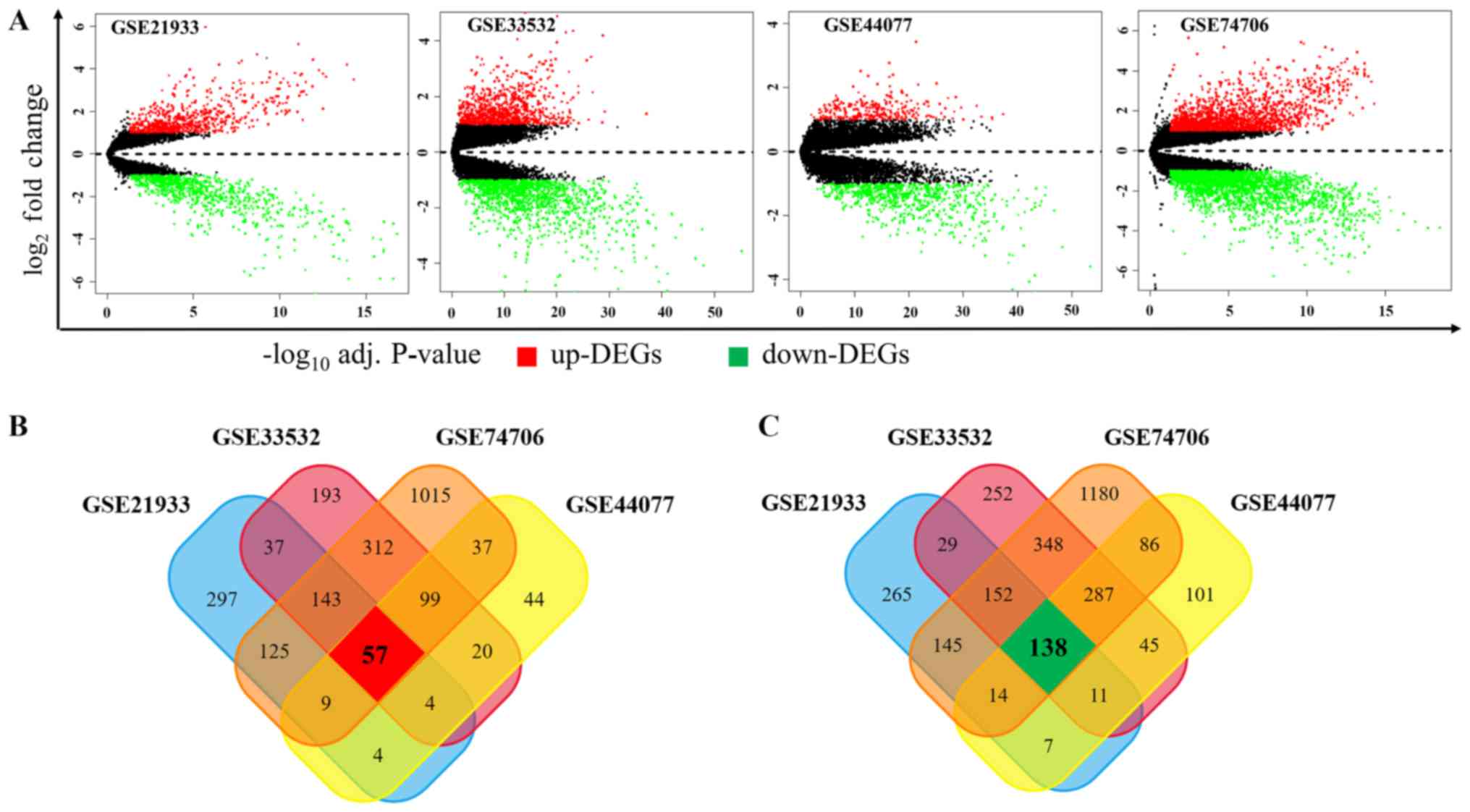
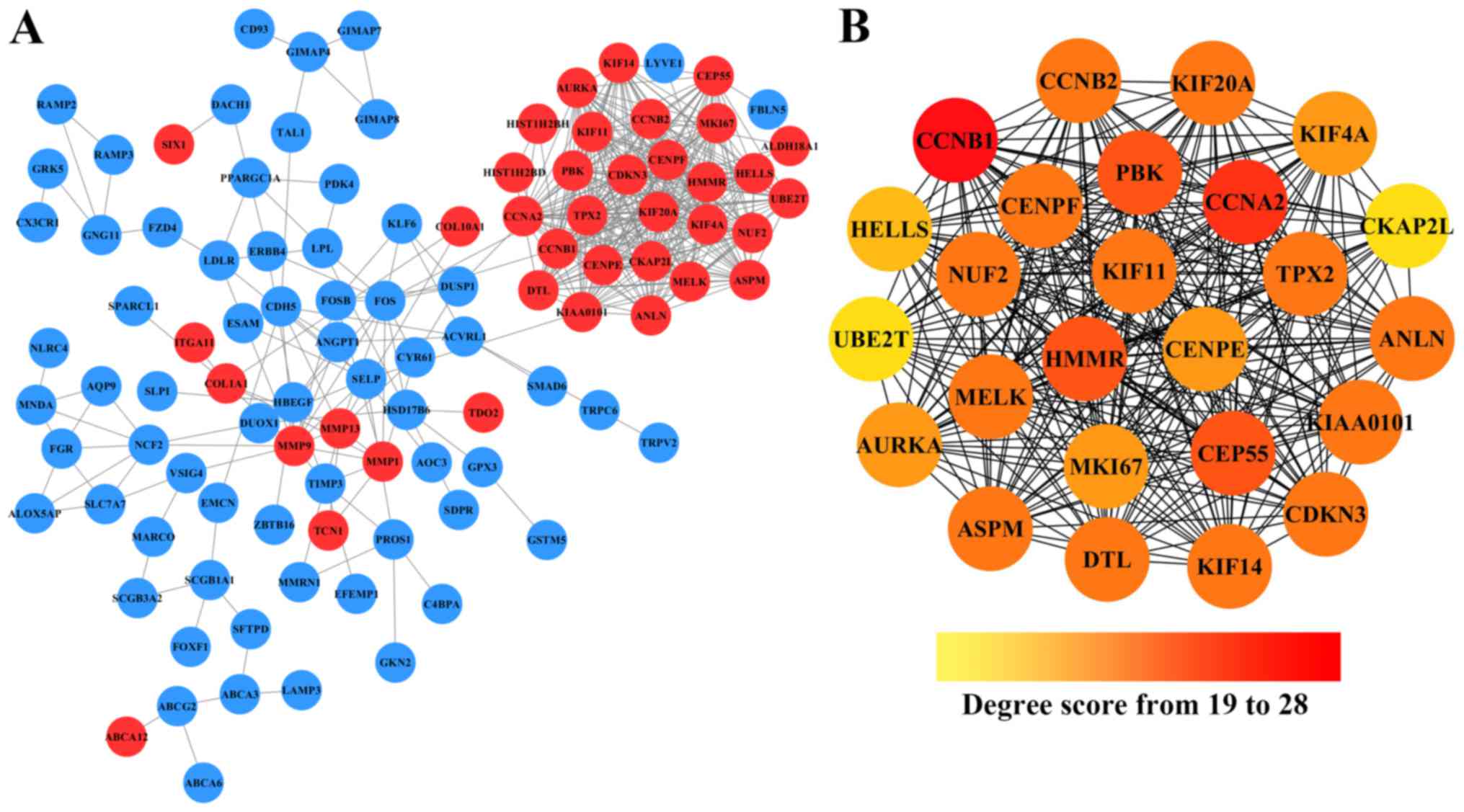
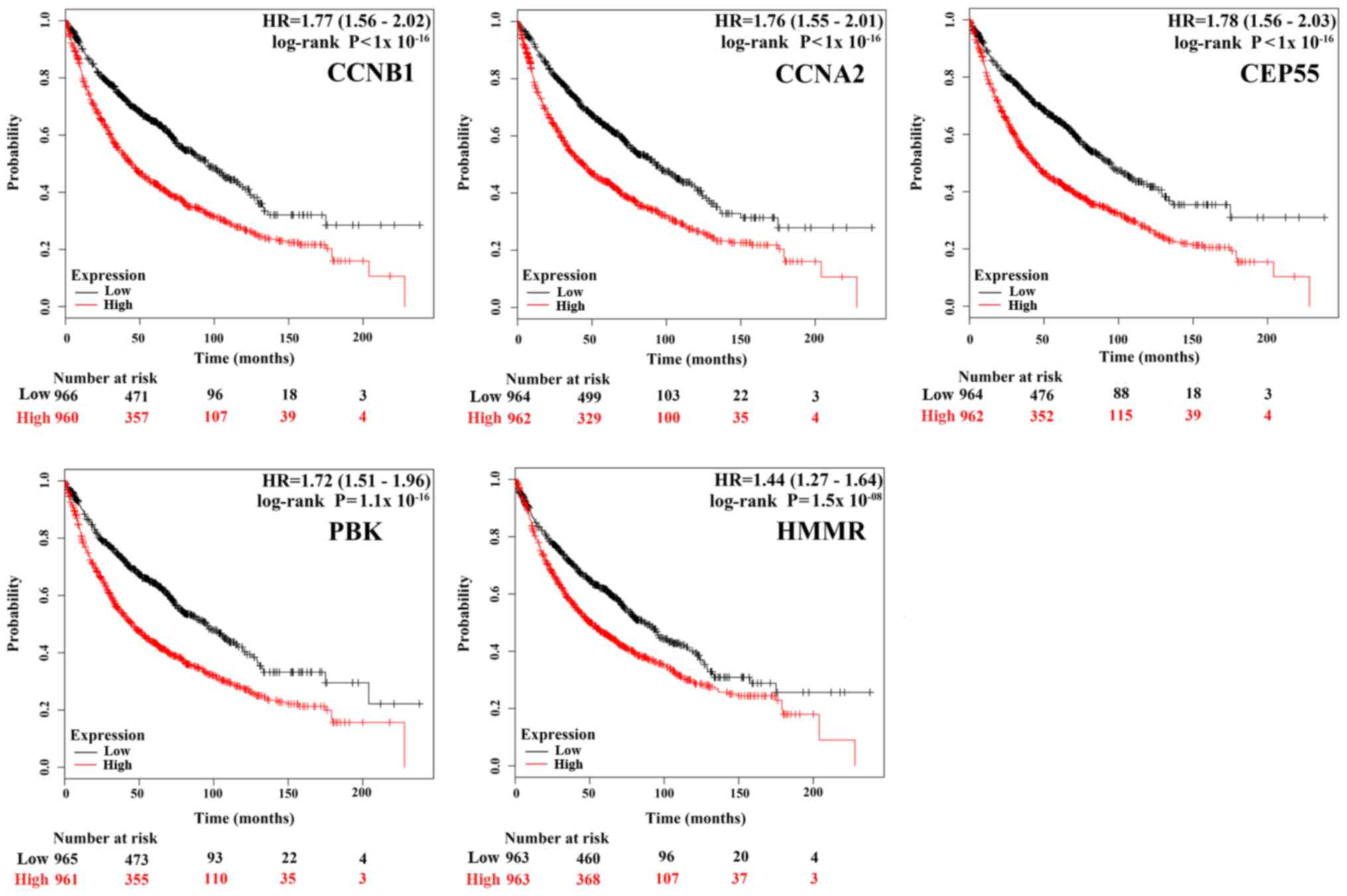
|
1
|
Semenova EA, Nagel R and Berns A: Origins,
genetic landscape, and emerging therapies of small cell lung
cancer. Genes Dev. 29:1447–1462. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Herbst RS, Heymach JV and Lippman SM: Lung
cancer. N Engl J Med. 359:1367–1380. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Danesi R, Pasqualetti G, Giovannetti E,
Crea F, Altavilla G, Del Tacca M and Rosell R: Pharmacogenomics in
non-small-cell lung cancer chemotherapy. Adv Drug Deliv Rev.
61:408–417. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shih MC, Chen JY, Wu YC, Jan YH, Yang BM,
Lu PJ, Cheng HC, Huang MS, Yang CJ, Hsiao M and Lai JM: TOPK/PBK
promotes cell migration via modulation of the PI3K/PTEN/AKT pathway
and is associated with poor prognosis in lung cancer. Oncogene.
