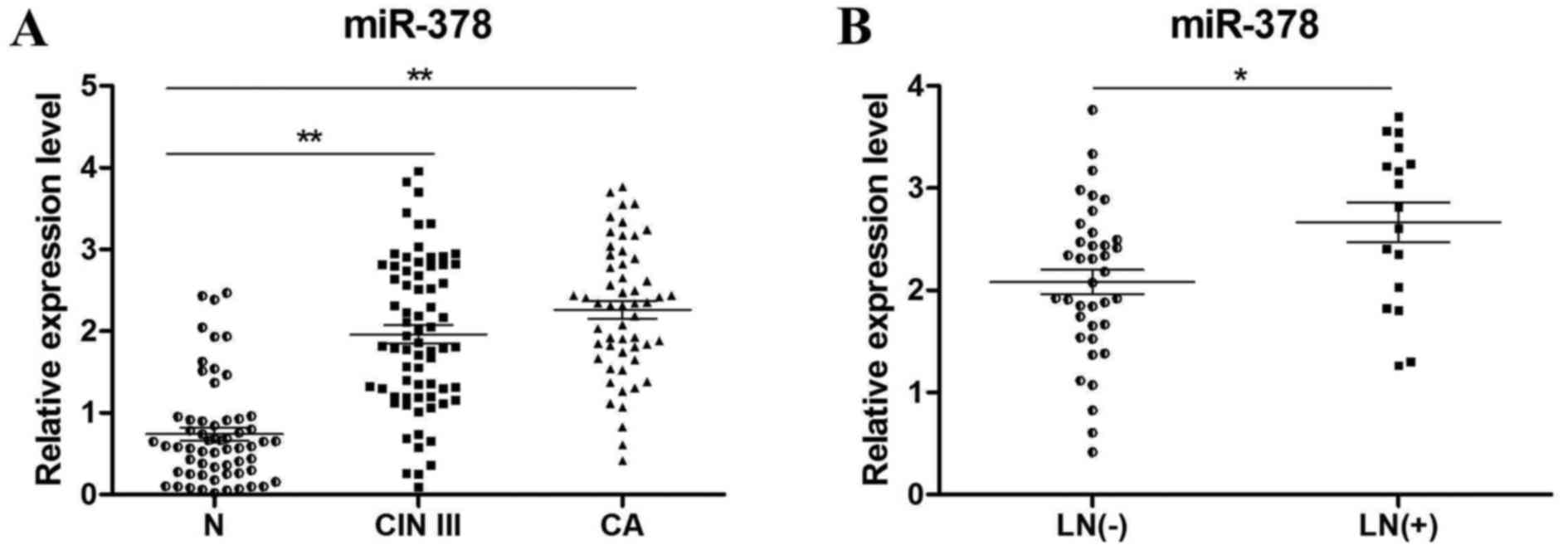
miR-378 is frequently upregulated in
CIN III and lymph node positive cervical cancer tissues
Patients enrolled in the present study included 3
subgroups, which were normal cervix (N), CIN III and cervical
carcinoma (CA). The clinicopathological characteristics of these
tissues were listed in Table I. To
determine the expression levels of miR-378, RT-qPCR of N (n=60),
CIN III (n=70) and cervical cancer tissues (n=55) was performed.
The results revealed that miR-378 expression was significantly
increased in CIN III (P<0.01) and CA tissues (P<0.01)
compared with N tissues (Fig. 1A).
In addition, miR-378 expression levels in lymph node positive
(n=17) and negative (n=38) cancer tissues were compared (Fig. 1B), which revealed that the positive
group with lymph node metastasis exhibited significantly a higher
expression of miR-378 (P<0.05).
