Introduction
Acute myeloid leukemia (AML) is one kind of
hematological malignant cancers and characterized by an incensement
of undifferentiated myeloid progenitor cells in peripheral blood
and bone marrow (1,2). AML cells were clinically
heterogeneous and their progenitor cells have a self-renewal
potential to generate the bulk of leukemia cells (3,4). The
incidence and mortality are gradually increasing in developing
countries (5). In China, the
incidence of AML is the highest among young patients (5). Chemotherapy consisting paclitaxel,
cisplatin or others is the main approach for AML treatment
(6). However, because of therapy
resistance, tumor metastasis and recurrence, the outcomes of AML
patients remain very poor (7). AML
has become a great problem for public health. Thus, it is critical
to elucidate the underlying molecular mechanism of AML development
and progression, which will contribute to the development of
effective therapeutic strategies for AML.
MicroRNAs (miRNAs/miRs) are a class of small
noncoding RNAs with a length of about 22 nucleotides and expressed
in nearly all kinds of cells and tissues (8). miRNAs have been demonstrated to
regulate gene expression post-transcriptionally via specifically
binding to the 3′-UTR region of target mRNAs for degradation
(9,10). In the past decades, a vast of
evidence indicates that miRNAs exert vital functions in various
biological processes, such as cell proliferation, apoptosis,
migration and invasion (11).
There was a close relationship between miRNA expression and human
cancers, including lung cancer (12), bladder cancer (13), hepatocellular carcinoma (14) and AML (15). miRNAs could regulate the
development and progression of cancers through serving as oncogenes
or tumor suppressors (16).
Aberrant expression of miRNAs is often observed in human cancer.
Therefore, it is important to define the function and mechanism of
miRNAs involved in cancers.
miR-135a has been reported to serve as tumor
suppressor or an oncogene in different cancers. However, the role
of miR-135a in AML remains largely unknown. In the present study,
we found that miR-135a was downregulated in AML samples and its low
expression predicted poor prognosis. Furthermore, we found that
miR-135a overexpression inhibited the proliferation and induced
apoptosis of AML cells. miR-135a could arrest AML cell cycle
progression. In mechanism, we showed that miR-135a directly
targeted HOXA10, whose restoration rescued the effects of miR-135a
mimic transfection in AML cells. Taken together, our study
identified miR-135a as a tumor suppressor in AML by targeting
HOXA10 and a prognostic marker for AML patients.
Materials and methods
Clinical specimens
All the blood samples from AML patients (n=29) and
the normal volunteers (n=11) were obtained from the Second
Affiliated Hospital of Harbin Medical University (Harbin, China).
All the patients were diagnosed as AML according to the
pathological and clinic features. The normal volunteers were the
healthy persons without any diseases. Informed consent was obtained
from each patient, and the research protocols were approved by the
Ethics Committee of the Second Affiliated Hospital of Harbin
Medical University (Harbin, China).
Cell culture and transfection
AML cell lines including HL60, AML193, AML2, AML5
and HS-5 normal cells from marrow stroma were primarily purchased
from ATCC and maintained in Eagle's Minimum Essential Medium,
supplemented with 10% fetal bovine serum (both from Gibco; Thermo
Fisher Scientific, Inc.) and 1% penicillin/streptomycin
(Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C, 5%
CO2. Cell transfection was conducted using
Lippofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol.
The coding sequence of HOXA10 was amplified from
human cDNA and inserted into pcDNA6.1 (Invitrogen; Thermo Fisher
Scientific, Inc.) vectors. miR-135a mimic
(5′-AGUGUAUCCUUAUUUUUCGGUAU-3′), miR-135a inhibitor
(5′-UCACAUAGGAAUAAAAAGCCAUA-3′) and mimic controls
(5′-AUUCUCCGAACGUGUCACGUAGU-3′) were purchased from Shanghai
GenePharma Co., Ltd. (Shanghai, China) and used at a final
concentration of 25 nmol/l. Cell transfection was performed by
using Lipo2000 (Invitrogen; Thermo Fisher Scientific, Inc.) in
accordance with the manufacturer's instructions.
Cell proliferation assay
AML5 and HL60 cells were transfected with miR-135a
mimics or the controls in 6-well plates for 24 h and then collected
to seed in 96-well plates incubating for 1, 3 and 5 days. 10 µl
CCK-8 reagents were added to cultures and incubated for 2 h;
measurement of absorbance of each well at 450 nm using a SUNRISE
Microplate Reader (Tecan Group, Ltd., Mannedorf, Switzerland) was
then performed.
Reverse transcription and quantitative
PCR
Peripheral blood mononuclear cells from AML patients
or normal individuals were separated using Ficoll
Hypaque® (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany)
gradient centrifugation, and total RNAs were extracted using TRIzol
Reagent (TransStart, Beijing, China) following the manufacturer's
instructions. The first strand cDNA was compounded using a
Tianscript RT kit (Tiangen Biotech, Beijing, China). PCR
amplification was performed using TaqMan Human MicroRNA Assay
(Applied Biosystems; Thermo Fisher Scientific, Inc.) and UltraSYBR
Mixture (CW0957; CWBio, Beijing, China) in LC 480 PCR System (Roche
Diagnostics, Indianapolis, IN, USA). U6 and 18S was employed as
reference genes to normalize the expression of miR-135a or HOXA10.
