Introduction
Breast cancer is the most commonly diagnosed cancer
and the leading cause of cancer death in females worldwide
(1,2). Incidence and mortality rates vary
internationally by >five-fold. The highest incidence rates were
reported in many European countries, while low rates were in Asia,
Africa and South America (3).
However, the incidence of breast cancer has increased by 3% per
year in China, which has threatened the health of women, and
created a great burden on the society (4). With the development of chemotherapy,
hormonal therapy, immunotherapy, gene therapy and other treatment
technologies, the long-term survival of breast cancer patients has
become possible. Breast cancer patients continue to succumb to the
disease due to tumor metastasis, drug resistance and other reasons,
including hemorrhage, infection and recurrence (5).
Long non-coding (lnc)RNAs are defined as RNA
transcripts which are >200 bp and lack open reading frames.
Thousands of lncRNAs have been identified in mammalian cells, many
with expression patterns specifically restricted by cell or
tissue-type and development stage (6,7). A
number of lncRNAs were initially identified through the
whole-genome tiling array and the next generation sequencing of
transcriptomes (8–10). These studies suggested that lncRNAs
have complicated structures and intrinsic origins, and therefore,
they can no longer be defined just by their length and
protein-coding incapability (11).
Generally, lncRNAs are linked to diverse
gene-regulatory roles, such as chromosome dosage compensation,
imprinting, epigenetic regulation, cell-cycle control, nuclear and
cytoplasmic trafficking, transcription, translation, splicing and
cell differentiation (11,12). Most importantly, aberrant
expression of lncRNAs is linked to several disease states,
including cancer (13).
A few studies have associated certain lncRNAs with
poor outcome and disease progression in different types of cancer:
High HOTAIR expression was identified in several types of cancer,
including breast and colorectal cancer (14–16),
overexpression of prostate cancer associated transcript-1 in
prostate cancer and overexpression of metastasis associated lung
adenocarcinoma transcript-1 in several types of cancer, including
early-stage non-small cell lung cancer (17,18).
Some research indicated that lncRNAs were associated
with chemoresistance: MEG3 and HOTAIR were considered to contribute
to the cisplatin resistance of human lung adenocarcinoma (19,20).
HOTTIP promotes progression and gemcitabine resistance by
regulating HOXA13 in pancreatic cancer (21). Takahashi et al (22) suggest that extracellular vesicle
lncRNA is a mediator of the chemotherapeutic response, and supports
targeting long intergenic non-protein coding RNA (LINC-ROR) to
enhance chemosensitivity in hepatocellular carcinoma.
Materials and methods
Cell culture
The human breast cancer MCF-7 cell line was obtained
from the American Type Culture Collection (Manassas, VA, USA).
Cells were cultured in RPMI-1640 medium (Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal
bovine serum (Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml
penicillin (Gibco; Thermo Fisher Scientific, Inc.) and 100 U/ml
streptomycin (Gibco; Thermo Fisher Scientific, Inc.) at 37°C in a
humidified atmosphere containing 5% CO2. MCF-7 cells
were pulse-selected with doxorubicin (10 pulses, once a week for 4
h, with 1 µM doxorubicin (Zhejiang Hisun Chemical Co., Ltd.,
Taizhou, China) to generate MCF-7/ADR after 6 months, as described
previously (23). Pulse
concentrations were decided based on changes in cell morphology and
clinical doxorubicin plasma concentration. MCF-7/ADR cells were
cultured in the continuous presence of doxorubicin (1 µg/ml) to
maintain the drug resistance phenotype and cultured in drug-free
medium for >2 weeks before subsequent experiments. The
experiments were independently reproduced twice, and each cell line
was tested in triplicate.
MTT assay
Doxorubicin-resistance was demonstrated in cell
lines by means of the MTT (Sigma-Aldrich; Merck KGaA, Darmstadt,
Germany) dye reduction assay. MCF-7 and MCF-7/ADR cells were seeded
into 96-well plates at a density of 1×104 cells per well
and incubated overnight in 100 µl 10% FCS medium (Gibco; Thermo
Fisher Scientific, Inc.). Cells were then exposed to varied
concentrations of doxorubicin and incubated at 37°C in a humidified
atmosphere containing 5% CO2 for 48 h. After this time,
cells were treated with MTT solution (5 mg/ml in phosphate-buffered
saline) for a further 4 h at 37°C. Following this incubation
period, the medium was rapidly removed and the MTT crystals were
solubilized using 150 µl DMSO (Sigma-Aldrich; Merck KGaA). The
number of viable cells was determined by measuring the absorbance
at 490 nm for each well using a microplate spectrophotometer.
Absorbance readings were subtracted from the value of blank wells;
the reduction in cell growth was calculated as a percentage of
control absorbance in the absence of any drug. Data presented the
mean of at least three independent experiments ± standard
deviation.
RNA extraction and quality
control
According to the manufacturer's protocol, total RNA
was extracted from the cells grown in monolayer with TRIzol reagent
(Invitrogen; Thermo Fisher Scientific, Inc.). Quantification and
quality checks were performed with the Nanodrop ND-1000 and Agilent
2100 Bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA,
USA), respectively. RNA Integrity and gDNA contamination were
assessed by standard denaturing agarose gel electrophoresis.
Microarray analysis
Sample preparation and microarray hybridization were
performed by Kangcheng Bio-tech (Aksomics Inc., Shanghai, China).
Briefly, RNA was purified from 1 µg total RNA following removal of
rRNA (mRNA-ONLY Eukaryotic mRNA Isolation kit; Epicentre; Illumina,
Inc., San Diego, CA, USA). Then, each sample was amplified and
transcribed into fluorescent cRNA along the entire length of the
transcripts without 39 bias utilizing a random priming method. The
labeled cRNAs were hybridized onto the Human LncRNA Array (version,
2.0; 8660 K; ArrayStar, Inc., Rockville, MD, USA). Following the
washing of the slides, the arrays were scanned by the Agilent
Scanner G2505B (Agilent Technologies, Inc., Santa Clara, CA, USA).
Agilent Feature Extraction software (version, 10.7.3.1; Agilent
Technologies, Inc.) was utilized to analyze acquired array images.
Quantile normalization and subsequent data processing were carried
out using the GeneSpring GX software package (version, 11.5.1;
Agilent Technologies, Inc.). Differentially expressed LncRNAs and
mRNAs were identified through fold change filtering (fold change
≥3.0 or ≤0.5), standard student t-test (P<0.05) and multiple
hypothesis testing (FDR<0.05). P-values and FDR were calculated
by Microsoft Excel (Microsoft Corporation, Redmond, WA, USA) and
MATLAB 8.2 (The MathWorks, Inc., Natick, MA, USA),
respectively.
Gene ontology and pathway
analysis
Pathway analysis and GO analysis were used to
determine the roles of these differentially expressed mRNAs in
these biological pathways or GO terms. Differentially regulated
mRNAs were uploaded into the Database for Annotation, Visualization
and Integrated Discovery (http://david.abcc.ncifcrf.gov/) to analyze the
enrichment of these coding genes. The result of pathway enrichment
analysis was confirmed by the online database of Kyoto Encyclopedia
of Genes and Genomes (KEGG; http://www.kegg.jp/). The potential functions of these
differentially expressed lncRNAs were identified by functional
annotation clustering and ranked by enrichment scores.
Validation of differentially expressed
lncRNA by reverse transcription-quantitative polymerase chain
reaction
Total RNA was isolated from tissues by the TRIzol
reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to
the manufacturer's instructions. A total of 2.5 µg RNA for each
sample was reversely transcribed into cDNA by using random hexamer
primer with PrimeScipt™ RT MASTER MIX (Perfect Real Time kit;
Takara Bio, Inc., Otsu, Japan). Primers for each lncRNA were
designed according to Primer 3 (http://sourceforge.net/projects/primer3/) online and
checked with Basic Local Alignment Search Tool of NCBI (https://blast.ncbi.nlm.nih.gov/Blast.cgi) to ensure a
unique amplification product. RT-qPCR was performed on an Applied
BiosystemsViiA™ 7 Dx (Thermo Fisher Scientific, Inc.) using the
SYBR Green (Invitrogen; Thermo Fisher Scientific, Inc.) method
according to the manufacturer's instructions. The PCR reaction
conditions were: Denaturation at 10 min at 95°C, followed by 40
cycles of 15 sec at 95°C and 30 sec at 60°C and 30 sec and then 30
sec at 72°C. Elative gene expression levels were quantified based
on the cycle threshold values and normalized to the internal
control gene GAPDH. The 2−∆∆Cq method was used to
comparatively quantify the levels of mRNA (24).
