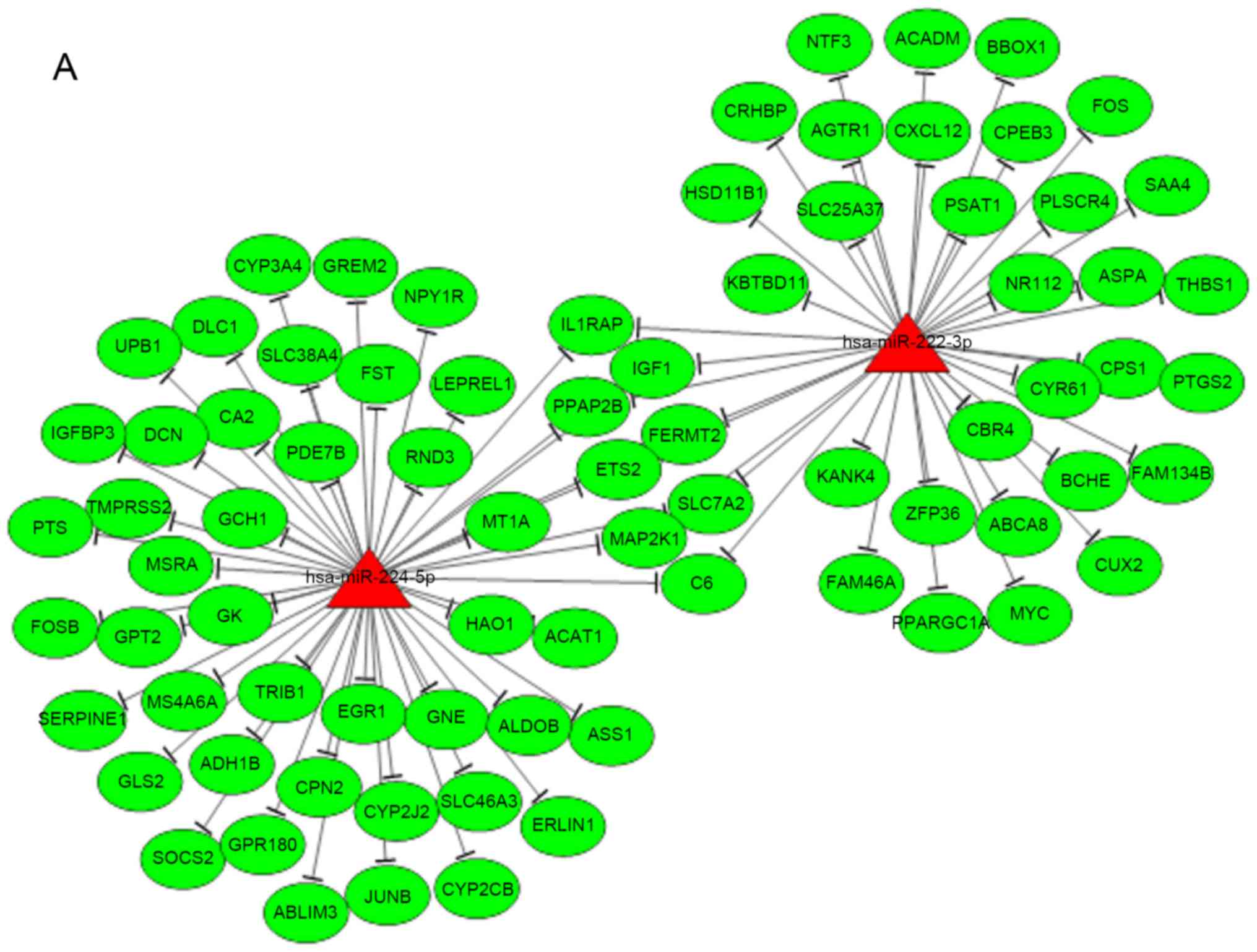
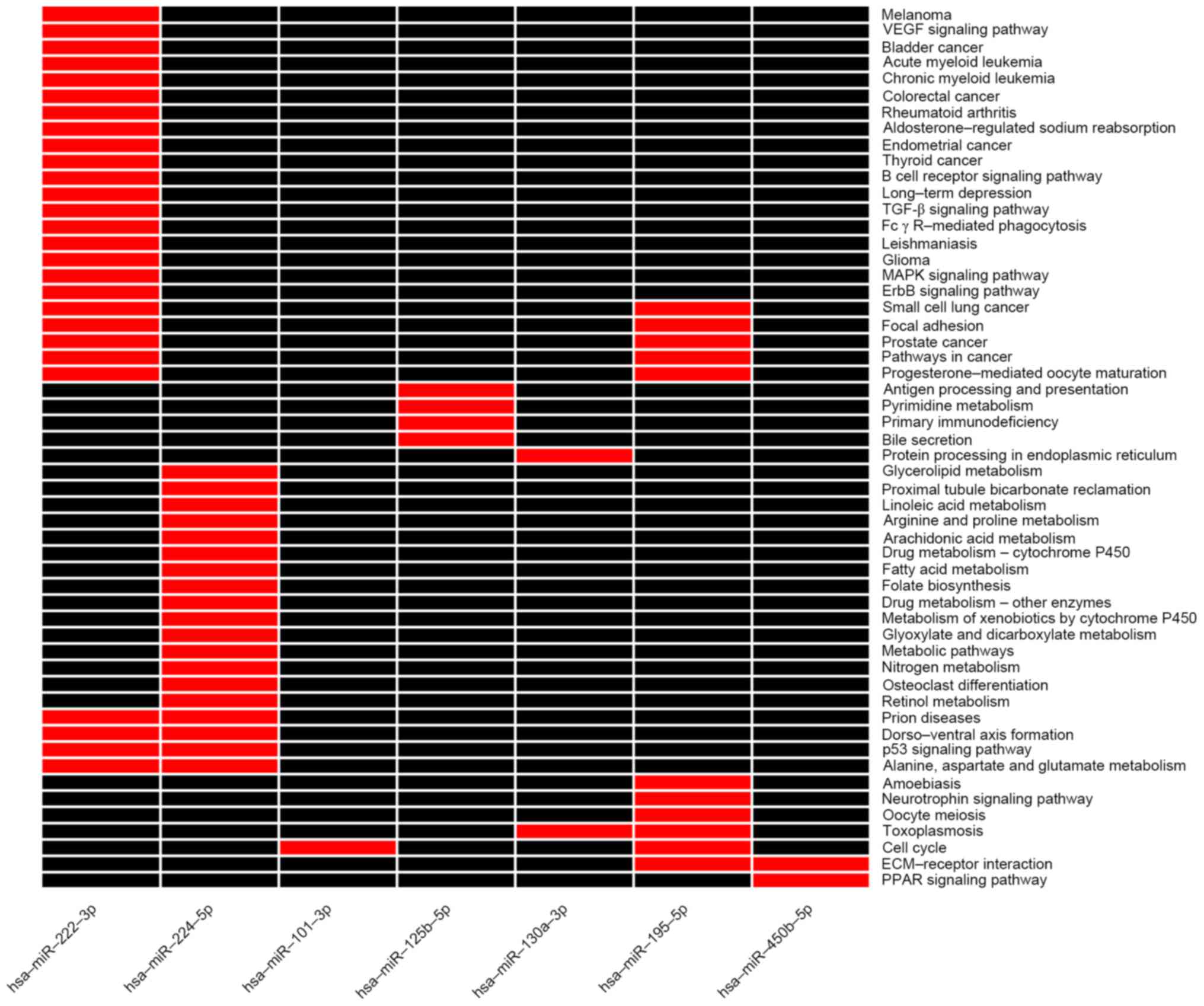
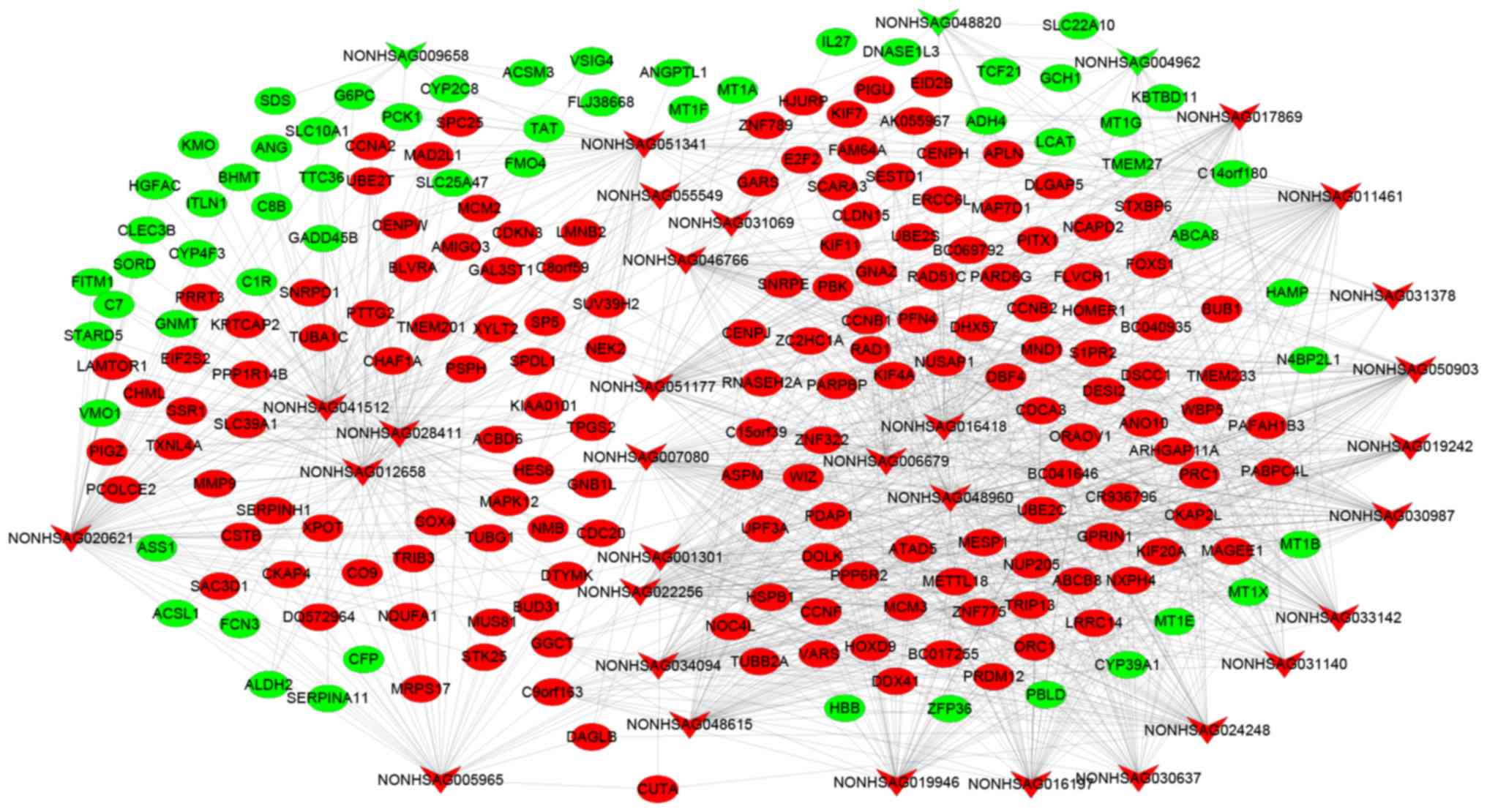
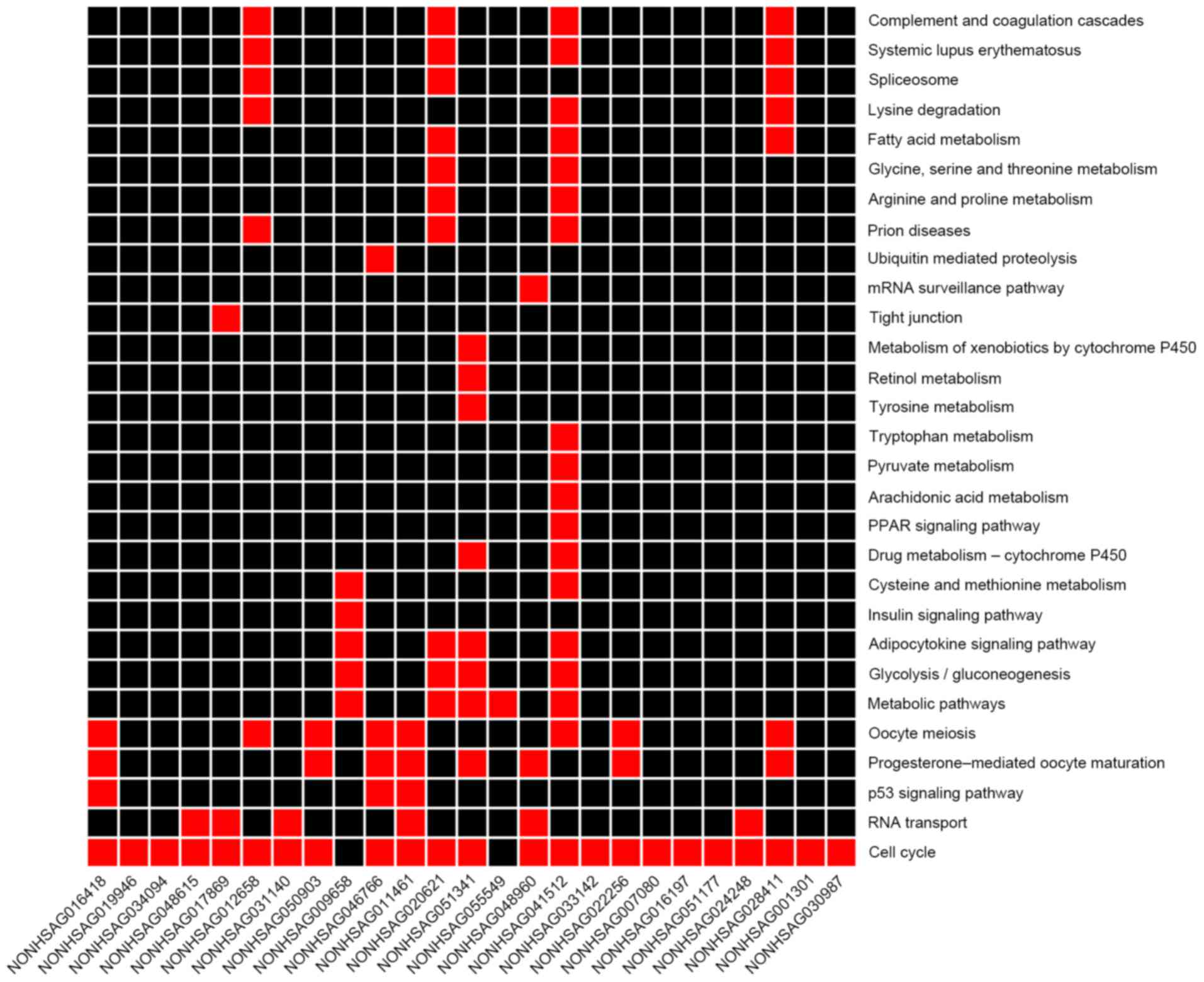
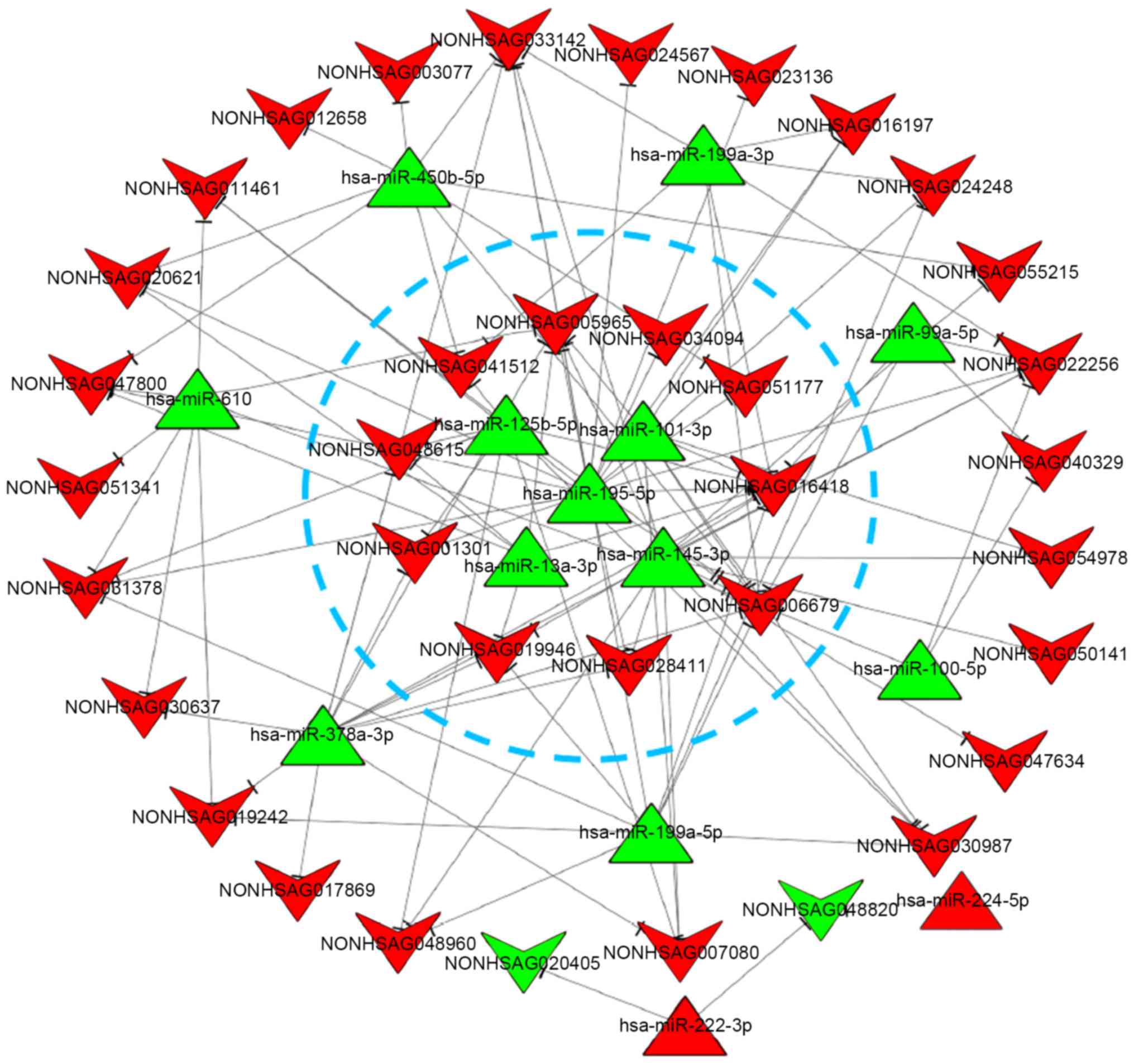
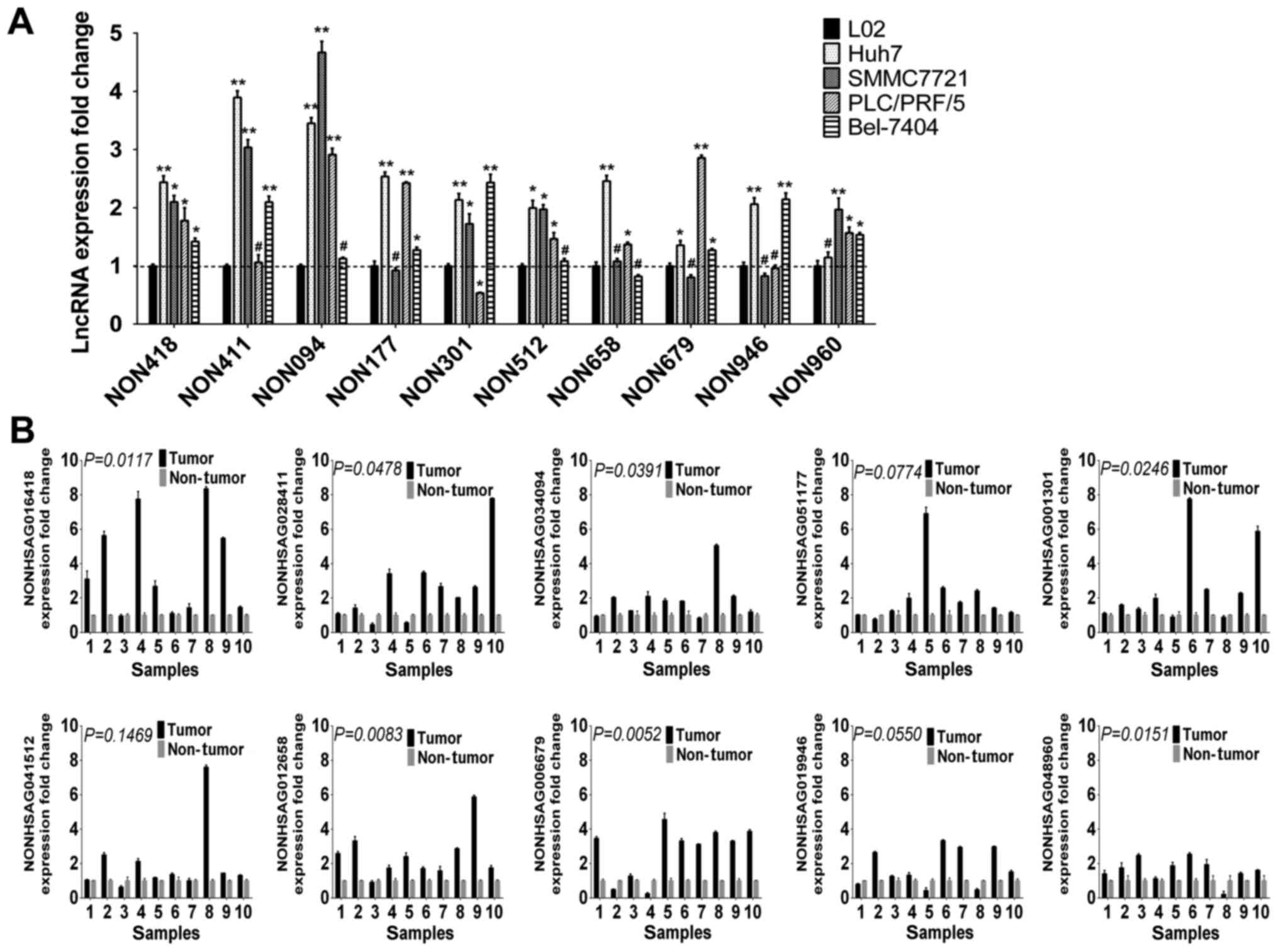
|
1
|
El-Serag HB and Rudolph KL: Hepatocellular
carcinoma: Epidemiology and molecular carcinogenesis.
Gastroenterology. 132:2557–2576. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bosetti C, Turati F and La Vecchia C:
Hepatocellular carcinoma epidemiology. Best Pract Res Clin
Gastroenterol. 28:753–770. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wallace MC, Preen D, Jeffrey GP and Adams
LA: The evolving epidemiology of hepatocellular carcinoma: A global
perspective. Expert Revi Gastroenterol Hepatol. 9:765–779.
2015.
|
|
4
|
Shi X, Sun M, Liu H, Yao Y and Song Y:
Long non-coding RNAs: A new frontier in the study of human
diseases. Cancer Lett. 339:159–166. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhao T, Xu J, Liu L, Bai J, Wang L, Xiao
Y, Li X and Zhang L: Computational identification of epigenetically
regulated lncRNAs and their associated genes based on integrating
genomic data. FEBS Lett. 589:521–531. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shahandeh A: Molecular mechanisms of
oncogenic long non-coding RNAs. Biosci Res. 10:38–54. 2013.
|
|
7
|
Marchese FP and Huarte M: Long non-coding
RNAs and chromatin modifiers: Their place in the epigenetic code.