Relative expression level was analyzed on the basis of the
2−ΔΔCq method (17).
The primers used were synthesized and purchased from Genecopoeia
(Guangzhou, China). The primer sequences were as follows: universal
miRNA qRT-PCR primer (5′-AACGAGACGACGACAGAC-3′), U6
(5′-GCAAATTCGTGAAGCGTTCCATA-3′), miR-135a
(5′-TATGGCTTTTTATTCCTATGT-3′), HOXA10 (forward,
5′-ATCTGCTCCCTTCGCCAAAT-3′ and reverse, 5′-CTGATGAGCGAGTCGACCAA-3′)
and 18S (forward, 5′-ATCAAAACCAACCCGGTCAG-3′ and reverse,
5′-CCCCGTCACCCGTGGTCACCA-3′).
Cell cycle and apoptosis assay
To determine the effect of miR-135a on cell
apoptosis, AML5 and HL60 cells were transfected with miR-135a
mimics or miR-NC for 36 h, then cells were harvested, washed with
PBS, and centrifuged at 500 g at 4°C for 10 min. Then cells were
incubated with Annexin V in the dark for 15 min, propidium iodide
(PI) was added 5 min before the end of incubation. After
centrifugation for 5 min, cells were resuspended using PBS and FACS
was performed on a FACSCalibur flow cytometer (BD Biosciences,
Franklin Lakes, NJ, USA). The effect of miR-135a on cell cycle was
further assessed as well. After transfection, cells were washed
with PBS, fixed with 75% ethanol for 1 h at −20°C. After washing
with PBS, cells were incubated in PI solution for 20 min at RT.
Then cells were analyzed with fluorescence-activated cell sorting
(FACS).
Western blot analysis
Proteins were extracted from whole cell lysates and
separated by sodium dodecyl sulfate-polyacrylamide gel
electrophoresis, then transferred to a polyvinylidene fluoride
(PVDF) membrane. The following primary antibodies were used:
anti-cyclin D1 (1:1,000; cat. no. 2978) and anti-cyclin E1
(1:1,000, cat. no. 20808; both from Cell Signaling Technology,
Inc., Danvers, MA, USA), anti-HOXA10 (1:1,000, cat. no. SAB2101061;
Sigma-Aldrich; Merck KGaA), anti-PCNA (1:3,000; cat. no. 13110) and
mouse anti-GAPDH (1:5,000; cat. no. 5174; both from Cell Signaling
Technology, Inc.). Membranes were incubated with primary antibodies
at 4°C overnight, followed by incubation with the horseradish
peroxidase-conjugated secondary anti-bodies (1:5,000; Abcam,
Cambridge, MA, USA) for 1 h at 25°C. Ultimately, the membrane was
treated with chemiluminescence reagents (Santa Cruz Biotechnology,
Inc., Dallas, TX, USA) according to the manufacturer's
instructions, and the image was analyzed.
Luciferase reporter assay
The potential miRNA binding sites of miR-135a
predicted by computer-aided algorithms were obtained from
microRNA.org target program (www.microRNA.org). The wild-type 3′-UTR of HOXA10
predicted to interact with miR-135a, together with mutant binding
sequence within the predicted target sites, were synthesized by
Shanghai GenePharma Co., Ltd., inserted into the pmirGLO
Dual-Luciferase vector (Promega Corporation, Madison, WI, USA). AML
cells were cotransfected with 500 ng of the luciferase construct
along with miR-135a and miR-135a control using Lipofectamine™ 2000
reagent, in accordance with the manufacturer's protocol. Following
cultivation for 48 h, the transfected cells were collected and
evaluated for luciferase activity using a Dual Luciferase Reporter
Assay kit (Promega Corporation) in accordance with the
manufacturer's manual. The activity of firefly luciferase was
normalized to that of Renilla luciferase. The results were
obtained from three independent experiments performed in
duplicate.
Statistical analysis
Each experiment was repeated at least three times.
All data are expressed in terms of means ± the SD. The Kaplan-Meier
method was used to calculate the survival curve, and log-rank test
to determine statistical significance. The differences between
groups were analyzed using two-tail Student's t-test or one-way
ANOVA followed by Tukey's post hoc test. P<.05 was considered
statistically significant.