31:2389–2400. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Keith RL and Miller YE: Lung cancer
chemoprevention: Current status and future prospects. Nat Rev Clin
Oncol. 10:334–343. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Oxnard GR, Lo PC, Nishino M, Dahlberg SE,
Lindeman NI, Butaney M, Jackman DM, Johnson BE and Jänne PA:
Natural history and molecular characteristics of lung cancers
harboring EGFR exon 20 insertions. J Thorac Oncol. 8:179–184. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gerber DE and Minna JD: ALK inhibition for
non-small cell lung cancer: From discovery to therapy in record
time. Cancer cell. 18:548–551. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Janku F, Stewart DJ and Kurzrock R:
Targeted therapy in non-small-cell lung cancer-is it becoming a
reality? Nat Rev Clin Oncol. 7:401–414. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lo FY, Chang JW, Chang IS, Chen YJ, Hsu
HS, Huang SF, Tsai FY, Jiang SS, Kanteti R, Nandi S, et al: The
database of chromosome imbalance regions and genes resided in lung
cancer from Asian and Caucasian identified by array-comparative
genomic hybridization. BMC Cancer. 12:2352012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Meister M, Belousov A, Xu EC, Schnabel PA,
Warth A, Hoffmann H, Dienemann H, Riedlinger J, Bodenmueller H,
Zolg W, et al: Intra-tumor heterogeneity of gene expression
profiles in early stage. Non-small cell lung cancer. 2014.
|
|
12
|
Kadara H, Fujimoto J, Yoo SY, Maki Y,
Gower AC, Kabbout M, Garcia MM, Chow CW, Chu Z, Mendoza G, et al:
Transcriptomic architecture of the adjacent airway field
cancerization in non-small cell lung cancer. J Natl Cancer Inst.
106:dju0042014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Marwitz S, Depner S, Dvornikov D, Merkle
R, Szczygieł M, Müller-Decker K, Lucarelli P, Wäsch M, Mairbäurl H,
Rabe KF, et al: Downregulation of the TGFβ Pseudoreceptor BAMBI in
non-small cell lung cancer enhances TGFβ signaling and invasion.
Cancer Res. 76:3785–3801. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Irizarry RA, Hobbs B, Collin F,
Beazer-Barclay YD, Antonellis KJ, Scherf U and Speed TP:
Exploration, normalization, and summaries of high density
oligonucleotide array probe level data. Biostatistics. 4:249–264.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate: A practical and powerful approach to
multiple testing. J Royal Statis Soc. 57:289–300. 1995.
|
|
16
|
Pathan M, Keerthikumar S, Ang CS, Gangoda
L, Quek CY, Williamson NA, Mouradov D, Sieber OM, Simpson RJ, Salim
A, et al: FunRich: An open access standalone functional enrichment
and interaction network analysis tool. Proteomics. 15:2597–2601.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gene Ontology Consortium: The gene
ontology (GO) project in 2006. Nucleic Acids Res. 34:(Database
Issue). D322–D326. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Dennis G Jr, Sherman BT, Hosack DA, Yang
J, Gao W, Lane HC and Lempicki RA: DAVID: Database for annotation,
visualization, and integrated discovery. Genome Biol. 4:P32003.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Szklarczyk D, Morris JH, Cook H, Kuhn M,
Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al:
The STRING database in 2017: Quality-controlled protein-protein
association networks, made broadly accessible. Nucleic Acids Res.
45(D1): D362–D368. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Szász AM, Lánczky A, Nagy Á, Förster S,
Hark K, Green JE, Boussioutas A, Busuttil R, Szabó A and Győrffy B:
Cross-validation of survival associated biomarkers in gastric
cancer using transcriptomic data of 1,065 patients. Oncotarget.
7:49322–49333. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Vogelstein B and Kinzler KW: Cancer genes
and the pathways they control. Nat Med. 10:789–799. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Saito M, Shiraishi K, Kunitoh H,
Takenoshita S, Yokota J and Kohno T: Gene aberrations for precision
medicine against lung adenocarcinoma. Cancer Sci. 107:713–720.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cardarella S and Johnson BE: The impact of
genomic changes on treatment of lung cancer. Am J Respir Crit Care
Med. 188:770–775. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liang B, Li C and Zhao J: Identification
of key pathways and genes in colorectal cancer using bioinformatics
analysis. Med Oncol. 33:1112016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kastan MB and Bartek J: Cell-cycle
checkpoints and cancer. Nature. 432:316–323. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Feng X, Ofstad W and Hawkins D:
Antiangiogenesis therapy: A new strategy for cancer treatment. US
Pharm. 35 Oncology Suppl:S4–S9. 2010.
|
|
28
|
Cavallaro U and Christofori G: Cell
adhesion and signalling by cadherins and Ig-CAMs in cancer. Nat Rev
Cancer. 4:118–132. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Petri ET, Errico A, Escobedo L, Hunt T and
Basavappa R: The crystal structure of human cyclin B. Cell Cycle.