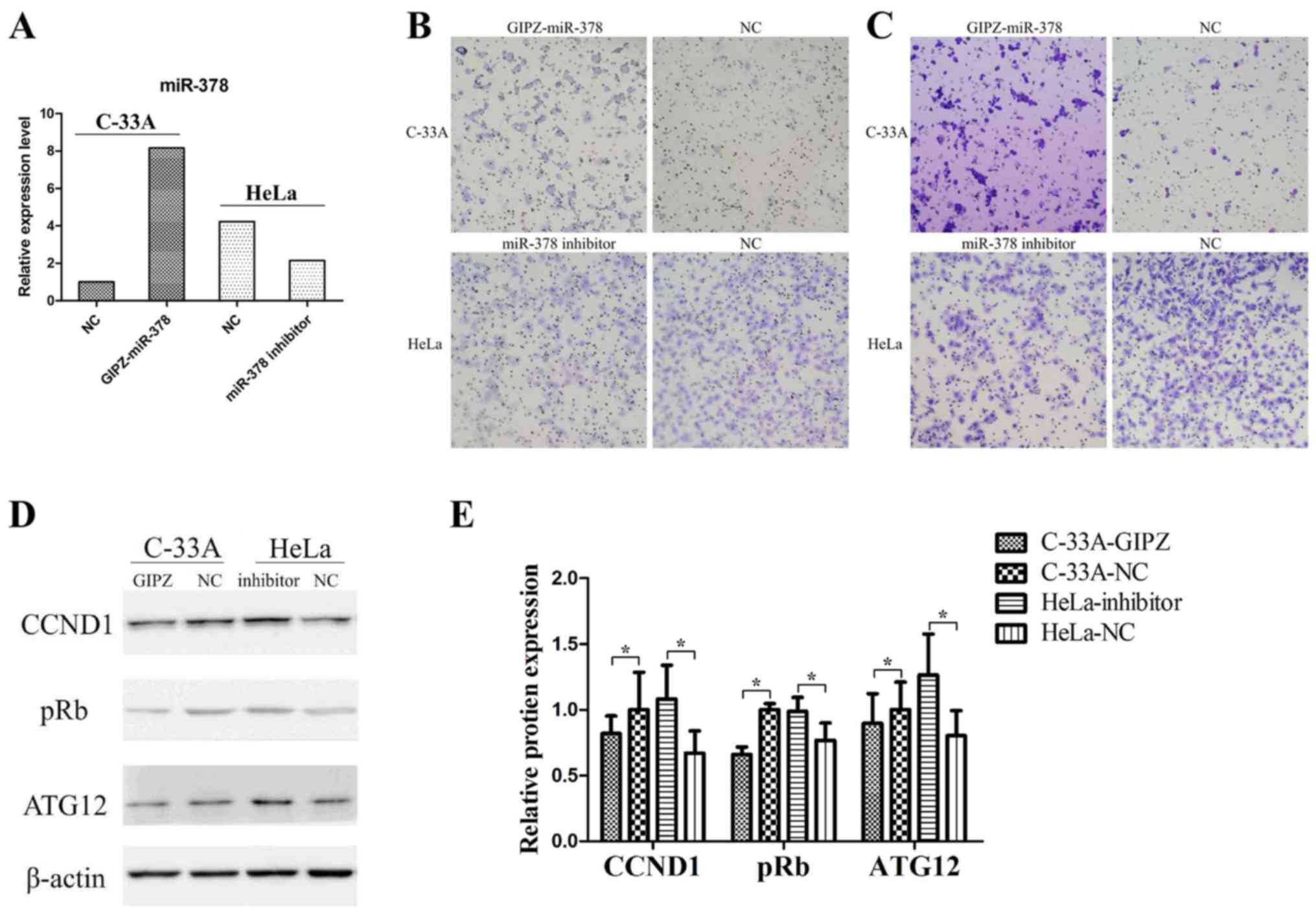
Overexpression of miR-378
significantly promotes cell migration and invasion in vitro
In the two cervical cancer cell lines, miR-378 basic
expression levels were detected by RT-qPCR analysis. The successful
overexpression or inhibition of miR-378 was determined by RT-qPCR
analysis (Fig. 2A). In the
Transwell assay, overexpression of miR-378 significantly promoted
cell migration and invasion in C-33A cells (Fig. 2B and C), and inhibition of miR-378
significantly suppressed migratory and invasive abilities in HeLa
cells (Fig. 2B and C).