Results
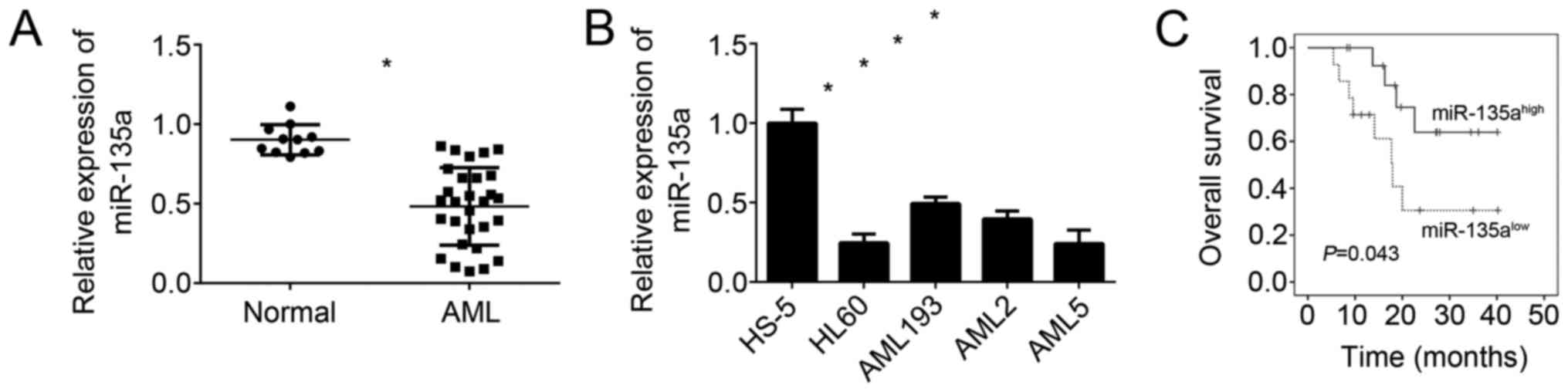
miR-135a is downregulated in AML
In order to explore the cellular functions of
miR-135a in AML, we firstly determined the expression of miR-135a
in blood samples with AML by qRT-PCR. We found that the expression
of miR-135a was significantly downregulated in AML samples compared
to normal samples (Fig. 1A).
Besides, miR-135a levels in AML cell lines were also lower than in
normal cell line (HS-5) (Fig. 1B).
Then we divided these AML samples into two subgroups base on
miR-135a levels and performed Kaplan-Meier curve analysis. We found
that higher expression of miR-135a in AML patients predicted better
prognosis (Fig. 1C). These results
suggested that miR-135a may exert a role in human AML progression
and serve as a predictor for patients' prognosis.
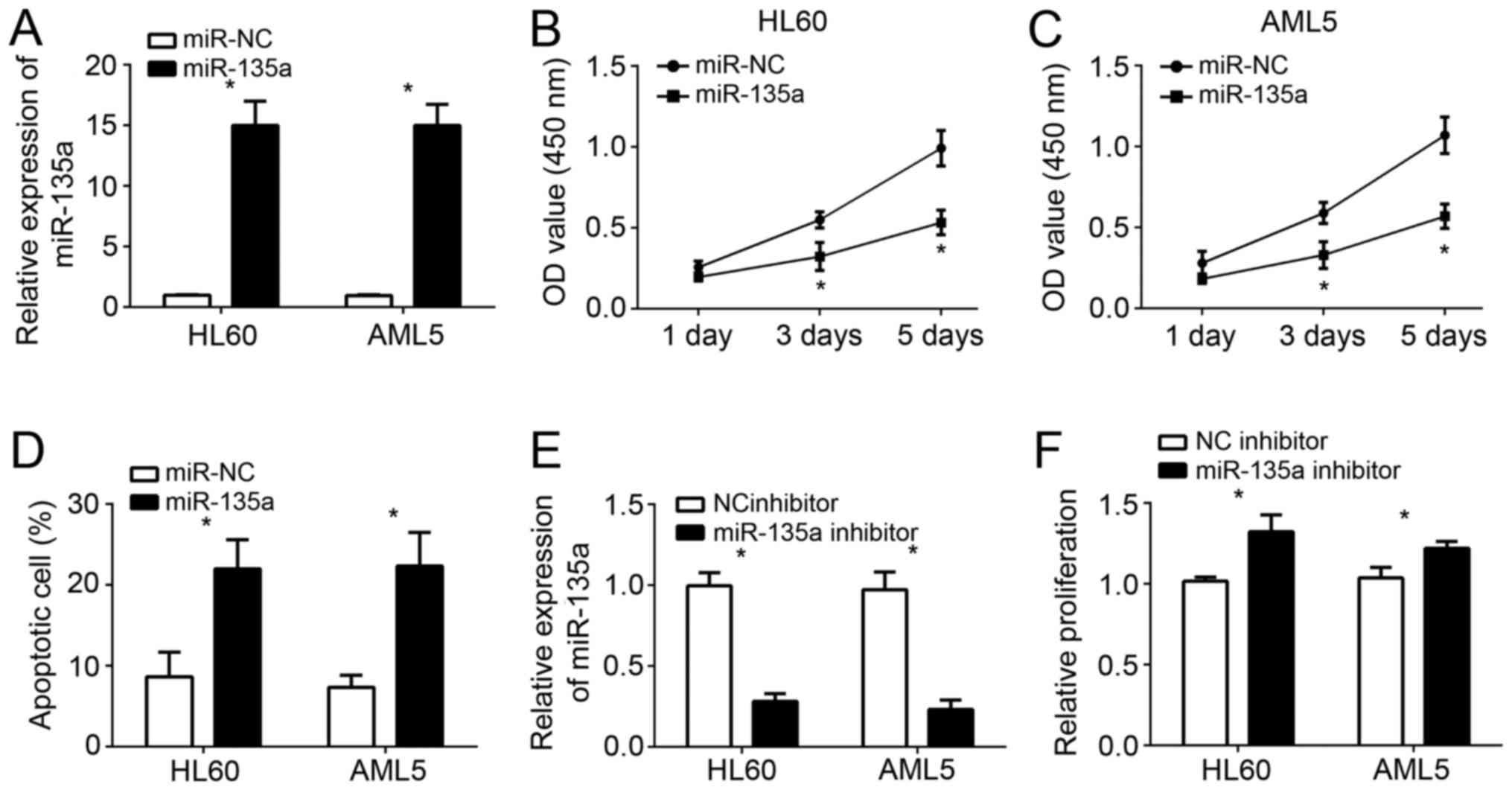
miR-135a inhibits cell proliferation
and induces apoptosis in AML
From Fig. 1B, we
knew that miR-135a levels were relative lower in HL60 and AML5
