Introduction
Fracture healing is an intricate biological process
that involves numerous events that occur during embryonic skeletal
development and requires the altered expression of thousands of
genes (1).
There have been numerous advances in the
understanding of the process of fracture healing. For example,
certain proinflammatory cytokines, including interleukin (IL)-1,
IL-6, IL-11, IL-18 and tumor necrosis factor-α (TNF-α), are
involved in the inflammatory response, which is essential for the
process of healing (2). Hypoxia
inducible factor-1α has been demonstrated to be critical for bone
repair, via the induction of vascular endothelial growth factor
(VEGF) in the revascularization process at the fracture site
(3). The deficiency of Fas
activity prolongs the life span of chondrocytes and Fas synergizes
with TNF-α signaling to modulate chondrocyte apoptosis, which
affects fracture healing (4). The
expression of α smooth muscle actin (αSMA) identifies mesenchymal
progenitor cells in bone marrow stromal cell cultures in
vitro (5), and osteoblast
precursors in the periodontium and bone marrow in vivo
(6,7). Using microarray analysis of
αSMA-labeled periosteal cells in mice, Matthews et al
(8) identified a series of
differentially expressed genes (DEGs) in fractured and unfractured
samples, and identified Notch signaling as an important signaling
pathway during bone healing. However, the protein-protein
interactions (PPIs) of DEGs, which are central to the majority of
biological processes and allow associations between genes to be
analyzed (9), were not
investigated.
The present study used the microarray data deposited
by Matthews et al (8) to
examine DEGs in fractured and unfractured samples. Following Gene
Ontology (GO) functional and Kyoto Encyclopedia of Genes and
Genomes (KEGG) pathway enrichment analyses, the PPIs of DEGs were
analyzed, and the PPI network was constructed. The results may
provide information for subsequent experimental studies, and
contribute to the understanding of the molecular mechanisms
underlying fracture healing.
Materials and methods
Illumina microarray data
The raw gene expression profile dataset GSE45156
(8) was obtained from Gene
Expression Omnibus (www.ncbi.nlm.nih.gov/geo/). The initial study was
performed on the platform of GPL6885 Illumina MouseRef-8 version
2.0 expression beadchip (Illumina, Inc., San Diego, CA, USA). A
total of nine αSMA-labeled periosteal cell samples from the tibia
of mice were included in this dataset, including three unfractured
controls collected two days following tamoxifen injections, which
labeled αSMA-expressing cells, and six samples isolated two (day 2;
n=3) and six (day 6; n=3) days following fracture.
In addition, CEL files and probe annotation files
were downloaded, and the gene expression data of all samples were
preprocessed by background correction, quantile normalization,
probe summarization and expression calculation using the linear
models for microarray data (LIMMA) package of Bioconductor
(bioconductor.org/packages/release/bioc/html/limma.html)
(10).
DEG screening
The LIMMA package was used to identify DEGs in day 2
and 6 fractured samples, compared with unfractured controls.
P-values for each gene were calculated using unpaired Student's
t-test, and genes with P<0.05 and fold-change ≥2 were designated
as DEGs.
Furthermore, the up and downregulated DEGs common to
day 2 and 6 fractured samples were identified.
Enrichment analysis of DEGs
To further reveal the functions of DEGs, GO
functional and KEGG pathway enrichment analyses of DEGs were
performed, via the Database for Annotation, Visualization and
Integrated Discovery (david.abcc.ncifcrf.gov/) (11). P<0.05 was set as the cut-off
criterion, other parameters were set as default.
Construction of PPI network
To investigate the interactions of DEGs, the Search
Tool for the Retrieval of Interacting Genes (string-db.org/), which integrates a variety of known
and predicted proteins associations (12), was used to identify the PPIs of
DEGs by calculating the combined score (threshold, score >0.4),
and the PPI network was visualized using Cytoscape (cytoscape.org/) (13).
Results
Identification of DEGs
Based on the cut-off criteria, a total of 774 DEGs
(371 upregulated and 403 downregulated) and 1,172 DEGs (636
upregulated and 536 downregulated) were identified in day 2 and 6
fractured samples, respectively, compared with unfractured
controls.