Statistical analysis
The differences of lncRNA levels were determined by
using a standard Student's t-test and multiple hypothesis testing.
The sensitivity and specificity were analyzed according to the
standard formulas. All the P-values are two-sided and P<0.05 was
considered to indicate a statistically significant difference.
Calculations of the mean ± standard deviation were conducted using
SPSS software (version 20.0; SPSS, Inc., Chicago, IL, USA).
Results
Cell viability of MCF-7 and MCF-7/ADR
cells after doxorubicin treatment
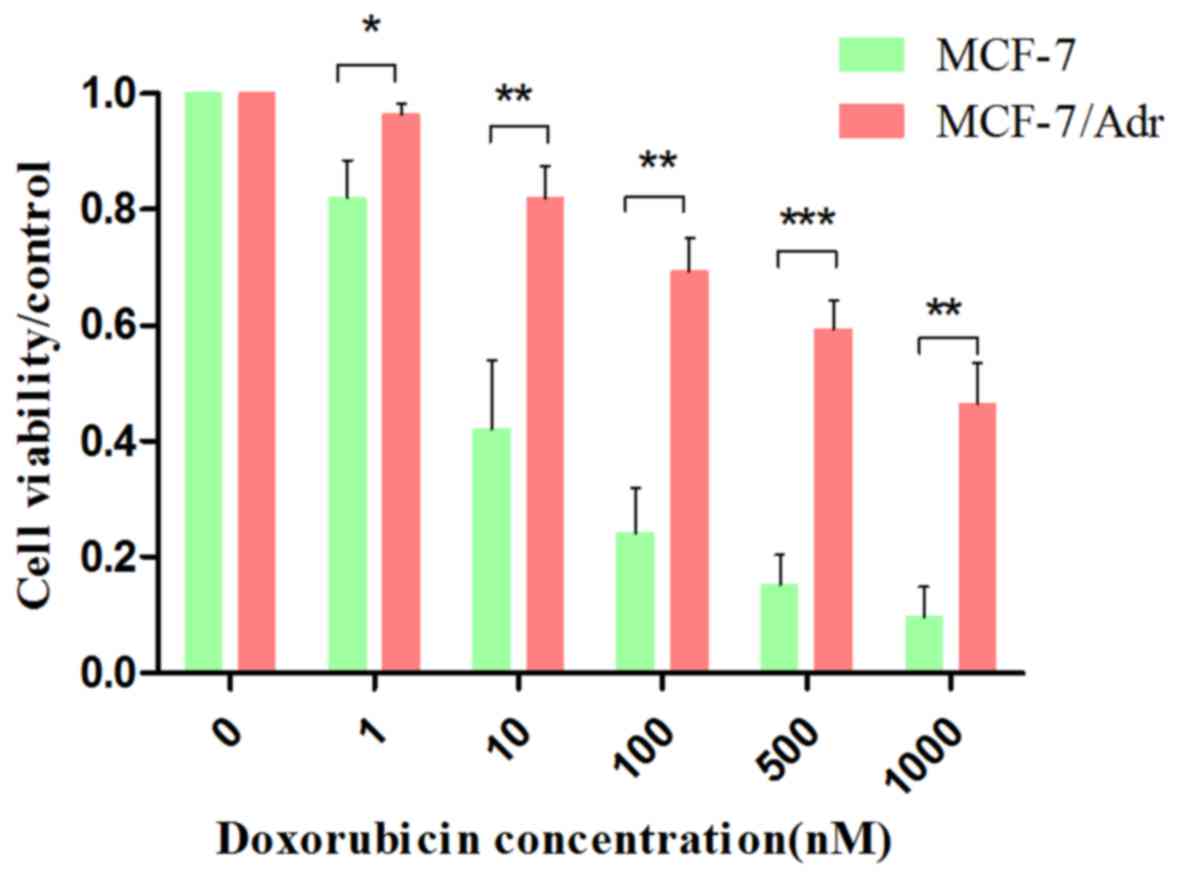
To determine whether MCF-7/ADR cells are resistant
to the chemotherapeutic agent doxorubicin, cell viability was
measured via an MTT assay. MCF-7 and MCF-7/ADR cells were treated
with different concentrations of doxorubicin (0, 1, 10, 100, 500
and 1,000 nM). The results indicated that the semi-effective
inhibitory concentration (IC50) of MCF-7 cells was 9.007
nM doxorubicin. However, the IC50 of the MCF-7/ADR cells
was 800.853 nM doxorubicin. When compared to the IC50 of
MCF-7 cells, the IC50 of MCF-7/ADR cells was elevated
88.91-fold (Fig. 1), which
indicated that MCF-7/ADR cells were resistant to doxorubicin.
LncRNA microarray data of MCF-7 and
MCF-7/ADR cells
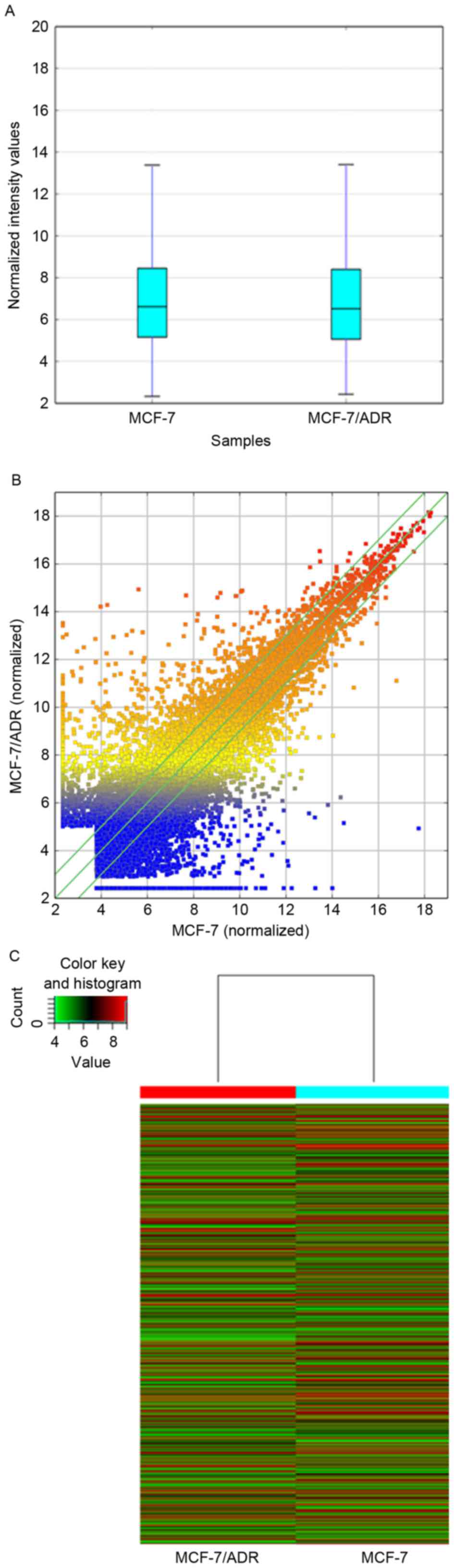
MCF-7 and MCF-7/ADR cells were collected to perform
a standard lncRNA microarray. The boxplot presented that after
normalization the distribution of expression values of each sample
was consistent to each other (Fig.