cells than other cell lines. Therefore, we chose these two cell
lines for further analysis. We overexpressed miR-135a in HL60 and
AML5 cells through transfection with miR-135a mimics. As shown by
qRT-PCR analysis, the levels of miR-135a were significantly
upregulated in HL60 and AML5 cells (Fig. 2A). CCK-8 assays were used to
examine the cellular proliferation. The results showed that
overexpression of miR-135a significantly suppressed the cell growth
in HL60 and AML5 cells at day 3 and 5 (Fig. 2B and C). Furthermore, we performed
fluorescence activated cell sorter (FACS) analysis to check cell
apoptosis. We found that miR-135a overexpression markedly elevated
the percentages of apoptotic cells (Fig. 2D). To further confirm it, we
inhibited miR-135a by transfection with miR-135a inhibitors in AML
cells (Fig. 2E). CCK-8 assays
indicated that miR-135a inhibition significantly promoted the
proliferation of HL60 and AML5 cells (Fig. 2F).
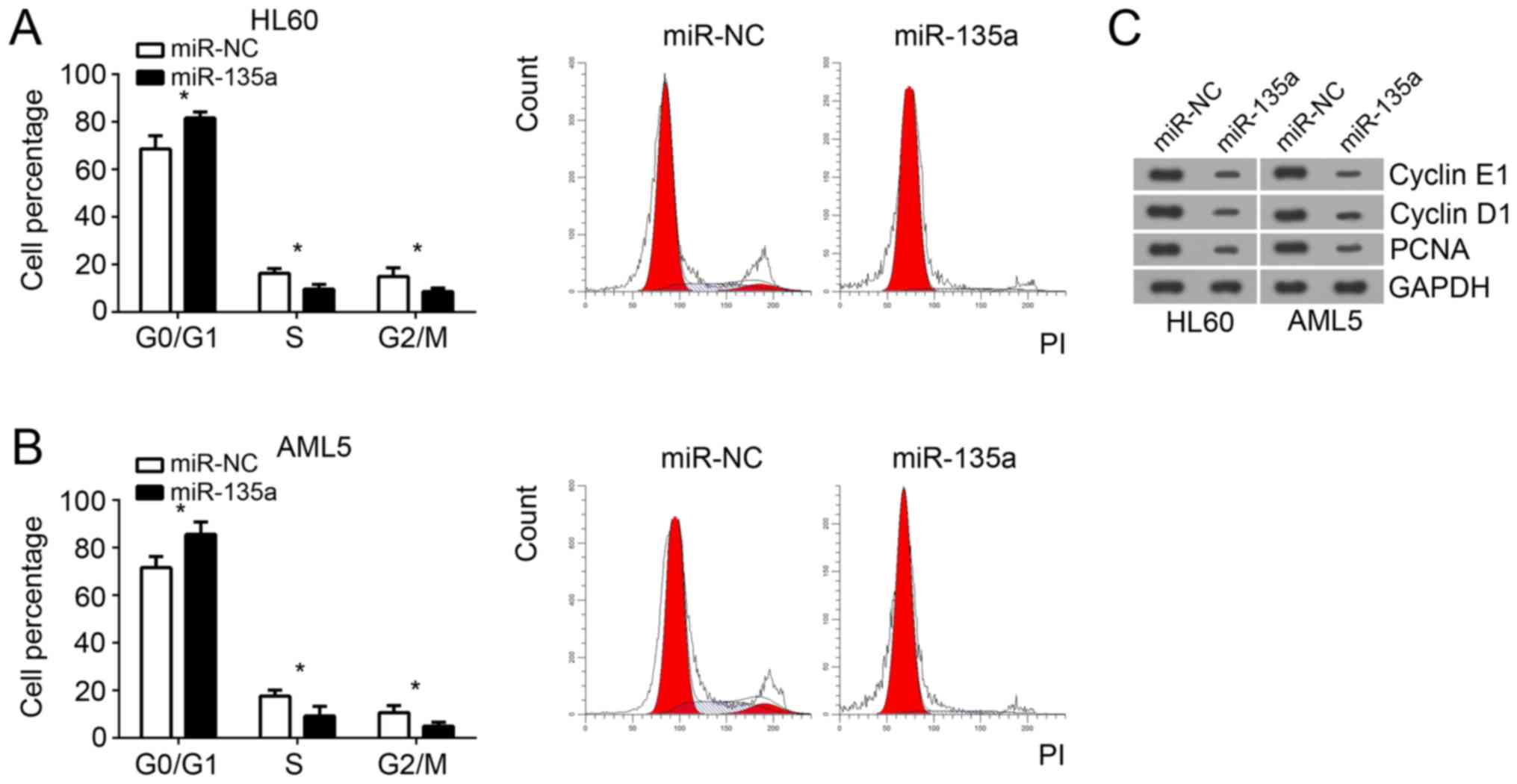
miR-135a suppresses cell cycle