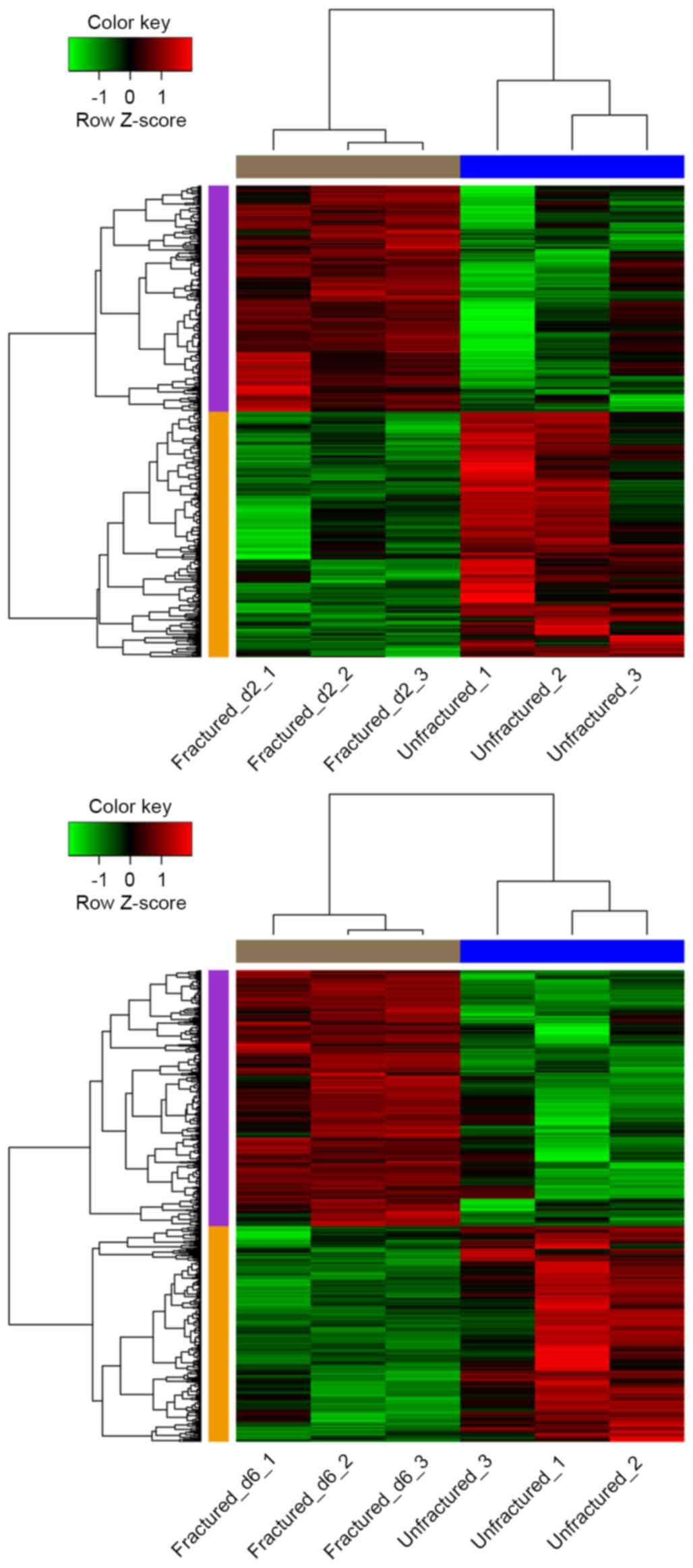
Hierarchical cluster analysis of the data suggested
that the DEGs may be used to accurately distinguish day 2 and 6
fractured samples from unfractured controls (Fig. 1).
Enrichment analysis of up and
downregulated DEGs
To examine the functions of DEGs, GO functional and
KEGG pathway enrichment analyses of DEGs common to day 2 and 6
fractured samples were performed.
Of the upregulated DEGs, various DEGs, including
protein kinase C α (Prkca), caspase 6 and B-cell lymphoma
antagonist/killer 1 were significantly enriched in GO terms
associated with cell death, including positive regulation of
apoptosis and positive regulation of programmed cell death. Various
other upregulated DEGs, including transcription factor B2,
mitochondrial and transcription factor B1, mitochondrial were
distinctly enriched in ribosomal RNA (adenine) methyltransferase
activity (Table I). Of the
downregulated DEGs, a set of genes, including fms-related tyrosine
kinase 1 (Flt1), nitric oxide synthase 3 (Nos3), bone morphogenetic
protein 4 (Bmp4) and Notch1 were markedly enriched in GO terms
associated with blood vessels, including angiogenesis and blood
vessel morphogenesis (Table
II).
 | Table I.GO terms with the greatest P-values in
BP, CC and MF for the upregulated genes differentially expressed by
day 2 and 6 fracture samples. |
Table I.