2A). The outcome of microarray demonstrated that 30,575 lncRNAs
and 23,682 mRNAs were detected in total. The LncRNA expression
patterns of the samples are presented as a hot-spot and cluster map
(Fig. 2B and C). Compared to the
MCF-7, 8,892 lncRNAs differentially expressed in MCF/ADR cells
absolute fold-change >2.0), among which 3,594 were upregulated
and 5,298 were downregulated.
Annotation of differentially expressed
lncRNAs in MCF-7/ADR cells
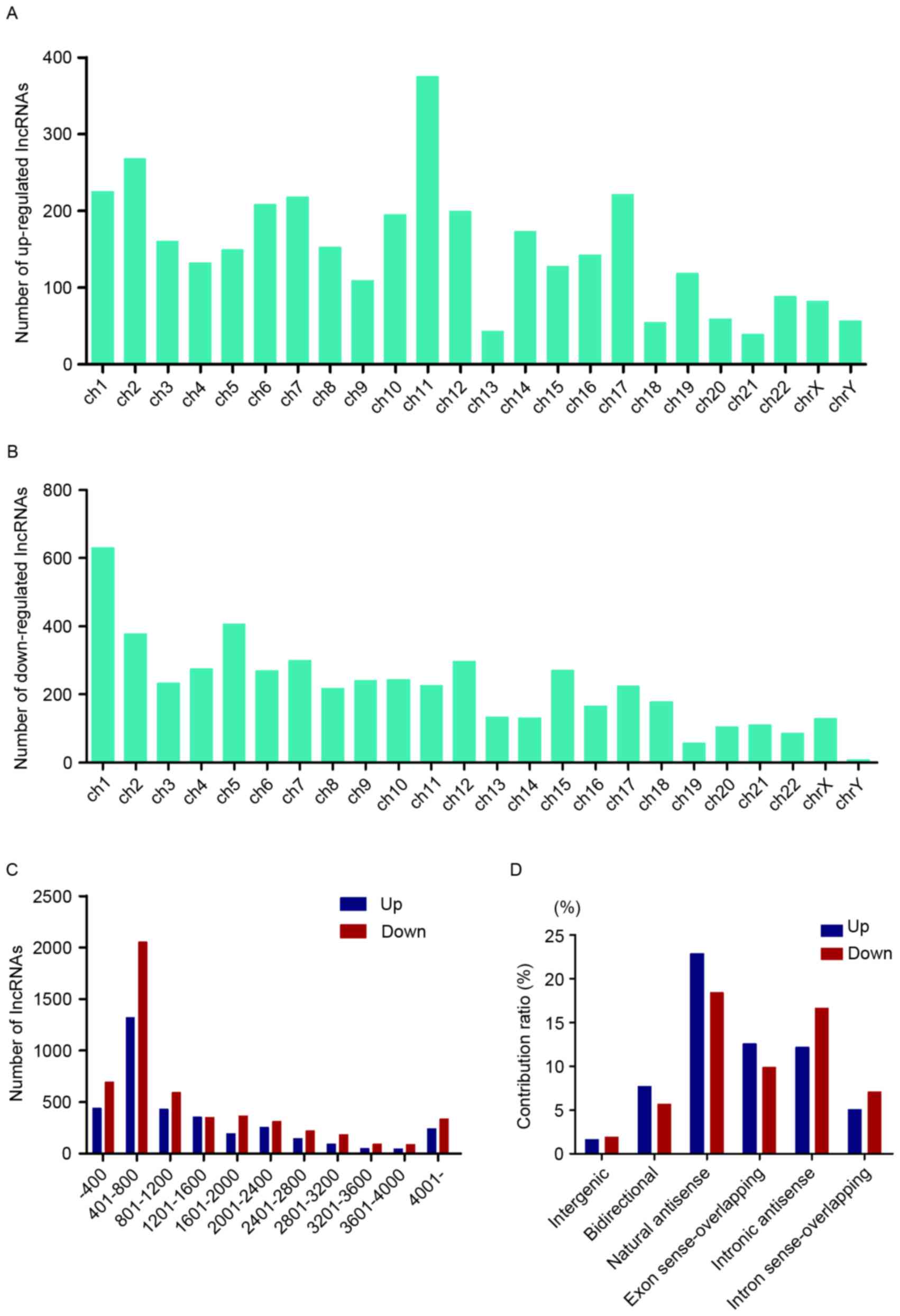
To make a further investigation into the expression
pattern of these differentially expressed lncRNAs, general
characteristics of these RNAs were taken into consideration,
including the chromosome location, length distribution and
classification. Chromosome 11 possessed the most upregulated
lncRNAs, while chromosome 1 contained the most downregulated ones
(Fig. 3A and B). The length of
these lncRNAs was between 400 and 2,400 nt (Fig. 3C). The relationship between these
lncRNAs and nearby coding genes included: i) Exon sense
overlapping, where the exon of lncRNA overlaps with coding
transcript exon on the same genomic strand; ii) intronic, where the
lncRNA overlaps with intron of a coding transcript on the same
genomic strand; iii) natural antisense, where the lncRNA is
transcribed from the antisense strand and overlaps with a coding
transcript; iv) non-overlapping antisense, where the lncRNA is
transcribed from the antisense strand without sharing overlapping
exons; v) bidirectional, where the lncRNA is oriented head to head
to a coding transcript within 1,000 bp; and vi) intergenic, where
there are no overlapping or bidirectional coding transcripts nearby
the lncRNA. Among them, natural antisense consisted >40% in
total differentially expressed lncRNAs (Fig. 3D).
GO and pathway analysis of
differentially expressed lncRNAs
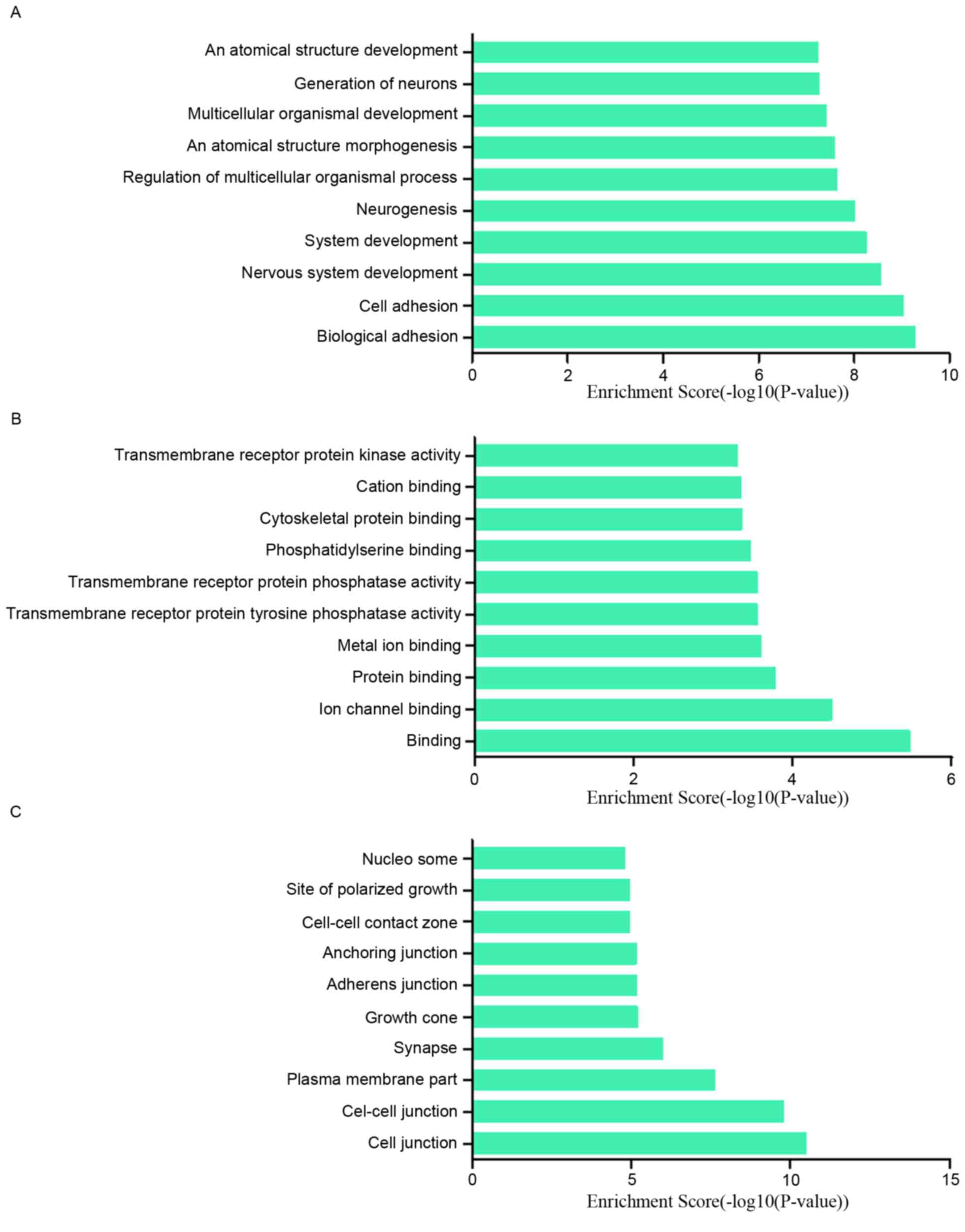
To research the potential functions of those
dysregulated lncRNAs in MCF-7/ADR cells preliminarily, the
associated target genes of those lncRNAs were predicted based on
the principles of chromosome location of nearby coding genes and
base-pairing. Subsequently, GO analysis was performed for the
lncRNAs and the target genes. The GO project (http://www.geneontology.org) mainly covers biological
process, molecular function and cellular component, as well as
providing controlled annotations to describe gene and gene product
attributed in any organism. In terms of the upregulated lncRNAs,
the GO analysis results inferred that the relative gene products
were primary involved in the biological process of biological
adhesion, cell adhesion, nervous system development and system
development (Fig. 4A); they were
mainly relative to the molecular functions such as binding, ion
channel binding and protein binding (Fig. 4B). Cell junction, cell-cell
junction, plasma membrane part were the main cell components those
lncRNAs associated with (Fig. 4C).