Epigenetics. 9:21–26. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yang G, Lu X and Yuan L: LncRNA: A link
between RNA and cancer. Biochim Biophys Acta. 1839:1097–1109. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lee NH and Saeed AI: MicroarraysProtocols
for Nucleic Acid Analysis by Nonradioactive Probes. Hilario E and
Mackay JF: 353. Humana Press; New York, NY: pp. 265–300. 2007,
View Article : Google Scholar
|
|
10
|
Butte A: The use and analysis of
microarray data. Nat Rev Drug Discov. 1:951–960. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Edgar R, Domrachev M and Lash AE: Gene
expression omnibus: NCBI gene expression and hybridization array
data repository. Nucleic Acids Res. 30:207–210. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Barrett T, Troup DB, Wilhite SE, Ledoux P,
Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M and
Edgar R: NCBI GEO: Mining tens of millions of expression
profiles-database and tools update. Nucleic Acids Res.
35:D760–D765. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The rosetta stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tay Y, Rinn J and Pandolfi PP: The
multilayered complexity of ceRNA crosstalk and competition. Nature.
505:344–352. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Rosenbloom KR, Armstrong J, Barber GP,
Casper J, Clawson H, Diekhans M, Dreszer TR, Fujita PA, Guruvadoo L
and Haeussler M: The UCSC genome browser database: 2015 update.
Nucleic Acids Res. 43:D670–D681. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu C, Bai B, Skogerbø G, Cai L, Deng W,
Zhang, Bu D, Zhao Y and Chen R: NONCODE: An integrated knowledge
database of non-coding RNAs. Nucleic Acids Res. 33:D112–D115. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: Limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Reczko M, Maragkakis M, Alexiou P, Grosse
I and Hatzigeorgiou AG: Functional microRNA targets in protein
coding sequences. Bioinformatics. 28:771–776. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Betel D, Koppal A, Agius P, Sander C and
Leslie C: Comprehensive modeling of microRNA targets predicts
functional non-conserved and non-canonical sites. Genome Biol.
11:R902010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang X: miRDB: A microRNA target
prediction and functional annotation database with a wiki
interface. RNA. 14:1012–1017. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kertesz M, Iovino N, Unnerstall U, Gaul U
and Segal E: The role of site accessibility in microRNA target
recognition. Nat Genet. 39:1278–1284. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Dweep H, Sticht C, Pandey P and Gretz N:
miRWalk-database: Prediction of possible miRNA binding sites by
‘walking’ the genes of three genomes. J Biomed Inform. 44:839–847.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xiao F, Zuo Z, Cai G, Kang S, Gao X and Li
T: miRecords: An integrated resource for microRNA-target
interactions. Nucleic Acids Res. 37:D105–D110. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cao C, Sun J, Zhang D, Guo X, Xie L, Li X,
Wu D and Liu L: The long intergenic noncoding RNA UFC1, a target of
microRNA 34a, interacts with the mRNA stabilizing protein HuR to
increase levels of β-catenin in HCC cells. Gastroenterology.
148(415–426): e4182015. View Article : Google Scholar
|
|
28
|
Chen J, Fu Z, Ji C, Gu P, Xu P, Yu N, Kan
Y, Wu X, Shen R and Shen Y: Systematic gene microarray analysis of
the lncRNA expression profiles in human uterine cervix carcinoma.
Biomed Pharmacother. 72:83–90. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yang F, Zhang L, Huo XS, Yuan JH, Xu D,
Yuan SX, Zhu N, Zhou WP, Yang GS, Wang YZ, et al: Long noncoding
RNA high expression in hepatocellular carcinoma facilitates tumor
growth through enhancer of zeste homolog 2 in humans. Hepatology.
54:1679–1689. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kohl M, Wiese S and Warscheid B:
Cytoscape: Software for visualization and analysis of biological
networks. Methods Mol Biol. 696:291–303. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wong QW, Ching AK, Chan AW, Choy KW, To
KF, Lai PB and Wong N: MiR-222 overexpression confers cell
migratory advantages in hepatocellular carcinoma through enhancing
AKT signaling. Clin Cancer Res. 16:867–875. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Goto Y, Kojima S, Nishikawa R, Kurozumi A,
Kato M, Enokida H, Matsushita R, Yamazaki K, Ishida Y, Nakagawa M,
et al: MicroRNA expression signature of castration-resistant
prostate cancer: The microRNA-221/222 cluster functions as a tumour
suppressor and disease progression marker. Br J Cancer.