GO terms with the greatest P-values in
BP, CC and MF for the upregulated genes differentially expressed by
day 2 and 6 fracture samples.
| Category | Term | P-value | Count | Genes |
|---|
| BP | GO:0043065-positive
regulation of apoptosis | 0.011428 | 7 | Prkca, Casp6,
Bak1, Nudt2, Mlh1, Dapk3, Il10 |
|
| GO:0043068-positive
regulation of programmed cell death | 0.011855 | 7 | Prkca, Casp6,
Bak1, Nudt2, Mlh1, Dapk3, Il10 |
|
| GO:0010942-positive
regulation of cell death | 0.012292 | 7 | Prkca, Casp6,
Bak1, Nudt2, Mlh1, Dapk3, Il10 |
|
|
GO:0005996-monosaccharide metabolic
process | 0.015052 | 6 | Pdk1, Galk1,
Ugt1A10, Pgam1, Gale, Eno1 |
|
|
GO:0006775-fat-soluble vitamin metabolic
process | 0.027270 | 3 | Rdh11, Vkorc1L1,
Crabp2 |
| CC |
GO:0005783-endoplasmic reticulum | 0.013554 | 14 | Scd2, Rrbp1, Alg3,
Tor2A, Ugt1A10, Rdh11, Bak1, Vrk2, Zdhhc16, Stx18… |
|
|
GO:0005739-mitochondrion | 0.027269 | 18 | Pdk1, Prkca,
Usp30, Nudt2, Acat2, Spryd4, Gmppb, Bak1, Nudt8, Tfb2M… |
| MF | GO:0000166-nucleotide
binding | 0.020428 | 28 | Acox2, Rac3, Tia1,
Tube1, Rhof, Prkca, Pdk1, Nudt2, U2Af1L4, Tor2A… |
|
| GO:0016433-rRNA
(adenine) methyltransferase activity | 0.024853 | 2 | Tfb2M,
Tfb1M |
|
| GO:0008649-rRNA
methyltransferase activity | 0.024853 | 2 | Tfb2M,
Tfb1M |
|
| GO:0000179-rRNA
(adenine-N6,N6-)-dimethyltransferase activity | 0.024853 | 2 | Tfb2M,
Tfb1M |
|
| GO:0017076-purine
nucleotide binding | 0.034672 | 24 | Pdk1, Acox2,
Prkca, Ephb4, Tor2A, Galk1, Rac3, Dhx37, Rhof, Nek6… |
 | Table II.GO terms with the greatest P-values
in BP, CC and MF for the downregulated genes differentially
expressed by day 2 and 6 fracture samples. |
Table II.
GO terms with the greatest P-values
in BP, CC and MF for the downregulated genes differentially
expressed by day 2 and 6 fracture samples.