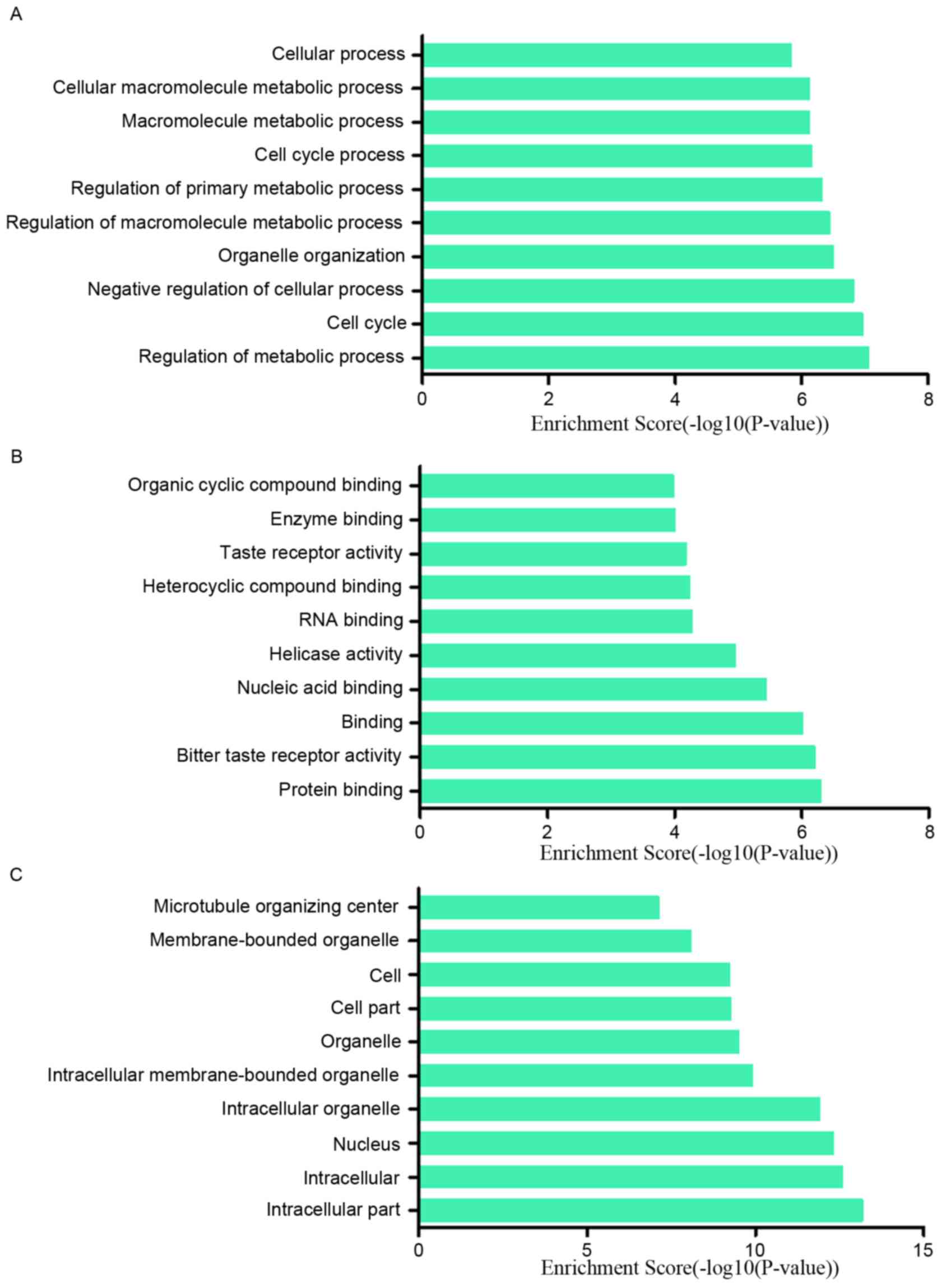
Among the downregulated lncRNAs, regulation of metabolic process,
cell cycle and negative regulation of cellular process ranked the
top three of ‘biological process’; protein binding, bitter taste
receptor activity and binding were the top three of ‘molecular
functions’. The majority of the cell components in which those
lncRNAs involved in were intracellular and in the nucleus (Fig. 5A-C). In addition, the pathway
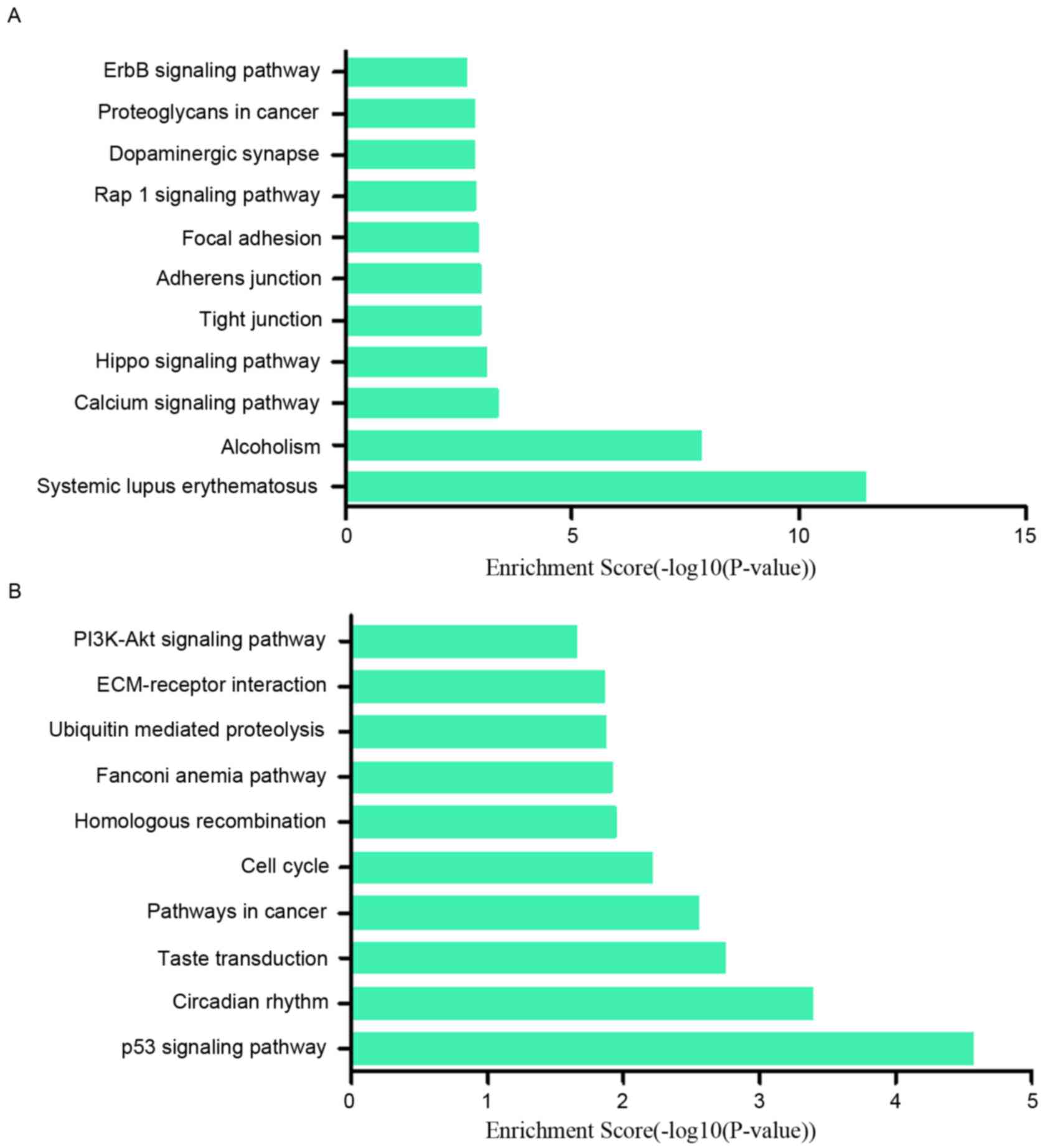
results suggested that the upregulated lncRNAs were part of several
signaling pathways, including systemic lupus erythematosus
(hsa05322), alcoholism (hsa05034), calcium signaling pathway
(hsa04020) and the Hippo signaling pathway (hsa04390). However, the
downregulated lncRNAs participated in the following pathways: p53
signaling pathway (hsa04115), circadian rhythm (hsa04710), taste
transduction (hsa04742), pathways in cancer (hsa05200) and cell
cycle (hsa04110) (Fig. 6A and B).
The P-value (EASE-score, Fisher P-value or hypergeometric P-value)