In addition, the relative expression levels of
ATG12, cyclin D1 (CCND1) and retinoblastoma (pRb) in C-33A cells
and HeLa cells were detected (Fig. 2D
and E). Overexpression of miR-378 in C-33A cells significantly
decreased the expression of ATG12, CCND1 and pRb. In addition, the
inhibition of miR-378 in HeLa cells increased the expression levels
of ATG12, CCND1 and pRb. These findings indicated that miR-378 may
have promoted cell migration and invasion in vitro and
regulated the expression of numerous key genes.
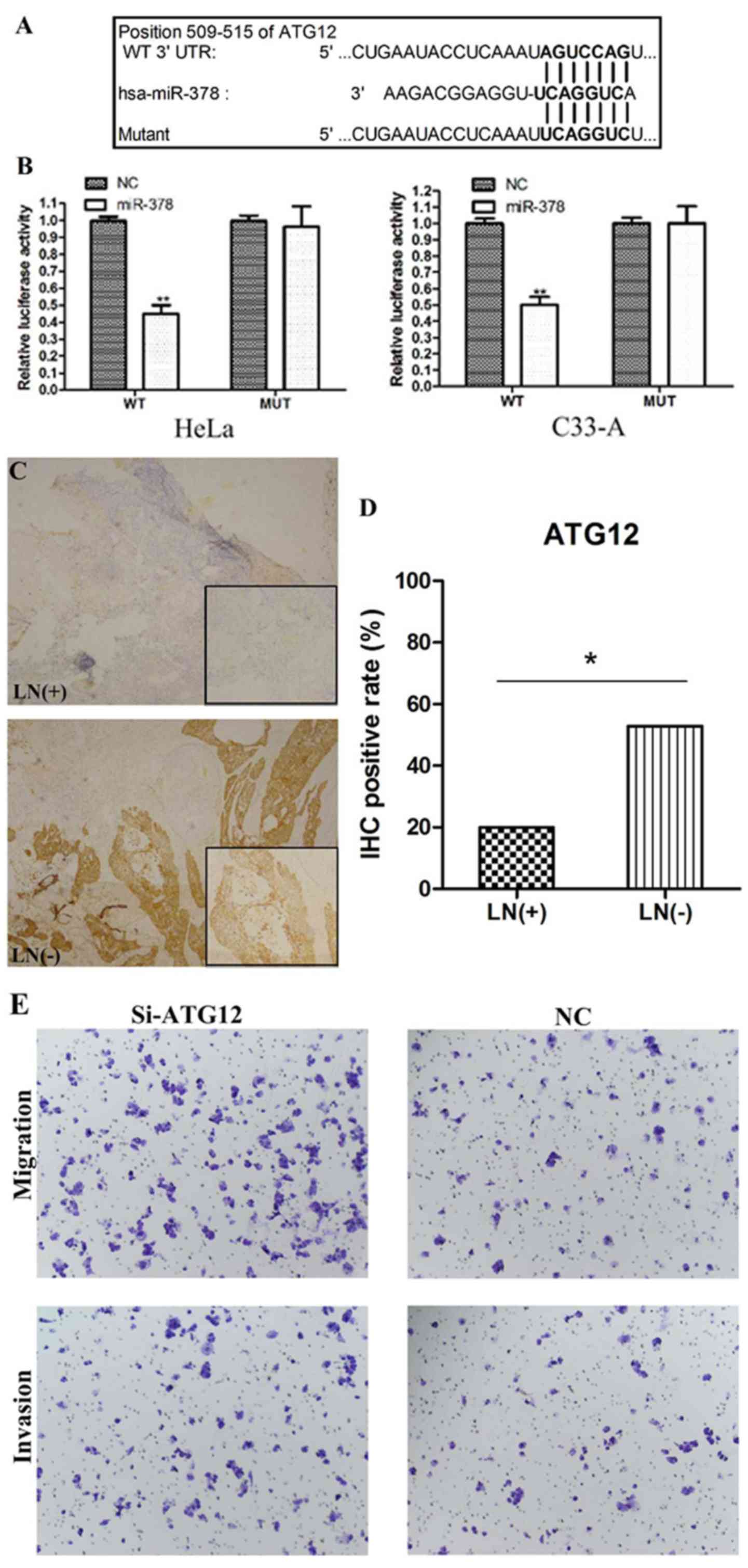
ATG12 is a direct target of miR-378
and regulates cell metastasis in cervical cancer
To investigate the downstream targets of miR-378,
putative targets were analyzed using TargetScan. One conserved
putative binding site was reported at 509–515 bp of ATG12 3′-UTR
(Fig. 3A). To verify the putative
target, a luciferase reporter assay was performed in C-33A and HeLa
cells. When cells were co-transfected with miR-378 mimic and
pmirGLO-ATG12-3′-UTR-WT, the relative luciferase activity was
significantly decreased compared with the miR-control (P<0.01)
(Fig. 3B). The luciferase activity
was rescued when cells were co-transfected with miR-378 mimic and
pmirGLO-ATG12-3′-UTR-MUT (Fig.
3B). These findings indicated that ATG12 may be a direct target