| Category | Term | P-value | Count | Genes (n≤10) |
|---|
| BP |
GO:0001525-angiogenesis | 3.09E-07 | 13 | Bmp4, Vegfc,
Notch1, Flt1, Epas1, Ovol2, Notch4, Edn1, Sox18, Nos3… |
|
| GO:0048514-blood
vessel morphogenesis | 6.42E-07 | 15 | Bmp4, Flt1,
Epas1, Notch1, Hey1, Ovol2, Notch4, Tgm2, Nos3, Sox18… |
|
| GO:0001568-blood
vessel development | 1.50E-06 | 16 | Bmp4, Flt1,
Epas1, Edn1, Notch1, Hey1, Ovol2, Notch4, Nos3, Sox18… |
|
|
GO:0001944-vasculature development | 2.03E-06 | 16 | Bmp4, Flt1,
Epas1, Edn1, Notch1, Hey1, Ovol2, Notch4, Nos3, Sox18… |
|
|
GO:0007242-intracellular signaling
cascade | 2.27E-05 | 30 | Adcy4, Rab9B,
Edn1, Cdc42Ep4, Pik3C2G, Bmx, Rps6Ka6, Prkcq, Adcy9,
Notch4… |
| CC | GO:0005886-plasma
membrane | 7.67E-05 | 63 | Adcy4, Eltd1,
Nos3, Prkcq, Gpr22, Flt1, Maob, Abca8A, Notch1, Adcy9… |
|
|
GO:0005576-extracellular region | 0.003702 | 37 | Edn1, Clu, Il15,
Timp3, Dspp, Ifna1, Apod, Tgm2, Itih5, Bmp4… |
|
| GO:0044459-plasma
membrane part | 0.007555 | 35 | Enpp5, Nos3,
Slc22A2, Gabrg1, F11R, Selp, Flt1, Prkcq, Notch1, Notch4… |
|
| GO:0016021-integral
to membrane | 0.013021 | 94 | Adcy4, Kcnc4,
Olfr883, Flt1, Gpr33, Rprml, Notch1, Adcy9, Plscr4,
Notch4… |
|
|
GO:0031224-intrinsic to membrane | 0.025857 | 95 | Adcy4, Kcnc4,
Olfr883, Flt1, Notch1, Notch4, Olfr917, Pecam1, Dsc3,
Adra1A… |
| MF | GO:0030552-cAMP
binding | 8.23E-04 | 4 | Pde1C, Rapgef3,
Hcn3, Cnga2 |
|
| GO:0016208-AMP
binding | 0.002427 | 4 | Pde1C, Rapgef3,
Hcn3, Cnga2 |
|
| GO:0030551-cyclic
nucleotide binding | 0.003213 | 4 | Pde1C, Rapgef3,
Hcn3, Cnga2 |
|
| GO:0009975-cyclase
activity | 0.004139 | 4 | Adcy4, Adcy9,
Gucy1A2, Npr1 |
|
|
GO:0016849-phosphorus-oxygen lyase
activity | 0.004139 | 4 | Adcy4, Adcy9,
Gucy1A2, Npr1 |
Additionally, according to the pathway enrichment
analysis, the upregulated DEGs GDP-mannose pyrophosphorylase B,
galactokinase 1, N-acetylneuraminate synthase and
UDP-galactose-4-epimerase were primarily enriched in the pathways
of amino and nucleotide sugar metabolism. The downregulated DEGs
were significantly enriched in certain pathways, including the
notch signaling pathway (hes family bHLH transcription factor 1,
Notch1 and MFNG O-fucosylpeptide
3-beta-N-acetylglucosaminyltransferase), leukocyte transendothelial
migration (F11 receptor, claudin 9 and platelet and endothelial
cell adhesion molecule 1), and vascular smooth muscle contraction
[protein kinase C θ, adenylate cyclase (Adcy) 4 and Adcy9; Table III].
 | Table III.Enriched pathways for the up and
downregulated genes differentially expressed in day 2 and 6
fractured samples. |
Table III.
Enriched pathways for the up and
downregulated genes differentially expressed in day 2 and 6
fractured samples.