indicates the significance of the GO term and pathway correlated to
the conditions. The lower the P-value, more significant is the GO
term and pathway (P<0.05 is recommended).
Validation of the dysregulated
lncRNAs
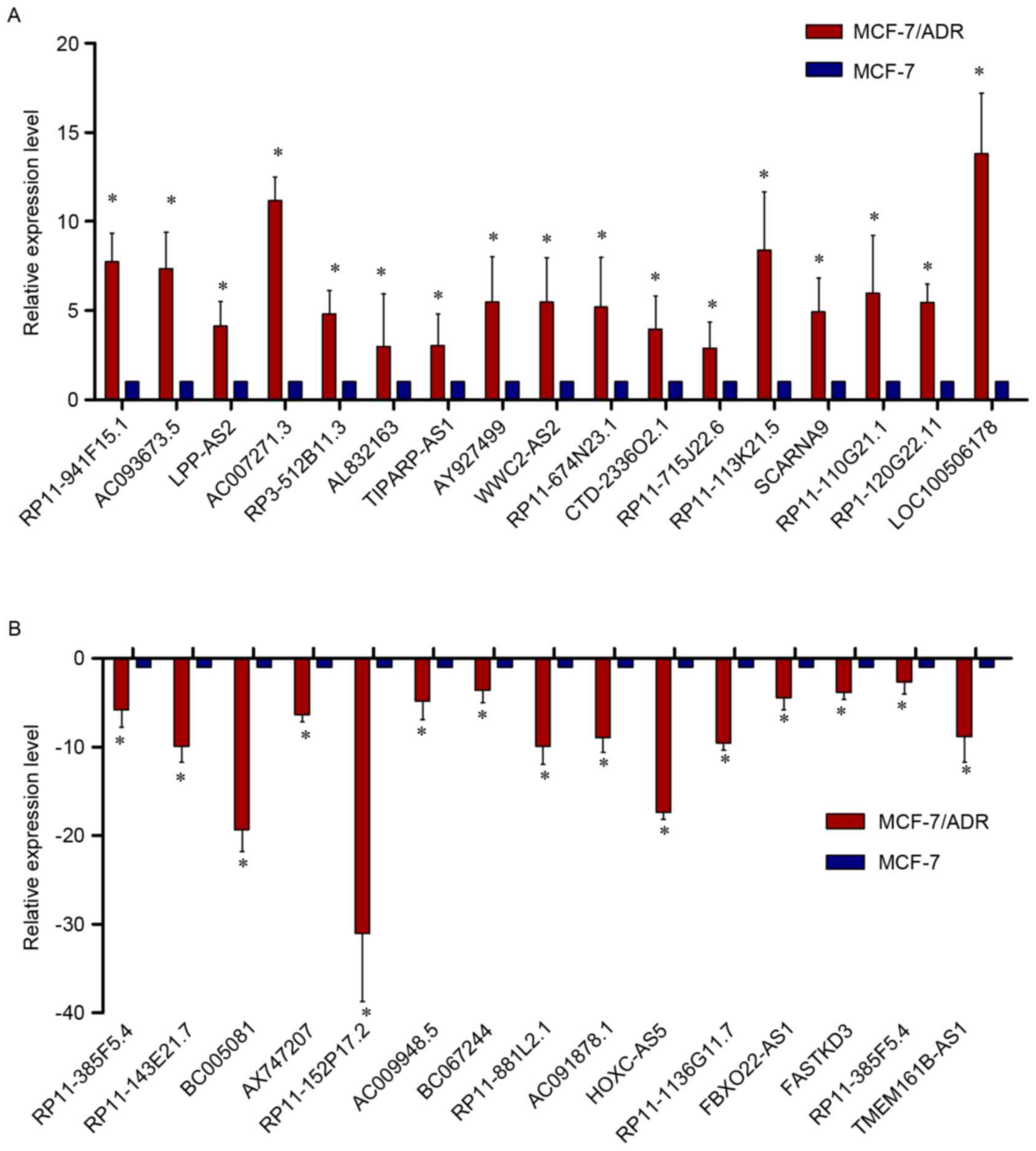
In order to verify the expression levels of the
dysregulated lncRNAs in the microarray pattern, 32 lncRNAs were
selected for (RT-qPCR) by fold-change filtering (absolute
fold-change >2.0), standard Student's t-test (P<0.05),
multiple hypothesis testing (FDR<0.05). Among the 32 lncRNAs, 17
were proven to be upregulated and 15 were downregulated. According
to the RT-qPCR results, LOC100506178, AC007271.3, AC093673.5,
RP11-113K21.5 and RP3-512B11.3 were upregulated in MCF-7/ADR cells
when compared with MCF-7 cells, while AX747207, HOXC-AS5,
RP11-152P17.2, RP11-1136G11.7, BC005081, RP11-143E21.7 and
AC091878.1 were downregulated. Basically, the results of the
RT-qPCR presented a considerable consistence with that of the
microarray (Fig. 7A and B).