progression
Cell proliferation was directly influenced by cell
cycle. Therefore, we analyzed the effect of miR-135a on cell cycle
by FACS. We found that overexpression of miR-135a significantly
increased the numbers of HL60 and AML5 cells in G0/G1 phase
(Fig. 3A and B). However, the HL60
and AML5 cells in S and G2/M phase were significantly decreased
after miR-135a overexpression (Fig. 3A
and B). Moreover, through western blot, we found that
overexpression of miR-135a significantly reduced the protein levels
of cyclin E1, cylcin D1 and PCNA in HL60 and AML5 cells (Fig. 3C), which indicated that miR-135a
overexpression significantly inhibited cell cycle progression.
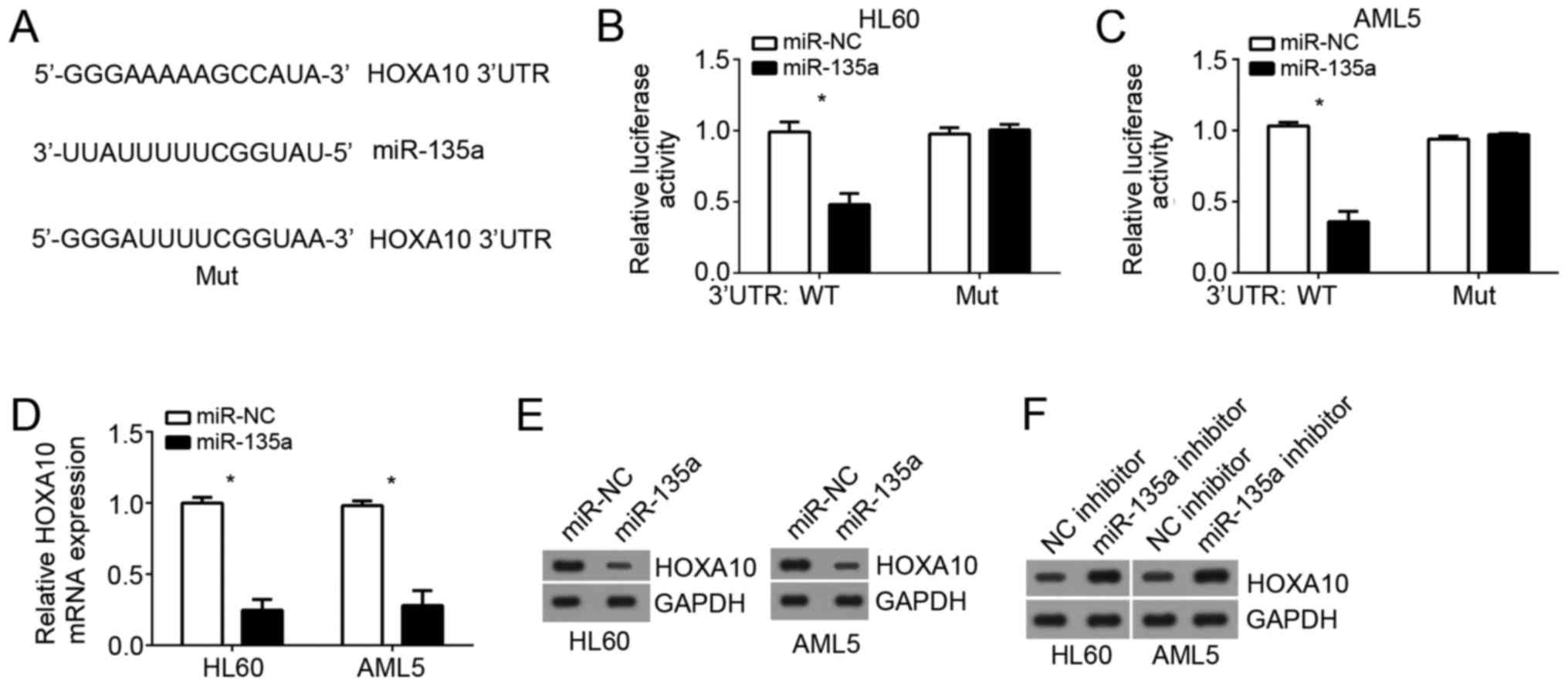
HOXA10 is a target of miR-135a
To search the target genes of miR-135a in AML cells,
we used online prediction software (TargetScan 7.0 and miRBase) to
look for the most possible target genes. Among all results, HOXA10
ranked top and has been reported to promote AML progression.