|
Up/downregulated | Term | P-value | Count | Genes |
|---|
| Upregulated | mmu00520-amino
sugar and nucleotide sugar metabolism | 0.011615 | 4 | Gmppb, Galk1,
Nans, Gale |
|
|
mmu05310-asthma | 0.048953 | 3 | Fcer1A, Prg2,
Il10 |
| Downregulated | mmu04330-Notch
signaling pathway | 0.010106 | 5 | Hes1, Notch1,
Mfng, Notch4, Dll1 |
|
| mmu04670-leukocyte
transendothelial migration | 0.015684 | 7 | F11R, Cldn9,
Pecam1, Cldn11, Rapgef3, Jam2, Ctnna3 |
|
| mmu04270-vascular
smooth muscle contraction | 0.016287 | 7 | Prkcq, Adcy4,
Adcy9, Gucy1A2, Adra1A, Prkch, Npr1 |
|
| mmu04530-tight
junction | 0.027350 | 7 | F11R, Prkcq,
Cldn9, Prkch, Cldn11, Jam2, Ctnna3 |
|
| mmu04514-cell
adhesion molecules | 0.047343 | 7 | F11R, Selp,
Cldn9, Pecam1, Cldn11, Jam2, Sele |
Analysis of the PPI network
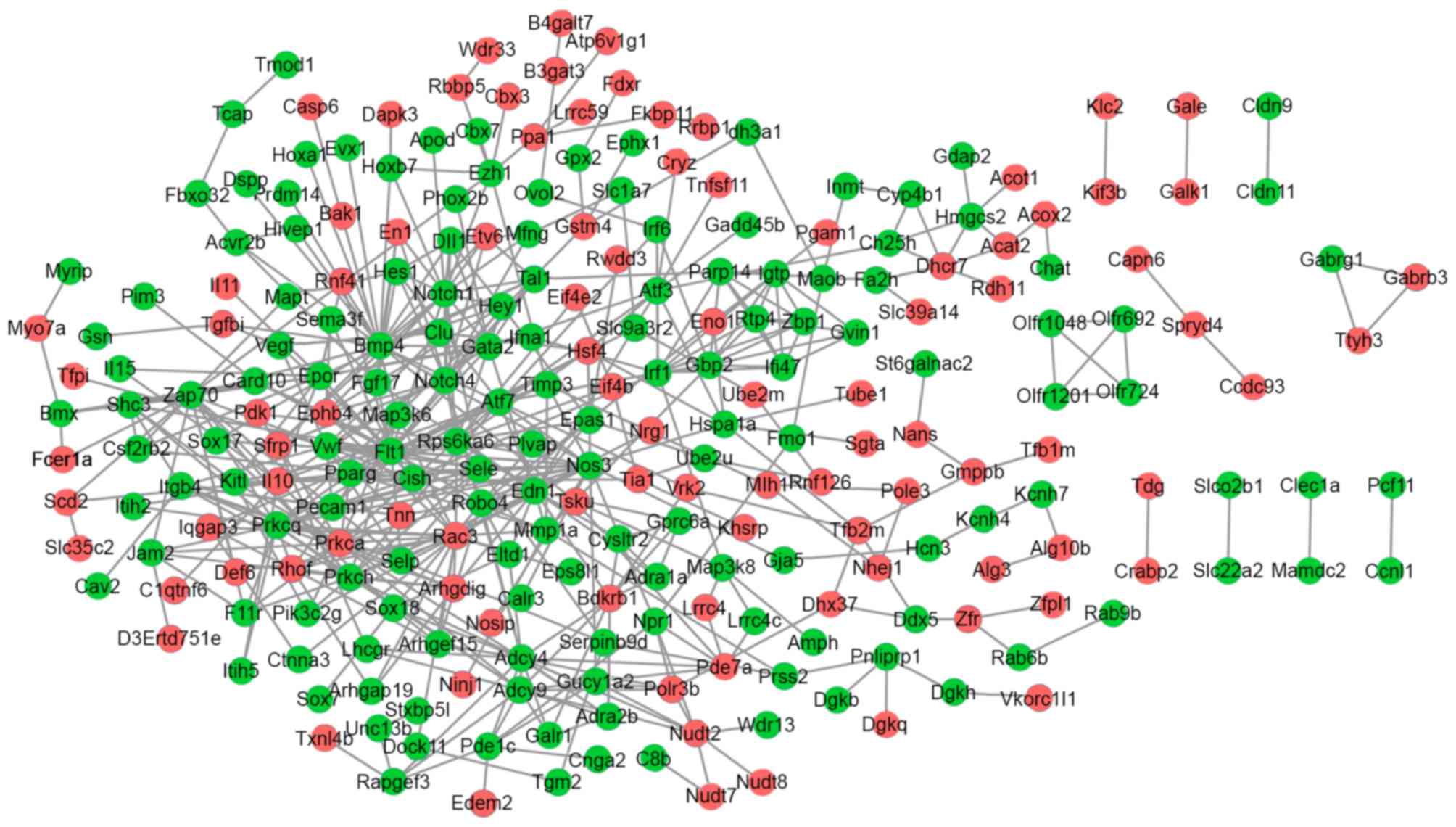
The PPI network for the up and downregulated DEGs
consisted of 249 genes and 512 interactions. Prkca and Il10
interacted with Nos3; Flt1, Nos3, Bmp4 and Notch1 interacted with
each other (Fig. 2).
Various genes had a high connectivity degree,
including Flt1 (degree=27), Nos3 (degree=23), Bmp4 (degree=22),
ras-related C3 botulinum toxin substrate 3 (Rac3; degree=21),
Notch1 (degree=18) and Prkca (degree=18).