of miR-378.
To further validate whether ATG12 is downregulated
by miR-378 in cancer tissues, the expression of ATG12 in lymph node
positive and negative cervical cancer samples was detected by IHC
analysis (Fig. 3C). ATG12
expression levels between the two groups were significantly
different as they were upregulated in the lymph node negative group
(52.9% positive) compared with in the lymph node positive group
(20% positive; P<0.05; Fig.
3D). Combined with the data presented in Fig. 1B, the results indicated that ATG12
expression levels were inversely associated with miR-378 expression
in cervical cancer cases with or without lymph node metastasis.
To evaluate the function of ATG12 in cell
metastasis, in vitro migration and invasion assays were
conducted using C33-A cells. Cells transfected with si-ATG12
exhibited decreased expression of ATG12 mRNA and protein, compared
with cells transfected with the negative control (data not shown).
Suppression of ATG12 notably promoted cell migration and invasion
in C-33A cells (Fig. 3E)
suggesting that ATG12 may participate in cell metastasis.
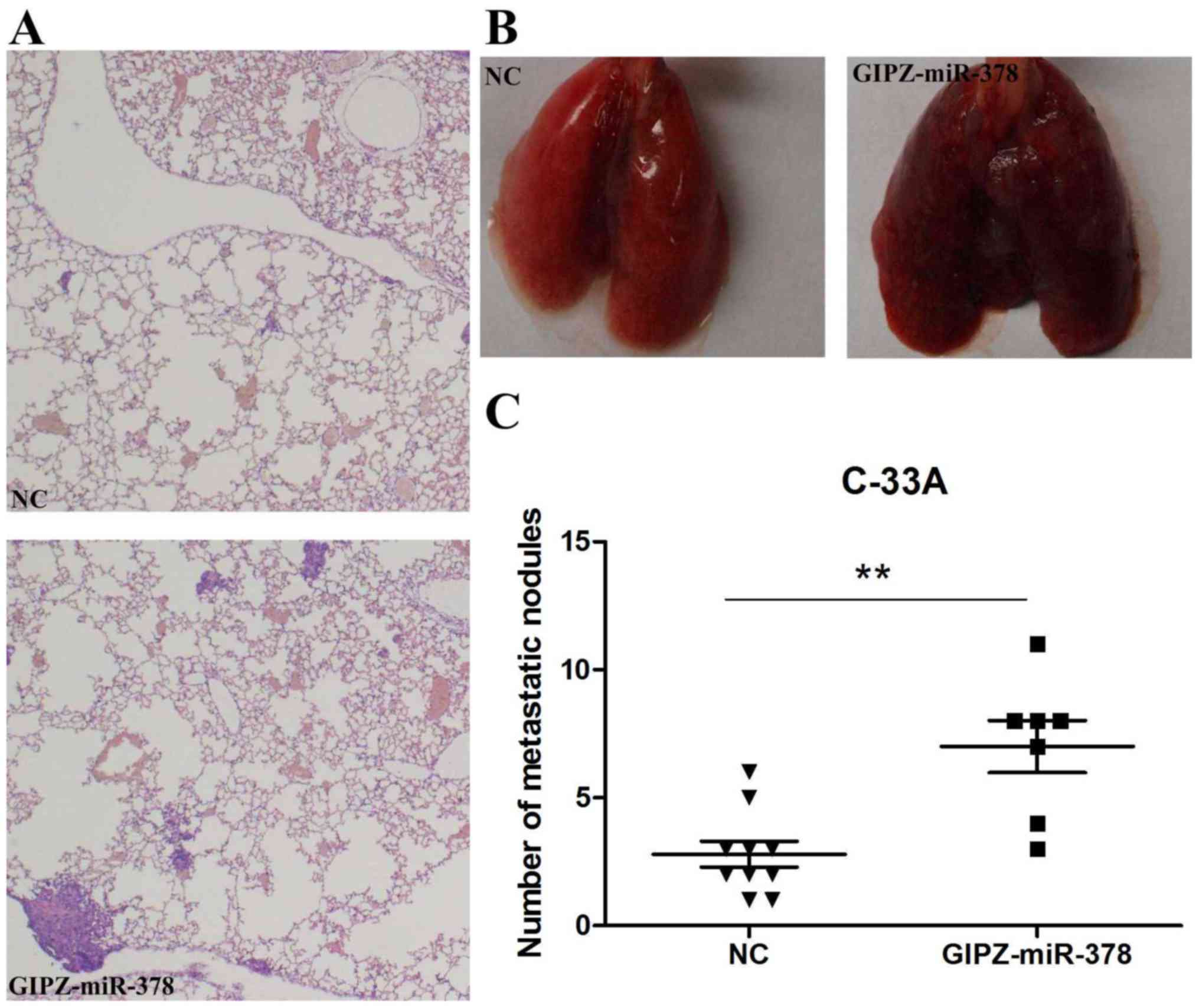
Overexpression of miR-378 promotes
metastasis of cervical cancer in vivo
To verify the promotion of miR-378 on cervical
cancer metastasis in vivo, C-33A cells transfected with
GIPZ-miR-378 or negative control were injected into mice via a tail
vein injection. Following 8 weeks incubation, lungs and tumor foci
were compared between the GIPZ-miR-378 and control groups.
Histological analysis of the lungs revealed that miR-378
overexpression promoted lung metastasis (Fig. 4A). Lungs with fewer metastasis
nodules were observed in the control group than the GIPZ-miR-378
group (Fig. 4B). In addition, the
number of tumor foci was significantly increased in the
GIPZ-miR-378 group when compared with in the control group
(Fig. 4C).
Discussion
In recent years, miRNAs have been reported to be
involved in tumor initiation, promotion and progression (6). In the present study, miR-378 was
observed to be frequently upregulated in CIN III and cervical
cancer tissues when compared with normal cervix tissues, and
overexpression of miR-378 significantly promoted cell metastasis
in vitro and in vivo, while the inhibition of miR-378
significantly decreased tumor metastasis in vitro; ATG12 was
identified as a functional and direct target of miR-378 in the
present study.
miR-378 has been identified to be aberrantly
expressed in a number of types of cancer and may be applied as an
early diagnostic biomarker (14).