Discussion
Breast cancer has become a major cancer in females
worldwide. According to international statistics, on a global scale
for women in 2013, breast cancer caused the highest incidence and
disability-adjusted life-years, and was regarded as the second
largest cause of cancer death (25). Depending on the tumor grade/stage
and the molecular characteristics of the malignancy, treatments
vary, ranging from surgery, chemotherapy, radiation, hormone
treatment and targeted therapy. Chemotherapy is the most commonly
used treatment where it is often used with other therapies, even
following surgery (26). However,
chemoresistance has become a major reason for treatment failure
(27,28).
Using lncRNA has been a novel field since the year
2000. Some research has been focused on the association between
lncRNA and chemoresistance. LncRNA MEG3 and HOTAIR were
demonstrated to contribute to cisplatin resistance of lung
adenocarcinoma (19,20). LncRNA HOTTIP was reported to
promote gemcitabine resistance by regulating HOXA13 in pancreatic
cancer (21). LncRNA linc-ROR was
confirmed to contribute to the effects of transforming growth
factor-β (TGF-β) on chemoresistance in hepatocellular carcinoma
(22). LncRNA UCA1 was proven to
increase the cisplatin resistance of bladder cancer cells by
enhancing the expression of Wnt6 (29). HOTAIR may regulate breast cancer
proliferation and chemoresistance using oncogenic lncRNA (30–33).
In the present study, the gene microarray results
were verified by RT-qPCR. Totally, 32 lncRNAs were screened for
analysis. Among the dysregulated lncRNAs, some were associated with
those genes that may be linked with chemoresistance of tumors.
AX747207 is a sequence of 2,874 bp located on chromosome 1, which
is relative to the gene RUNX3. The RUNX3 gene serves a critical
role in the regulation of cell proliferation, apoptosis and
angiogenesis, as well as cell adhesion and invasion (34,35).
Additionally, RUNX3 functions as a tumor suppressor involved in the
TGF-β signaling pathway in breast, colon, gastric and ovarian
cancers (36). Previously, some
research indicated that RUNX3 was associated with the
chemoresistance of tumors. Barghout et al (37) reported that overexpression of RUNX3
rendered epithelial ovarian cancer cells more resistant to
carboplatin, whereas inhibition of RUNX3 increased the sensitivity
of epithelial ovarian cancer cells to carboplatin. In addition, Guo
et al (38) indicated that
overexpression of Runx3 in gastric cancer cells sensitized the
cells to chemotherapeutic drugs, while blocking Runx3 expression in
immortalized gastric epithelial cells or gastric cancer cells
conferred the cells multidrug resistance. Zhang et al
(39) demonstrated that, compared
with human gastric adenocarcinoma cell line SGC7901, RUNX3 was
significantly decreased in two multidrug resistance variants,
SGC7901/ADR and SGC7901/VCR cells. Although the function of RUNX3
in breast cancer still remains unknown, this gene deserves further
research to reveal its association with chemoresistance in
malignant tumors.
GO analysis and KEGG pathway analysis were both
conducted among the differentially expressed lncRNAs. These lncRNAs
are involved in regulating several biological process (Figs. 4A and 5A), and the top three included regulation
of metabolic process, cell cycle and negative regulation of
cellular process for downregulated lncRNAs and biological adhesion,
cell adhesion and nervous system development for upregulated
lncRNAs, which are closely relative to the malignancy of the tumor.
In addition, the authors classified the potential function into 10
categories through analyzing the target gene pool (Figs. 4B and 5B), and the top five for upregulated and
downregulated lncRNAs respectively involved binding, ion channel
binding, protein binding, metal ion binding, transmembrane receptor
protein tyrosine phosphatase activity, protein binding, bitter
taste receptor activity, binding, nucleic acid binding and helicase
activity.
Moreover, pathway analysis results indicated that
these dysregulated lncRNAs primarily participated in the signaling
pathways in Fig. 6A and B, and
some were demonstrated to participate in chemoresistance of the
tumor. The phosphoinositide 3-kinase (PI3K)-protein kinase B (Akt)
signaling pathway (hsa04151) was identified to have a close
connection with chemoresistance, not only in breast cancer, but
also in pancreatic cancer, glioma, colon cancer, ovarian cancer,
gastric cancer and hepatocellular carcinoma (40–48).
The Hippo signaling pathway (hsa04390) serves
critical roles not only in mammary gland development but also in
breast cancer. Besides, the Yes-associated proteins (YAP, YAP1), an
effector of the Hippo signaling pathway, was reported to serve an
important role in the chemoresistance of malignant tumors,
including hepatocellullar carcinoma and gastric cancer (49–53).
Whether the Hippo-YAP signaling pathway participates in the
chemoresistance of breast cancer requires further studies.
The p53 signaling pathway (hsa04115) was considered
to take part in the chemoresistance process by interacting with
other pathways. The role that p53 pathway plays in chemoresistance
has been validated in breast cancer, renal cancer, glioma, lung
cancer and colorectal cancer (54–58).
The ErbB signaling pathway (hsa04012) was also
involved in the pathway analysis results. In this pathway, ErbB2
has been prominent in research as ~20% of human breast cancers
overexpress the human epidermal growth factor receptor 2 (HER2)
(59). HER2 belongs to the type I
receptor tyrosine kinase family that includes four members: EGFR,
HER2 (ErbB2/neu), HER3 and HER4 (60). HER2-positivity confers aggressive
tumor growth, early metastases, worse prognosis and variable
response to conventional chemotherapy (61,62).
Zhang et al (63)
demonstrated that HER2 overexpression led to an increased
resistance of MCF7 cells to multiple antitumor drugs, such as
paclitaxel, cisplatin, etoposide, doxorubicin, mitoxantrone and
5-fluorouracil (5-FU), which was consistent to previous research
(64–67). However, there are some
discrepancies regarding chemosensitization by overexpression of
HER2 in both laboratory and clinical studies. Coley's (68) study presented no link between
HER2-overexpression or HER2 amplification and resistance to
cytoxan/methotrexate/fluorouracil or to
fluorouracil/epirubicin/cytoxan (68). Furthermore, further study is
required to resolve the apparent contradictions between ErbB2 and
chemotherapy resistance.
In the present study, microarray analysis was
performed on MCF-7 and MCF-7/ADR cells to screen differentially
expressed lncRNAs. The results indicted a potential
chemoresistance-related lncRNA, AX747207, together with its
associated target gene, RUNX3. In addition, the pathway analysis
results provided some pathways that may potentially participate in
the chemoresistance network, such as PI3K-Akt, p53, Hippo and ErbB
signaling pathways. More future studies are required to confirm the
roles that lncRNA AX747207 and the target gene RUNX3 serve and the
relative pathways in chemoresistance of breast cancer.
Acknowledgements
The authors thank the following individuals for
their assistance: Hongxia Shen (Aksomics Inc., Shanghai, China), Dr
Zhengchao Li (Southern Medical University, Guangzhou, China), Dr
Lei Yang (Nanjing Maternity and Child Health Medical Institute).
The authors would like to thank Aksomics Inc. (Shanghai, China) for
the technical support.
Funding
The present study was financially supported by the
National Natural Science Foundation of China (grant nos. 81302304,
81302306, 81402139 and 81202007) and the Science and Technology
Development Foundation of Nanjing Medical University (grant nos.
2012NJMU201 and 2012NJMU185).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
LH, LZ, JC participated in data acquisition,
analysis and interpretation of data, and the writing and editing of
the manuscript. ZF, HX and SW participated in the conception and
design of the study. PX, ML, JX, JW, WL, LW and XW assisted with
the analysis and interpretation of the data and the writing and
editing of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
World Health Organization, . Cancer. Fact
sheet no. 297. https://www.who.int/mediacentre/factsheets/fs297/en/February.
2011
|
|
2
|
Kolonel L and Wilkens L: Migrant
studiesSchottenfeld D and Fraumeni JF Jr: Cancer epidemiology and
prevention. 3rd edition. Oxford: Oxford University Press; pp.