Discussion
The present study identified a total of 774 DEGs
(371 upregulated and 403 downregulated) and 1,172 DEGs (636
upregulated and 536 downregulated) from day 2 and 6 fractured
samples, respectively, compared with unfractured controls.
According to the analysis of the PPI network, various downregulated
DEGs with a high degree were revealed to interact with each other,
including Flt1, Nos3, Bmp4 and Notch1. Furthermore, based on the
enrichment analysis, all of these genes were significantly enriched
in angiogenesis and blood vessel morphogenesis.
Flt1, is also known as vascular endothelial growth
factor receptor 1 (Vegfr-1) (14).
VEGF is an essential regulator during angiogenesis, which is
critical for bone growth, remodeling and repair (15). A previous study observed Flt1
expression in vascular endothelial cells at the fracture site 8 h
to 8 weeks following fracture (16). Endothelial nitric oxide synthase
(eNOS), encoded by Nos3 in endothelial cells, is the predominant
NOS isoform expressed in bone (17). A previous study has demonstrated
that mice with eNOS deficiency have reduced bone mineral density,
compared with wild-type controls (18). In addition, Nos3 was detected to be
differentially expressed in lymph node lymphocytes and endothelial
cells in patients with bone fracture (19). Nitric oxide is associated with
vascular smooth muscle relaxation, and modulates VEGF-induced
angiogenesis (20). Thus, Flt1 and
Nos3 may be closely associated with angiogenesis during fracture
healing.
Bmp4 is a member of the transforming growth factor-β
superfamily (21). Bone
morphogenetic proteins (BMPs) are important in the initiation of
endochondral bone formation in humans. Types I and II, the BMP
receptors, bind BMPs and act in collaboration to phosphorylate
mothers against decapentaplegic (SMAD) 1 and SMAD5, which
translocate to the nucleus in cooperation with SMAD4 to initiate
BMP responses including fracture healing (22). There is evidence that rat
adipose-derived stromal cells expressing Bmp4 may induce bone
formation in vitro and in vivo (23), indicating that Bmp4 may be key for
bone repair. Furthermore, Notch1 was significantly enriched in the
Notch signaling pathway in the present study. Genetically inducible
inhibition of Notch signaling extends the inflammatory phase of
fracture healing and alters cartilage formation (24). Matthews et al (8) reported that downregulation of Notch
signaling in αSMA-labeled progenitor cells contributes to fracture
callus formation. A recent study demonstrated that transient
inhibition of Notch signaling and gamma secretase activity
temporarily promotes osteoclastogenesis and accelerates bone
remodeling (25). In the present
study, the PPI network revealed that Notch1 interacts with Flt1 and
Bmp4. Notch1 may modulate angiogenesis (26,27),
and functional Notch signaling is essential for BMP-induced
osteoblast differentiation (28).
Taken together, these results suggested that Notch1 may be crucial
in fracture healing, via interactions with Flt1 and Bmp4.
Of the upregulated DEGs, Rac3 and Prkca have a high
degree in the PPI network, and interacted with Nos3. Rac3 encodes a
GTPase belonging to the ras superfamily of small GTP-binding
proteins, which are involved in the regulation of cell growth, the
activation of protein kinases and cytoskeletal reorganization
(29,30). To date, there is no evidence that
Rac3 is associated with bone; it does however interact with Nos3,
and therefore may be involved in fracture healing via Nos3. It has
been demonstrated that Rac1 deficiency increases vertebral
osteoclast-mediated bone quality compared with wild-type bones in a
murine ovariectomy model (31).
Therefore, Rac3 may be additionally implicated in bone quality.
Prkca, a serine- and threonine-specific protein kinase, was
markedly enriched in positive regulation of apoptosis in the
present study. Apoptosis is active during the phase of callus
remodeling (32). In addition,
Prkca has been observed to be upregulated during fracture repair
(33). Furthermore, during
fracture healing accelerated by thrombin peptide TP508, a series of
genes involved in apoptosis, including Prkca, were upregulated
(34). Therefore, Prkca may be
important in fracture repair.