In gastric cancer, miR-378 functions as a tumor suppressor by
inhibiting tumor growth via the suppression of cyclin dependent
kinase (CDK) 6 and vascular epithelial growth factor signaling
(15). In colorectal cancer,
miR-378 inhibited tumor growth and invasion (16), and the plasma levels of miR-378 may
be used as a diagnostic biomarker to discriminate colorectal cancer
patients from healthy individuals (16). Downregulated miR-378 correlated
with tumor invasiveness and poor prognosis of patients with glioma
(17). In addition to functioning
as a tumor suppressor, miR-378 also acts as an onco-miRNA. In liver
cancer, miR-378 enhanced cell survival, tumor growth and metastasis
by downregulating suppressor of fused homolog and FUS RNA binding
protein expression (18,19). Upregulation of miR-378 markedly
promoted cell proliferation, colony formation, migration and
invasion in vitro, as well as tumor growth in vivo by
downregulating transducer of receptor tyrosine-protein kinase
erbB-2 (TOB2) (20). The increased
serum levels of miR-378 served as powerful non-invasive diagnostic
and prognostic biomarkers in renal cell carcinoma (21). In cervical cancer, Wang et
al (4) demonstrated that
miR-378 expression levels were significantly increased in
HPV-infected tissues, which was consistent with the findings in the
present study, that miR-378 was significantly upregulated in CIN
III and cervical cancers. In the present study, miR-378 was
demonstrated to serve as an onco-RNA in cervical cancer as the
overexpression of miR-378 promoted cell migration and invasion
in vitro. To the best of our knowledge, the present study is
the first to investigate the functional role of miR-378 in the
migration and invasion of cervical cancer.
It has been reported that miR-378 is a direct target
of the c-Myc oncoprotein, which functions as a transcription factor
that serves as a master regulator in various biological processes
(22). However, a potential
association between miR-378 and autophagy was revealed as ATG12 was
identified to be a direct target of miR-378 as determined by a
luciferase reporter assay in the present study. It is generally
accepted that ATG12 is an important factor in apoptosis vacuole
formation (23) by conjugating to
ATG5 or ATG3. ATG12 induces mitophagy, mitochondrial fusion and
activates mitochondrial apoptosis by conjugating to ATG5, and
promotes mitochondrial fusion and restricts mitochondrial mass by
conjugating to ATG3 (11). Free
ATG12 promotes mitochondrial apoptosis in a similar manner to
proapoptotic B-cell lymphoma-2 homology 3-only proteins (24). Studies have demonstrated that
downregulation of ATG12 promoted tumor growth by inhibiting cell
autophagy (25,26). In the present study, the
overexpression of miR-378 reduced ATG12 expression in C-33A cells,
and miR-378 inhibitor transfection increased ATG12 expression in
HeLa cells. In addition to ATG12, CCND1 and pRb were also
downregulated by miR-378. The CCND1 gene encodes the cyclin D1
protein, which is required for progression through the G1 phase of
the cell cycle. It was demonstrated that the Myc-miR-378-transducer
of Erb-B receptor kinase 2, 2 (TOB2) signaling pathway converges on
the CCND1 level to promote transformation, and TOB2 acts to inhibit
CCND1 expression (27). pRb, which
controls the G1-S checkpoint of the cell cycle, was inhibited by
the CCND1-CDK4 complex in the promotion of passage via the G1
phase. It has been reported that in gastric cancer, aberrantly
expressed pRb and CCND1 were associated with lymph node metastases
(28). Thus, miR-378 may modulate
cervical cancer metastasis by regulating ATG12, CCND1 and pRb.
In conclusion, the present study demonstrated that
miR-378 significantly promoted cervical cancer metastasis in
vitro and in vivo. ATG12 was identified as a direct and
functional target of miR-378. The findings of the present study
suggested that miR-378 may provide novel insights into the
molecular mechanism of the pathology and therapeutic targets in
cervical cancer patients.
Acknowledgements
Not applicable.
Funding
The present study was conducted at Qilu Hospital,
Shandong University and was supported by the National Natural
Science Foundation of China (grant no. 81572559), The Key Research
Project of Shandong Province (grant no. 2017CXGC1210), The Science
and Technology Development Plan of Shandong Province (grant no.