113:1055–1065. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhang Y, Takahashi S, Tasaka A, Yoshima T,
Ochi H and Chayama K: Involvement of microRNA-224 in cell
proliferation, migration, invasion and anti-apoptosis in
hepatocellular carcinoma. J Gastroenterol Hepatol. 28:565–575.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Bi Q, Tang S, Xia L, Du R, Fan R, Gao L,
Jin J, Liang S, Chen Z, Xu G, et al: Ectopic expression of MiR-125a
inhibits the proliferation and metastasis of hepatocellular
carcinoma by targeting MMP11 and VEGF. PLoS One. 7:e401692012.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ding J, Huang S, Wang Y, Tian Q, Zha R,
Shi H, Wang Q, Ge C, Chen T, Zhao Y, et al: Genome-wide screening
reveals that miR-195 targets the TNF-α/NF-κB pathway by
down-regulating IκB kinase alpha and TAB3 in hepatocellular
carcinoma. Hepatology. 58:654–666. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yang X, Yu J, Yin J, Xiang Q, Tang H and
Lei X: MiR-195 regulates cell apoptosis of human hepatocellular
carcinoma cells by targeting LATS2. Pharmazie. 67:645–651.
2012.PubMed/NCBI
|
|
38
|
Um SH, Mulhall C, Alisa A, Ives AR, Karani
J, Williams R, Bertoletti A and Behboudi S: Alpha-fetoprotein
impairs APC function and induces their apoptosis. J Immunol.
173:1772–1778. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Duvoux C, Roudotâ-'Thoraval F, Decaens T,
Pessione F, Badran H, Piardi T, Francoz C, Compagnon P, Vanlemmens
C, Dumortier J, et al: Liver transplantation for hepatocellular
carcinoma: A model including α-fetoprotein improves the performance
of Milan criteria. Gastroenterology. 143(986–994): e32012.
View Article : Google Scholar
|
|
40
|
Xue Y, Bowen B, Orr A, Koral K, Haynes M,
Bell A, Paranjpe S, Mars W and Michalopoulos G: GPC3-CD81 axis in
the HCV mediated liver carcinogenesis. FASEB J. 29:611–619.
2015.PubMed/NCBI
|
|
41
|
Lei CJ, Yao C, Pan QY, Long HC, Li L,
Zheng SP, Zeng C and Huang JB: Lentivirus vectors construction of
SiRNA targeting interference GPC3 gene and its biological effects
on liver cancer cell lines Huh-7. Asian Pac J Trop Med. 7:780–786.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Koo CY, Muir KW and Lam EW: FOXM1: From
cancer initiation to progression and treatment. Biochim Biophys
Acta. 1819:28–37. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhao J, Liu Y, Zhang W, Zhou Z, Wu J, Cui
P, Zhang Y and Huang G: Long non-coding RNA Linc00152 is involved
in cell cycle arrest, apoptosis, epithelial to mesenchymal
transition, cell migration and invasion in gastric cancer. Cell
Cycle. 14:3112–3123. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Pickard M, Mourtada-Maarabouni M and
Williams G: Long non-coding RNA GAS5 regulates apoptosis in
prostate cancer cell lines. Biochim Biophys Acta. 1832:1613–1623.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Xu C, Shao Y, Xia T, Yang Y, Dai J, Luo L,
Zhang X, Sun W, Song H, Xiao B and Guo J: lncRNA-AC130710 targeting
by miR-129-5p is upregulated in gastric cancer and associates with
poor prognosis. Tumour Biol. 35:9701–9706. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Braconi C, Kogure T, Valeri N, Huang N,
Nuovo G, Costinean S, Negrini M, Miotto E, Croce CM and Patel T:
microRNA-29 can regulate expression of the long non-coding RNA gene
MEG3 in hepatocellular cancer. Oncogene. 30:4750–4756. 2011.
View Article : Google Scholar : PubMed/NCBI
|