In conclusion, the present study identified 774 and
1,172 DEGs in day 2 and 6 fractured samples, respectively, compared
with unfractured controls. Various upregulated DEGs (for example,
Rac3 and Prkca) and downregulated DEGs (for example, Flt1, Nos3,
Bmp4 and Notch1) with a high degree in the PPI network may be
critical for fracture healing via involvement in angiogenesis or
apoptosis regulation. These results require confirmation by further
studies, which is a limitation of the present study. However, the
results of the present study may provide useful information for
subsequent studies, and contribute to an improved understanding of
the molecular mechanisms underlying fracture healing.
References
|
1
|
Marsell R and Einhorn TA: The biology of
fracture healing. Injury. 42:551–555. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gerstenfeld LC, Cullinane DM, Barnes GL,
Graves DT and Einhorn TA: Fracture healing as a post-natal
developmental process: Molecular, spatial, and temporal aspects of
its regulation. J Cell Biochem. 88:873–884. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wan C, Shao J, Gilbert SR, Riddle RC, Long
F, Johnson RS, Schipani E and Clemens TL: Role of HIF-1alpha in
skeletal development. Ann N Y Acad Sci. 1192:322–326. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Al-Sebaei MO, Daukss DM, Belkina AC, Kakar
S, Wigner NA, Cusher D, Graves D, Einhorn T, Morgan E and
Gerstenfeld LC: Role of Fas and Treg cells in fracture healing as
characterized in the Fas-Deficient (lpr) mouse model of lupus. J
Bone Miner Res. 29:1478–1491. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Grcevic D, Pejda S, Matthews BG, Repic D,
Wang L, Li H, Kronenberg MS, Jiang X, Maye P, Adams DJ, et al: In
vivo fate mapping identifies mesenchymal progenitor cells. Stem
cells. 30:187–196. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kalajzic Z, Li H, Wang LP, Jiang X,
Lamothe K, Adams DJ, Aguila HL, Rowe DW and Kalajzic I: Use of an
alpha-smooth muscle actin GFP reporter to identify an
osteoprogenitor population. Bone. 43:501–510. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Roguljic H, Matthews B, Yang W, Cvija H,
Mina M and Kalajzic I: In vivo identification of periodontal
progenitor cells. J Dent Res. 92:709–715. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Matthews BG, Grcevic D, Wang L, Hagiwara
Y, Roguljic H, Joshi P, Shin DG, Adams DJ and Kalajzic I: Analysis
of αSMA-labeled progenitor cell commitment identifies notch
signaling as an important pathway in fracture healing. J Bone Miner
Res. 29:1283–1294. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shen J, Zhang J, Luo X, Zhu W, Yu K, Chen
K, Li Y and Jiang H: Predicting protein-protein interactions based
only on sequences information. Proc Natl Acad Sci USA.
104:4337–4341. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Smyth GK: Limma: linear models for
microarray dataBioinformatics and computational biology solutions
using R and Bioconductor. Springer; New York, NY: pp. 397–420.
2005, View Article : Google Scholar
|
|
11
|
Huang DW, Sherman BT, Tan Q, Collins JR,
Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC and Lempicki
RA: The DAVID gene functional classification tool: A novel
biological module-centric algorithm to functionally analyze large
gene lists. Genome Biol. 8:R1832007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43:D447–D452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kohl M, Wiese S and Warscheid B:
Cytoscape: Software for visualization and analysis of biological
networks. Methods Mol Biol. 696:291–303. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sawano A, Takahashi T, Yamaguchi S and
Shibuya M: The phosphorylated 1169-tyrosine containing region of
flt-1 kinase (VEGFR-1) is a major binding site for PLCgamma.
Biochem Biophys Res Commun. 238:487–491. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yang YQ, Tan YY, Wong R, Wenden A, Zhang
LK and Rabie AB: The role of vascular endothelial growth factor in
ossification. Int J Oral Sci. 4:64–68. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chu TW, Liu YG, Wang ZG, Zhu PF and Liu
LD: Vascular endothelial growth factor and its receptor expression
during the process of fracture healing. Chin J Traumatol.
11:161–164. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Marsden PA, Schappert KT, Chen HS, Flowers
M, Sundell CL, Wilcox JN, Lamas S and Michel T: Molecular cloning
and characterization of human endothelial nitric oxide synthase.