2014GH218029) and the National Science and Technology Project of
China (grant no. 2015BAI13B05).
Availability of data and materials
The analyzed data sets generated during the study
are available from the corresponding author on reasonable
request.
Authors' contributions
DT, CZ and SH designed the study and performed the
experiments. XH and SK performed the statistical analysis and YZ
was involved in patient recruitment. All authors reviewed and
approved the final manuscript.
Ethics approval and consent to
participate
Written informed consent was obtained from all
patients and the present study was approved by the Ethics Committee
of Qilu Hospital of Shandong University (Jinan, China).
All animal experiments were approved by the Ethics
Committee of Qilu Hospital of Shandong University (Jinan,
China).
Consent for publication
Written informed consent was obtained from all
patients.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Muñoz N, Bosch FX, de Sanjosé S, Herrero
R, Castellsagué X, Shah KV, Snijders PJ and Meijer CJ:
International Agency for Research on Cancer Multicenter Cervical
Cancer Study Group: Epidemiologic classification of human
papillomavirus types associated with cervical cancer. N Engl J Med.
348:518–527. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sakuragi N: Up-to-date management of lymph
node metastasis and the role of tailored lymphadenectomy in
cervical cancer. Int J Clin Oncol. 12:165–175. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang X, Wang HK, Li Y, Hafner M, Banerjee
NS, Tang S, Briskin D, Meyers C, Chow LT, Xie X, et al: microRNAs
are biomarkers of oncogenic human papillomavirus infections. Proc
Natl Acad Sci USA. 111:pp. 4262–4267. 2014; View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tsujiura M, Ichikawa D, Komatsu S,
Shiozaki A, Takeshita H, Kosuga T, Konishi H, Morimura R, Deguchi
K, Fujiwara H, et al: Circulating microRNAs in plasma of patients
with gastric cancers. Br J Cancer. 102:1174–1179. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K,
Guo J, Zhang Y, Chen J, Guo X, et al: Characterization of microRNAs
in serum: A novel class of biomarkers for diagnosis of cancer and
other diseases. Cell Res. 18:997–1006. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Heneghan HM, Miller N, Lowery AJ, Sweeney
KJ, Newell J and Kerin MJ: Circulating microRNAs as novel minimally
invasive biomarkers for breast cancer. Ann Surg. 251:499–505. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wen SY, Lin Y, Yu YQ, Cao SJ, Zhang R,
Yang XM, Li J, Zhang YL, Wang YH, Ma MZ, et al: miR-506 acts as a
tumor suppressor by directly targeting the hedgehog pathway
transcription factor Gli3 in human cervical cancer. Oncogene.
34:717–725. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li BH, Zhou JS, Ye F, Cheng XD, Zhou CY,
Lu WG and Xie X: Reduced miR-100 expression in cervical cancer and
precursors and its carcinogenic effect through targeting PLK1
protein. Eur J Cancer. 47:2166–2174. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xu J, Li Y, Wang F, Wang X, Cheng B, Ye F,
Xie X, Zhou C and Lu W: Suppressed miR-424 expression via
upregulation of target gene Chk1 contributes to the progression of
cervical cancer. Oncogene. 32:976–987. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Haller M, Hock AK, Giampazolias E, Oberst
A, Green DR, Debnath J, Ryan KM, Vousden KH and Tait SW:
Ubiquitination and proteasomal degradation of ATG12 regulates its
proapoptotic activity. Autophagy. 10:2269–2278. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Murrow L, Malhotra R and Debnath J:
ATG12-ATG3 interacts with Alix to promote basal autophagic flux and
late endosome function. Nat Cell Biol. 17:300–310. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li ZZ, Shen LF, Li YY, Chen P and Chen LZ:
Clinical utility of microRNA-378 as early diagnostic biomarker of
human cancers: A meta-analysis of diagnostic test. Oncotarget.