Therefore, we chose them for following validation. There was a
potential binding site of miR-135a in the 3′-UTR region of HOXA10
mRNA (Fig. 4A). Then we performed
luciferase reporter assays. Results showed that the luciferase
activity of WT-3′-UTR HOXA10 in HL60 and AML5 cells was
significantly downregulated by overexpression of miR-135a while no
change was observed in the mutated 3′UTR transfection (Fig. 4B and C). Furthermore, qRT-PCR and
western blot analysis indicated that overexpression of miR-135a
significantly downregulated the mRNA and protein levels of HOXA10
in HL60 and AML5 cells (Fig. 4D and
E), whereas miR-135a knockdown promoted HOXA10 expression
(Fig. 4F).
miR-135a is negatively associated with
HOXA10 expression in AML
To further confirm the relationship of miR-135a and
HOXA10 in AML cells, we analyzed the expression patterns of HOXA10
in AML sample cells by qRT-PCR. We found that HOXA10 expression was
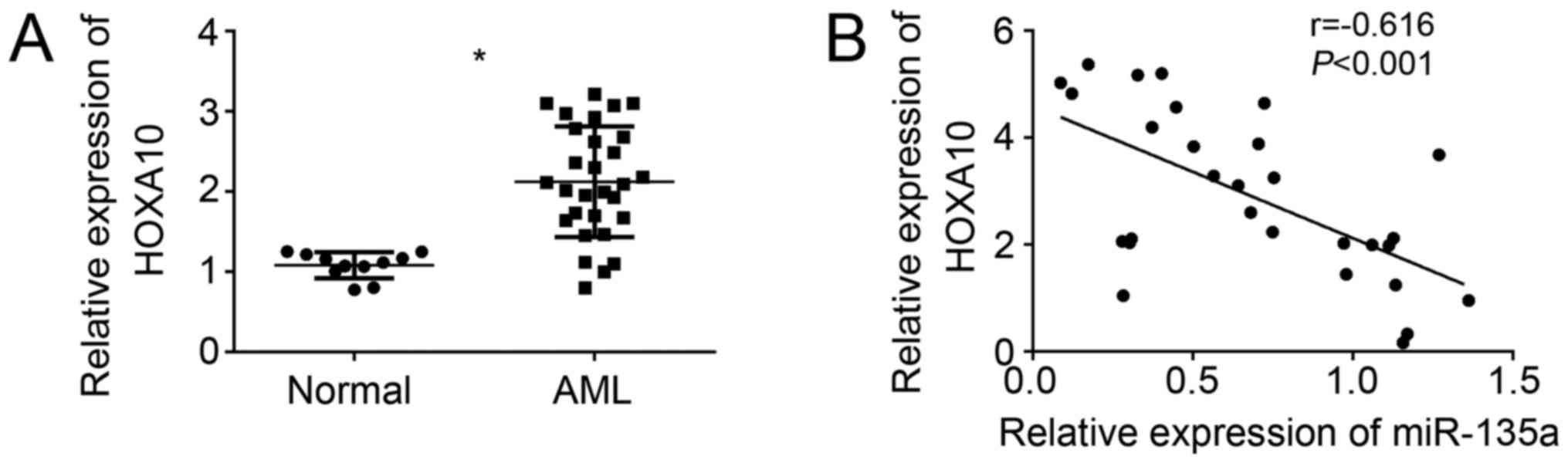
upreuglated in AML sample cells compared to normal cells (Fig. 5A). Moreover, we found that HOXA10
mRNA level was negatively correlated with miR-135a expression in
AML samples (Fig. 5B). These data
further confirmed that miR-135a directly targeted HOXA10 in AML
cells.
Restoration of HOXA10 reversed the
effects of miR-135a overexpression
We have demonstrated HOXA10 was a direct target of
miR-135a in AML cells. In order to determine whether miR-135a
suppressed the proliferation and promoted apoptosis of AML cells
through HOXA10, HL60 and AML5 cells were transduced with HOXA10.