FEBS Lett. 307:287–293. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Armour KE, Armour KJ, Gallagher ME,
Gödecke A, Helfrich MH, Reid DM and Ralston SH: Defective bone
formation and anabolic response to exogenous estrogen in mice with
targeted disruption of endothelial nitric oxide synthase.
Endocrinology. 142:760–766. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Szczesny G, Olszewski WL and Zaleska M:
Limb lymph node response to bone fracture. Lymphat Res Biol.
2:155–164. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Suganthalakshmi B, Anand R, Kim R,
Mahalakshmi R, Karthikprakash S, Namperumalsamy P and Sundaresan P:
Association of VEGF and eNOS gene polymorphisms in type 2 diabetic
retinopathy. Mol Vis. 12:336–341. 2006.PubMed/NCBI
|
|
21
|
Shore EM, Xu M, Shah PB, Janoff HB, Hahn
GV, Deardorff MA, Sovinsky L, Spinner NB, Zasloff MA, Wozney JM and
Kaplan FS: The human bone morphogenetic protein 4 (BMP-4) gene:
Molecular structure and transcriptional regulation. Calcif Tissue
Int. 63:221–229. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Reddi A: Initiation of fracture repair by
bone morphogenetic proteins. Clin Orthop Relat Res. 355
Suppl:S66–S72. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lin L, Fu X, Zhang X, Chen LX, Zhang JY,
Yu CL, Ma KT and Zhou CY: Rat adipose-derived stromal cells
expressing BMP4 induce ectopic bone formation in vitro and in vivo.
Acta Pharmacol Sin. 27:1608–1615. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Dishowitz MI, Mutyaba PL, Takacs JD, Barr
AM, Engiles JB, Ahn J and Hankenson KD: Systemic inhibition of
canonical notch signaling results in sustained callus inflammation
and alters multiple phases of fracture healing. PLoS One.
8:e687262013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang C, Shen J, Yukata K, Inzana JA,
O'Keefe RJ, Awad HA and Hilton MJ: Transient gamma-secretase
inhibition accelerates and enhances fracture repair likely via
Notch signaling modulation. Bone. 73:77–89. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Okamura H, Proia T, Bell A, Liu Q,
Siddiquee Z, Lin J and Gyuris J: Notch1 monoclonal antibody
inhibits tumor growth and modulates angiogenesis. Cancer Res.
74:2990. 2014. View Article : Google Scholar
|
|
27
|
Zhu J, Liu Q, Jiang Y, Wu L, Xu G and Liu
X: Enhanced angiogenesis promoted by human umbilical mesenchymal
stem cell transplantation in stroked mouse is Notch1 signaling
associated. Neuroscience. 290:288–299. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Nobta M, Tsukazaki T, Shibata Y, Xin C,
Moriishi T, Sakano S, Shindo H and Yamaguchi A: Critical regulation
of bone morphogenetic protein-induced osteoblastic differentiation
by Delta1/Jagged1-activated Notch1 signaling. J Biol Chem.
280:15842–15848. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Haataja L, Groffen J and Heisterkamp N:
Characterization of RAC3, a novel member of the Rho family. J Biol
Chem. 272:20384–20388. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hajdo-Milasinovic A, van der Kammen RA,
Moneva Z and Collard JG: Rac3 inhibits adhesion and differentiation
of neuronal cells by modifying GIT1 downstream signaling. J Cell
Sci. 122:2127–2136. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Magalhaes JK, Grynpas MD, Willett TL and
Glogauer M: Deleting Rac1 improves vertebral bone quality and
resistance to fracture in a murine ovariectomy model. Osteoporosis
Int. 22:1481–1492. 2011. View Article : Google Scholar
|
|
32
|
Li G, White G, Connolly C and Marsh D:
Cell proliferation and apoptosis during fracture healing. J Bone
Miner Res. 17:791–799. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li X, Wang H, Touma E, Rousseau E, Quigg
RJ and Ryaby JT: Genetic network and pathway analysis of
differentially expressed proteins during critical cellular events
in fracture repair. J Cell Biochem. 100:527–543. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li X, Wang H, Touma E, Qi Y, Rousseau E,
Quigg RJ and Ryaby JT: TP508 accelerates fracture repair by
promoting cell growth over cell death. Biochem Biophys Res Commun.
364:187–193. 2007. View Article : Google Scholar : PubMed/NCBI
|