7:58569–58578. 2016.PubMed/NCBI
|
|
15
|
Deng H, Guo Y, Song H, Xiao B, Sun W, Liu
Z, Yu X, Xia T, Cui L and Guo J: MicroRNA-195 and microRNA-378
mediate tumor growth suppression by epigenetical regulation in
gastric cancer. Gene. 518:351–359. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zanutto S, Pizzamiglio S, Ghilotti M,
Bertan C, Ravagnani F, Perrone F, Leo E, Pilotti S, Verderio P,
Gariboldi M and Pierotti MA: Circulating miR-378 in plasma: A
reliable, haemolysis-independent biomarker for colorectal cancer.
Br J Cancer. 110:1001–1007. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li B, Wang Y, Li S, He H, Sun F, Wang C,
Lu Y, Wang X and Tao B: Decreased expression of miR-378 correlates
with tumor invasiveness and poor prognosis of patients with glioma.
Int J Clin Exp Pathol. 8:7016–7021. 2015.PubMed/NCBI
|
|
18
|
Lee DY, Deng Z, Wang CH and Yang BB:
MicroRNA-378 promotes cell survival, tumor growth, and angiogenesis
by targeting SuFu and Fus-1 expression. Proc Natl Acad Sci USA.
104:pp. 20350–20355. 2007; View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ma J, Lin J, Qian J, Qian W, Yin J, Yang
B, Tang Q, Chen X, Wen X, Guo H and Deng Z: MiR-378 promotes the
migration of liver cancer cells by down-regulating Fus expression.
Cell Physiol Biochem. 34:2266–2274. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yu BL, Peng XH, Zhao FP, Liu X, Lu J, Wang
L, Li G, Chen HH and Li XP: MicroRNA-378 functions as an onco-miR
in nasopharyngeal carcinoma by repressing TOB2 expression. Int J
Oncol. 44:1215–1222. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Fedorko M, Stanik M, Iliev R,
Redova-Lojova M, Machackova T, Svoboda M, Pacik D, Dolezel J and
Slaby O: Combination of MiR-378 and MiR-210 serum levels enables
sensitive detection of renal cell carcinoma. Int J Mol Sci.
16:23382–23389. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Dang CV: c-Myc target genes involved in
cell growth, apoptosis, and metabolism. Mol Cell Biol. 19:1–11.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Metlagel Z, Otomo C, Takaesu G and Otomo
T: Structural basis of ATG3 recognition by the autophagic
ubiquitin-like protein ATG12. Proc Natl Acad Sci USA. 110:pp.
18844–18849. 2013; View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Rubinstein AD, Eisenstein M, Ber Y, Bialik
S and Kimchi A: The autophagy protein Atg12 associates with
antiapoptotic Bcl-2 family members to promote mitochondrial
apoptosis. Mol Cell. 44:698–709. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cordani M, Oppici E, Dando I, Butturini E,
Dalla Pozza E, Nadal-Serrano M, Oliver J, Roca P, Mariotto S,
Cellini B, et al: Mutant p53 proteins counteract autophagic
mechanism sensitizing cancer cells to mTOR inhibition. Mol Oncol.
10:1008–1029. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Pan B, Feng B, Chen Y, Huang G, Wang R,
Chen L and Song H: MiR-200b regulates autophagy associated with
chemoresistance in human lung adenocarcinoma. Oncotarget.
6:32805–32820. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Feng M, Li Z, Aau M, Wong CH, Yang X and
Yu Q: Myc/miR-378/TOB2/cyclin D1 functional module regulates
oncogenic transformation. Oncogene. 30:2242–2251. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Feakins RM, Nickols CD, Bidd H and Walton
SJ: Abnormal expression of pRb, p16, and cyclin D1 in gastric
adenocarcinoma and its lymph node metastases: Relationship with
pathological features and survival. Hum Pathol. 34:1276–1282. 2003.
View Article : Google Scholar : PubMed/NCBI
|