6:1342–1349. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yuan J, Yan R, Krämer A, Eckerdt F, Roller
M, Kaufmann M and Strebhardt K: Cyclin B1 depletion inhibits
proliferation and induces apoptosis in human tumor cells. Oncogene.
23:5843–5852. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Soria JC, Jang SJ, Khuri FR, Hassan K, Liu
D, Hong WK and Mao L: Overexpression of cyclin B1 in early-stage
non-small cell lung cancer and its clinical implication. Cancer
Res. 60:4000–4004. 2000.PubMed/NCBI
|
|
32
|
Pagano M, Pepperkok R, Verde F, Ansorge W
and Draetta G: Cyclin A is required at two points in the human cell
cycle. EMBO J. 11:961–971. 1992.PubMed/NCBI
|
|
33
|
Yam CH, Fung TK and Poon RY: Cyclin A in
cell cycle control and cancer. Cell Mol Life Sci. 59:1317–1326.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chen CH, Lu PJ, Chen YC, Fu SL, Wu KJ,
Tsou AP, Lee YC, Lin TC, Hsu SL, Lin WJ, et al: FLJ10540-elicited
cell transformation is through the activation of PI3-kinase/AKT
pathway. Oncogene. 26:4272–4283. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Jeffery J, Sinha D, Srihari S, Kalimutho M
and Khanna KK: Beyond cytokinesis: The emerging roles of CEP55 in
tumorigenesis. Oncogene. 35:683–690. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Tao J, Zhi X, Tian Y, Li Z, Zhu Y, Wang W,
Xie K, Tang J, Zhang X, Wang L and Xu Z: CEP55 contributes to human
gastric carcinoma by regulating cell proliferation. Tumour Biol.
35:4389–4399. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhu F, Zykova TA, Kang BS, Wang Z, Ebeling
MC, Abe Y, Ma WY, Bode AM and Dong Z: Bidirectional signals
transduced by TOPK-ERK interaction increase tumorigenesis of HCT116
colorectal cancer cells. Gastroenterology. 133:219–231. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zykova TA, Zhu F, Lu C, Higgins L, Tatsumi
Y, Abe Y, Bode AM and Dong Z: Lymphokine-activated killer
T-cell-originated protein kinase phosphorylation of histone H2AX
prevents arsenite-induced apoptosis in RPMI7951 melanoma cells.
Clin Cancer Res. 12:6884–6893. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zlobec I, Molinari F, Kovac M, Bihl MP,
Altermatt HJ, Diebold J, Frick H, Germer M, Horcic M, Montani M, et
al: Prognostic and predictive value of TOPK stratified by KRAS and
BRAF gene alterations in sporadic, hereditary and metastatic
colorectal cancer patients. Br J Cancer. 102:151–161. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lei B, Liu S, Qi W, Zhao Y, Li Y, Lin N,
Xu X, Zhi C, Mei J, Yan Z, et al: PBK/TOPK expression in
non-small-cell lung cancer: Its correlation and prognostic
significance with Ki67 and p53 expression. Histopathology.
63:696–703. 2013.PubMed/NCBI
|
|
41
|
Pujana MA, Han JD, Starita LM, Stevens KN,
Tewari M, Ahn JS, Rennert G, Moreno V, Kirchhoff T, Gold B, et al:
Network modeling links breast cancer susceptibility and centrosome
dysfunction. Nat Genet. 39:1338–1349. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Hamilton SR, Fard SF, Paiwand FF, Tolg C,
Veiseh M, Wang C, McCarthy JB, Bissell MJ, Koropatnick J and Turley
EA: The hyaluronan receptors CD44 and Rhamm (CD168) form complexes
with ERK1,2 that sustain high basal motility in breast cancer
cells. J Biol Chem. 282:16667–16680. 2007. View Article : Google Scholar : PubMed/NCBI
|