189–201. 2006, View Article : Google Scholar
|
|
3
|
Jemal A, Center MM, DeSantis C and Ward
EM: Global patterns of cancer incidence and mortality rates and
trends. Cancer Epidemiol Biomarkers Prev. 19:1893–1907. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Fan L, Strasser-Weippl K, Li JJ, St Louis
J, Finkelstein DM, Yu KD, Chen WQ, Shao ZM and Goss PE: Breast
cancer in China. Lancet Oncol. 15:e279–e289. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lianos GD, Vlachos K, Zoras O, Katsios C,
Cho WC and Roukos DH: Potential of antibody-drug conjugates and
novel therapeutics in breast cancer management. Onco Targets Ther.
7:491–500. 2014.PubMed/NCBI
|
|
6
|
Rinn JL, Kertesz M, Wang JK, Squazzo SL,
Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E and
Chang HY: Functional demarcation of active and silent chromatin
domains in human HOX loci by noncoding RNAs. Cell. 129:1311–1323.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Loewer S, Cabili MN, Guttman M, Loh YH,
Thomas K, Park IH, Garber M, Curran M, Onder T, Agarwal S, et al:
Large intergenic non-coding RNA-RoR modulates reprogramming of
human induced pluripotent stem cells. Nat Genet. 42:1113–1117.
2010. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hah N and Kraus WL: Hormone-regulated
transcriptomes: Lessons learned from estrogen signaling pathways in
breast cancer cells. Mol Cell Endocrinol. 382:652–664. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ilott NE and Ponting CP: Predicting long
non-coding RNAs using RNA sequencing. Methods. 63:50–59. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Spizzo R, Almeida MI and Colombatti A:
Long non-coding RNAs and cancer: A new frontier of translational
research. Oncogene. 31:4577–4587. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Guttman M, Amit I, Garber M, French C, Lin
MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al:
Chromatin signature reveals over a thousand highly conserved large
non-coding RNAs in mammals. Nature. 458:223–227. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Huarte M and Rinn JL: Large non-coding
RNAs: missing links in cancer? Hum Mol Genet. 19:R152–R161. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kogo R, Shimamura T, Mimori K, Kawahara K,
Imoto S, Sudo T, Tanaka F, Shibata K, Suzuki A, Komune S, et al:
Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin
modification and is associated with poor prognosis in colorectal
cancers. Cancer Res. 71:6320–6326. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sørensen KP, Thomassen M, Tan Q, Bak M,
Cold S, Burton M, Larsen MJ and Kruse TA: Long non-coding RNA
HOTAIR is an independent prognostic marker of metastasis in
estrogen receptor-positive primary breast cancer. Breast Cancer Res
Treat. 142:529–536. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Prensner JR, Iyer MK, Balbin OA,
Dhanasekaran SM, Cao Q, Brenner JC, Laxman B, Asangani IA, Grasso
CS, Kominsky HD, et al: Transcriptome sequencing across a prostate
cancer cohort identifies PCAT-1, an unannotated lincRNA implicated
in disease progression. Nat Biotechnol. 29:742–749. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ji P, Diederichs S, Wang W, Böing S,
Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et
al: MALAT-1, a novel noncoding RNA and thymosin beta4 predict
metastasis and survival in early-stage non-small cell lung cancer.
Oncogene. 22:8031–8041. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu J, Wan L, Lu K, Sun M, Pan X, Zhang P,
Lu B, Liu G and Wang Z: The long noncoding RNA MEG3 contributes to
cisplatin resistance of human lung adenocarcinoma. PLoS One.
10:e01145862015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu Z, Sun M, Lu K, Liu J, Zhang M, Wu W,
De W, Wang Z and Wang R: The long noncoding RNA HOTAIR contributes
to cisplatin resistance of human lung adenocarcinoma cells via
downregualtion of p21(WAF1/CIP1) expression. PLoS One.
8:e772932013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li Z, Zhao X, Zhou Y, Liu Y, Zhou Q, Ye H,
Wang Y, Zeng J, Song Y, Gao W, et al: The long non-coding RNA
HOTTIP promotes progression and gemcitabine resistance by
regulating HOXA13 in pancreatic cancer. J Transl Med. 13:842015.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Takahashi K, Yan IK, Kogure T, Haga H and
Patel T: Extracellular vesicle-mediated transfer of long non-coding
RNA ROR modulates chemosensitivity in human hepatocellular cancer.
FEBS Open Bio. 4:458–467. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lv J, Xia K, Xu P, Sun E, Ma J, Gao S,
Zhou Q, Zhang M, Wang F, Chen F, et al: miRNA expression patterns
in chemoresistant breast cancer tissues. Biomed Pharmacother.
68:935–942. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C (T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Global Burden of Disease Cancer
Collaboration, . Fitzmaurice C, Dicker D, Pain A, Hamavid H,
Moradi-Lakeh M, MacIntyre MF, Allen C, Hansen G, Woodbrook R, et
al: The Global Burden of Cancer 2013. JAMA Oncol. 1:505–527. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Martin HL, Smith L and Tomlinson DC:
Multidrug-resistant breast cancer: Current perspectives. Breast
Cancer (Dove Med Press). 6:1–13. 2014.PubMed/NCBI
|
|
27
|
Amiri-Kordestani L, Basseville A, Kurdziel
K, Fojo AT and Bates SE: Targeting MDR in breast and lung cancer:
Discriminating its potential importance from the failure of drug
resistance reversal studies. Drug Resist Updat. 15:50–61. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jia H, Truica CI, Wang B, Wang Y, Ren X,
Harvey HA, Song J and Yang JM: Immunotherapy for triple-negative
breast cancer: Existing challenges and exciting prospects. Drug
Resist Updat. 32:1–15. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Fan Y, Shen B, Tan M, Mu X, Qin Y, Zhang F
and Liu Y: Long non-coding RNA UCA1 increases chemoresistance of
bladder cancer cells by regulating Wnt signaling. FEBS J.
281:1750–1758. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang L, Song X, Wang X, Xie Y, Wang Z, Xu
Y, You X, Liang Z and Cao H: Circulating DNA of HOTAIR in serum is
a novel biomarker for breast cancer. Breast Cancer Res Treat.
152:199–208. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gökmen-Polar Y, Vladislav IT, Neelamraju
Y, Janga SC and Badve S: Prognostic impact of HOTAIR expression is
restricted to ER-negative breast cancers. Sci Rep. 5:87652015.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang YL, Overstreet AM, Chen MS, Wang J,
Zhao HJ, Ho PC, Smith M and Wang SC: Combined inhibition of EGFR
and c-ABL suppresses the growth of triple-negative breast cancer
growth through inhibition of HOTAIR. Oncotarget. 6:11150–11161.
2015.PubMed/NCBI
|
|
33
|
Hajjari M and Salavaty A: HOTAIR: An
oncogenic long non-coding RNA in different cancers. Cancer Biol
Med. 12:1–9. 2015.PubMed/NCBI
|
|
34
|
Subramaniam MM, Chan JY, Yeoh KG, Quek T,
Ito K and Salto-Tellez M: Molecular pathology of RUNX3 in human
carcinogenesis. Biochim Biophys Acta. 1796:315–331. 2009.PubMed/NCBI
|
|
35
|
Lund AH and van Lohuizen M: RUNX: A
trilogy of cancer genes. Cancer Cell. 1:213–215. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Levanon D, Negreanu V, Bernstein Y, Bar-Am
I, Avivi L and Groner Y: AML1, AML2 and AML3, the human members of
the runt domain gene-family: cDNA structure, expression and
chromosomal localization. Genomics. 23:425–432. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Barghout SH, Zepeda N, Vincent K, Azad AK,
Xu Z, Yang C, Steed H, Postovit LM and Fu Y: RUNX3 contributes to
carboplatin resistance in epithelial ovarian cancer cells. Gynecol
Oncol. 138:647–655. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Guo C, Ding J, Yao L, Sun L, Lin T, Song
Y, Sun L and Fan D: Tumor suppressor gene Runx3 sensitizes gastric
cancer cells to chemotherapeutic drugs by downregulating Bcl-2,
MDR-1 and MRP-1. Int J Cancer. 116:155–160. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang Y, Lu Q and Cai X: MicroRNA-106a
induces multidrug resistance in gastric cancer by targeting RUNX3.