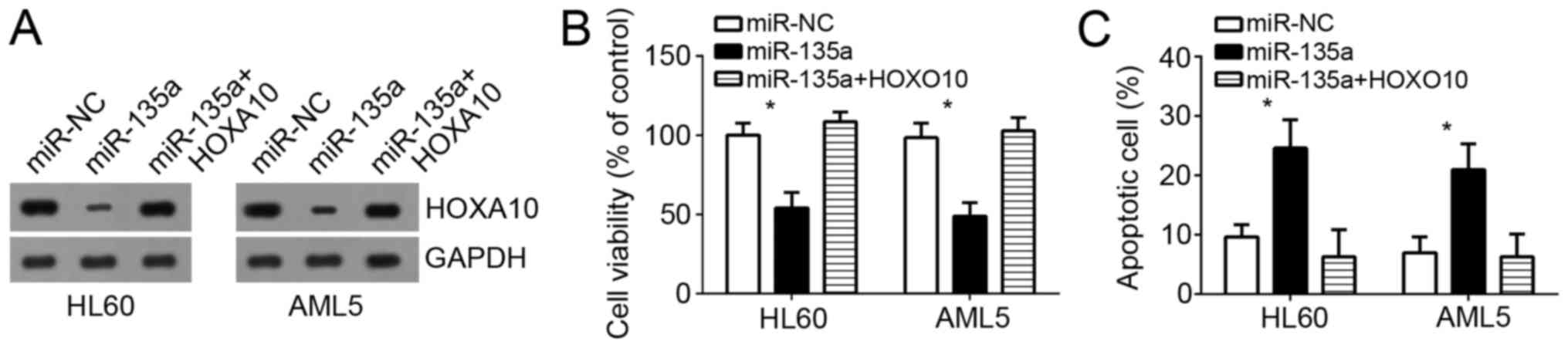
Western blot analysis results indicated that HOXA10 expression was
significantly upregulated in HL60 and AML5 cells (Fig. 6A). Then cell survival ability was
examined by CCK-8 assays in HL60 and AML5 cells. Results indicated
that miR-135a overexpression decreased the survival ability and
promoted cell apoptosis in HL60 and AML5 cells while restoration of
HOXA10 significantly abrogated these effects of miR-135a
overexpression (Fig. 6B and C).
Taken together, our data demonstrated that miR-135a suppressed AML
cell proliferation and induced apoptosis through directly targeting
HOXA10.
Discussion
AML is one the most malignant cancers and the
pathogenesis of AML need to be elucidated. miRNAs have been
reported to participate in the development and progression of
almost all kinds of human cancers, including breast cancer
(18), cervical cancer (19), gastric cancer (20) and AML (15). Therefore, elucidating the mechanism
of miRNA function will be crucial for the development of
therapeutic strategies. Several miRNAs have been demonstrated to
exert important roles on AML progression. For example, Wan et
al reported that miR-103 confers the resistance to
long-treatment of adriamycin to human leukemia cells by regulation
of COP1 (21). Liu et al
showed that miR-34a induced apoptosis and suppressed autophagy
through targeting HMGB1 in AML cells (22). Wang et al reported that
upregulation of miR-125b suppresses the invasion and proliferation
of human AML cells, and induced cellular apoptosis through
regulating NF-κB pathway (23). Ke
et al showed that miR-192 suppressed the proliferation, cell
cycle transition of AML cells by targeting CCNT2 (24). In addition, Wang et al
demonstrated that miR-183 enhanced the proliferation of pediatric
AML cells by targeting programmed cell death 6 (25). miR-135a has been shown to
aberrantly expressed in several cancer, including non-small cell
lung cancer (26), thyroid
carcinoma (27), glioblastoma
(28), pancreatic cancer (29), hepatocellular carcinoma (30) and gastric cancer (31). However, the function of miR-135a in
AML remains elusive. In this study, we showed that miR-135a
expression was significantly downregulated in AML samples and cell
lines. Moreover, the expression levels of miR-135a were correlated
with the prognosis of AML patients. Furthermore, we demonstrated
that miR-135a overexpression suppressed the proliferation and
induced apoptosis of AML cells by targeting HOXA10.
HOXA10 belongs to the HOX gene superfamily encoding
a class of transcription factors that regulate cell proliferation,
differentiation and migration (32,33).
Dysregulation of HOXA10 has been observed in several cancers,
including ovarian cancer (34,35),
colorectal cancer (36), oral
squamous cell carcinoma (37),
hepatocellular carcinoma (38),
prostate carcinoma (39) and
pancreatic cancer (40). As an
oncogene, HOXA10 has also been demonstrated to promote the
progression of human AML (41). In
addition, Wang et al reported that constitutively active
SHP2 cooperates with HoxA10 overexpression to induce AML (42). However, how HOXA10 expression was
regulated by miRNAs in AML cells remains largely unknown. In our
study, we found that HOXA10 was a direct target of miR-135a in AML
cells. We showed that overexpression of miR-135a suppressed the
luciferase activity of HOXA10 3′UTR in HL60 and AML5 cells. And
miR-135a overexpression significantly reduced the mRNA and protein
levels of HOXA10 in HL60 and AML5 cells. Furthermore, we also
showed that HOXA10 expression was upregulated and negatively
correlated with that of miR-135a in AML sample. Then through
functional experiments, we found that overexpression of HOXA10
significantly reversed the effects of miR-135a transfection on AML
cell proliferation and apoptosis. These results indicated that
HOXA10 serves as an oncogene and is critical for the function of
miR-135a in AML.