FEBS Lett. 587:3069–3075. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Bezler M, Hengstler JG and Ullrich A:
Inhibition of doxorubicin-induced HER3-PI3K-AKT signalling enhances
apoptosis of ovarian cancer cells. Mol Oncol. 6:516–529. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Gao AM, Ke ZP, Shi F, Sun GC and Chen H:
Chrysin enhances sensitivity of BEL-7402/ADM cells to doxorubicin
by suppressing PI3K/Akt/Nrf2 and ERK/Nrf2 pathway. Chem Biol
Interact. 206:100–108. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Li Y, Jia L, Ren D, Liu C, Gong Y, Wang N,
Zhang X and Zhao Y: Axl mediates tumor invasion and
chemosensitivity through PI3K/Akt signaling pathway and is
transcriptionally regulated by slug in breast carcinoma. IUBMB
Life. 66:507–518. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yang XL, Lin FJ, Guo YJ, Shao ZM and Ou
ZL: Gemcitabine resistance in breast cancer cells regulated by
PI3K/AKT-mediated cellular proliferation exerts negative feedback
via the MEK/MAPK and mTOR pathways. Onco Targets Ther. 7:1033–1042.
2014.PubMed/NCBI
|
|
44
|
Zhu Y, Yu J, Wang S, Lu R, Wu J and Jiang
B: Overexpression of CD133 enhances chemoresistance to
5-fluorouracil by activating the PI3K/Akt/p70S6K pathway in gastric
cancer cells. Oncol Rep. 32:2437–2444. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Li Y, Chen K, Li L, Li R, Zhang Z and Ren
W: Overexpression of SOX2 is involved in paclitaxel resistance of
ovarian cancer via the PI3K/Akt pathway. Tumour Biol. 36:9823–9828.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Xiao ZM, Wang XY and Wang AM: Periostin
induces chemoresistance in colon cancer cells through activation of
the PI3K/Akt/survivin pathway. Biotechnol Appl Biochem. 62:401–406.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zhang LH, Yin AA, Cheng JX, Huang HY, Li
XM, Zhang YQ, Han N and Zhang X: TRIM24 promotes glioma progression
and enhances chemoresistance through activation of the PI3K/Akt
signaling pathway. Oncogene. 34:600–610. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Li J, Liang X and Yang X: Ursolic acid
inhibits growth and induces apoptosis in gemcitabine-resistant
human pancreatic cancer via the JNK and PI3K/Akt/NF-κB pathways.
Oncol Rep. 28:501–510. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Fujimoto D, Ueda Y, Hirono Y, Goi T and
Yamaguchi A: PAR1 participates in the ability of multidrug
resistance and tumorigenesis by controlling Hippo-YAP pathway.
Oncotarget. 6:34788–34799. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Huo X, Zhang Q, Liu AM, Tang C, Gong Y,
Bian J, Luk JM, Xu Z and Chen J: Overexpression of Yes-associated
protein confers doxorubicin resistance in hepatocellullar
carcinoma. Oncol Rep. 29:840–846. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Lin L, Sabnis AJ, Chan E, Olivas V, Cade
L, Pazarentzos E, Asthana S, Neel D, Yan JJ, Lu X, et al: The Hippo
effector YAP promotes resistance to RAF- and MEK-targeted cancer
therapies. Nat Genet. 47:250–256. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Shi P, Feng J and Chen C: Hippo pathway in
mammary gland development and breast cancer. Acta Biochim Biophys
Sin (Shanghai). 47:53–59. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Zhao Y and Yang X: The Hippo pathway in
chemotherapeutic drug resistance. Int J Cancer. 137:2767–2773.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Knappskog S, Berge EO, Chrisanthar R,
Geisler S, Staalesen V, Leirvaag B, Yndestad S, de Faveri E,
Karlsen BO, Wedge DC, et al: Concomitant inactivation of the p53-
and pRB-functional pathways predicts resistance to DNA damaging
drugs in breast cancer in vivo. Mol Oncol. 9:1553–1564. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Lim SJ, Choi HG, Jeon CK and Kim SH:
Increased chemoresistance to paclitaxel in the MCF10AT series of
human breast epithelial cancer cells. Oncol Rep. 33:2023–2030.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Chen J, Zhu H, Zhang Y, Cui MH, Han LY,
Jia ZH, Wang L, Teng H and Miao LN: Low expression of phosphatase
and tensin homolog in clearcell renal cell carcinoma contributes to
chemoresistance through activating the Akt/HDM2 signaling pathway.
Mol Med Rep. 12:2622–2628. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Weiler M, Blaes J, Pusch S, Sahm F,
Czabanka M, Luger S, Bunse L, Solecki G, Eichwald V, Jugold M, et
al: mTOR target NDRG1 confers MGMT-dependent resistance to
alkylating chemotherapy. Proc Natl Acad Sci USA. 111:409–414. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Yang L, Zhou Y, Li Y, Zhou J, Wu Y, Cui Y,
Yang G and Hong Y: Mutations of p53 and KRAS activate NF-κB to
promote chemoresistance and tumorigenesis via dysregulation of cell
cycle and suppression of apoptosis in lung cancer cells. Cancer
Lett. 357:520–526. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Ross JS, Slodkowska EA, Symmans WF,
Pusztai L, Ravdin PM and Hortobagyi GN: The HER-2 receptor and
breast cancer: Ten years of targeted anti-HER-2 therapy and
personalized medicine. Oncologist. 14:320–368. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Roskoski R Jr: The ErbB/HER family of
protein-tyrosine kinases and cancer. Pharmacol Res. 79:34–74. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Slamon DJ, Clark GM, Wong SG, Levin WJ,
Ullrich A and McGuire WL: Human breast cancer: Correlation of
relapse and survival with amplification of the HER-2/neu oncogene.
Science. 235:177–182. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Nguyen PL, Taghian AG, Katz MS, Niemierko
A, Raad Abi RF, Boon WL, Bellon JR, Wong JS, Smith BL and Harris
JR: Breast cancer subtype approximated by estrogen receptor,
progesterone receptor and HER-2 is associated with local and
distant recurrence after breast-conserving therapy. J Clin Oncol.
26:2373–2378. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Zhang W, Ding W, Chen Y, Feng M, Ouyang Y,
Yu Y and He Z: Up-regulation of breast cancer resistance protein
plays a role in HER2-mediated chemoresistance through PI3K/Akt and
nuclear factor-kappa B signaling pathways in MCF7 breast cancer
cells. Acta Biochim Biophys Sin (Shanghai). 43:647–653. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Ejlertsen B, Jensen MB, Nielsen KV,
Balslev E, Rasmussen BB, Willemoe GL, Hertel PB, Knoop AS,
Mouridsen HT and Brünner N: TIMP-1 and responsiveness to adjuvant
anthracycline-containing chemotherapy in high-risk breast cancer
patients. J Clin Oncol. 28:984–990. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Pritchard KI, Shepherd LE, O'Malley FP,
Andrulis IL, Tu D, Bramwell VH and Levine MN: National Cancer
Institute of Canada Clinical Trials Group: HER2 and responsiveness
of breast cancer to adjuvant chemotherapy. N Engl J Med.
354:2103–2111. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Knuefermann C, Lu Y, Liu B, Jin W, Liang
K, Wu L, Schmidt M, Mills GB, Mendelsohn J and Fan Z:
HER2/PI-3K/Akt activation leads to a multidrug resistance in human
breast adenocarcinoma cells. Oncogene. 22:3205–3212. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Wang S, Huang X, Lee CK and Liu B:
Elevated expression of erbB3 confers paclitaxel resistance in
erbB2-overexpressing breast cancer cells via upregulation of
Survivin. Oncogene. 29:4225–4236. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Coley HM: Mechanisms and strategies to
overcome chemotherapy resistance in metastatic breast cancer.
Cancer Treat Rev. 34:378–390. 2008. View Article : Google Scholar : PubMed/NCBI
|