In conclusion, our study elucidated the essential
role of miR-135a in AML progression and demonstrated its functional
mechanism that miR-135a suppressed AML proliferation and induced
apoptosis of AML cell through targeting HOXA10. Our study indicated
that HOXA10 could act as a prognostic predictor for AML patients
and might be a promising therapeutic target for AML treatment.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
HX initiated, designed this work, analyzed,
interpreted the results and wrote this manuscript. QW performed the
experiments. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
For the use of human samples, the protocol for this
study was approved by the Institutional Ethics Committee of The
Second Affiliated Hospital of Harbin Medical University and all
enrolled patients signed a written informed consent document.
Consent for publication
All patients within this study provide consent for
the publication of their data.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Estey EH: Acute myeloid leukemia: 2013
update on risk-stratification and management. Am J Hematol.
88:318–327. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Passegue E, Jamieson CH, Ailles LE and
Weissman IL: Normal and leukemic hematopoiesis: Are leukemias a
stem cell disorder or a reacquisition of stem cell characteristics?
Proc Natl Acad Sci USA. 100 Suppl 1:S11842–S11849. 2003. View Article : Google Scholar
|
|
3
|
Eppert K, Takenaka K, Lechman ER, Waldron
L, Nilsson B, van Galen P, Metzeler KH, Poeppl A, Ling V, Beyene J,
et al: Stem cell gene expression programs influence clinical
outcome in human leukemia. Nat Med. 17:1086–1093. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kreso A and Dick JE: Evolution of the
cancer stem cell model. Cell Stem Cell. 14:275–291. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Xu XH, Zhang L, Cao XX, Li J, Zhang W, Zhu
TN, Cai HC, Chen M, Han X, Yang C, et al: Evaluation of the
implementation rate of primary antifungal prophylaxis and the
prognosis of invasive fungal disease in acute leukemia patients in
China. J Infect Chemother. 23:360–367. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Marcucci G, Mrozek K, Radmacher MD, Garzon
R and Bloomfield CD: The prognostic and functional role of
microRNAs in acute myeloid leukemia. Blood. 117:1121–1129. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Khalaj M, Tavakkoli M, Stranahan AW and
Park CY: Pathogenic microRNA's in myeloid malignancies. Front
Genet. 5:3612014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liao Q, Wang B, Li X and Jiang G: miRNAs
in acute myeloid leukemia. Oncotarget. 8:3666–3682. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zavala-Yoe R, Ramirez-Mendoza RA and
Cordero LM: Entropy measures to study and model long term
simultaneous evolution of children in Doose and Lennox-Gastaut
syndromes. J Integr Neurosci. 15:205–221. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bu W and Luo T: miR-1297 promotes cell
proliferation of non-small cell lung cancer cells: Involving in
PTEN/Akt/Skp2 signaling pathway. DNA Cell Biol. 36:976–982. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kang M, Li Y, Zhao Y, He S and Shi J:
miR-33a inhibits cell proliferation and invasion by targeting CAND1
in lung cancer. Clin Transl Oncol. 20:457–466. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xiong Y, Wang L, Li Y, Chen M, He W and Qi
L: The long non-coding RNA XIST interacted with MiR-124 to modulate
bladder cancer growth, invasion and migration by targeting androgen
receptor (AR). Cell Physiol Biochem. 43:405–418. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tran DDH, Kessler C, Niehus SE, Mahnkopf
M, Koch A and Tamura T: Myc target gene, long intergenic noncoding
RNA, Linc00176 in hepatocellular carcinoma regulates cell cycle and
cell survival by titrating tumor suppressor microRNAs. Oncogene.
37:75–85. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang S, Zhang Q, Shi G and Yin J:
MiR-182-5p regulates BCL2L12 and BCL2 expression in acute myeloid
leukemia as a potential therapeutic target. Biomed Pharmacother.
97:1189–1194. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang TJ, Lin J, Zhou JD, Li XX, Zhang W,
Guo H, Xu ZJ, Yan Y, Ma JC and Qian J: High bone marrow miR-19b
level predicts poor prognosis and disease recurrence in de novo
acute myeloid leukemia. Gene. 640:79–85. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ye P, Shi Y, An N, Zhou Q, Guo J and Long
X: miR-145 overexpression triggers alteration of the whole
transcriptome and inhibits breast cancer development. Biomed
Pharmacother. 100:72–82. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu YL, Wang GQ, Cui HX, Li XX, Xu ZL and
Wang XY: miRNA211 induces apoptosis of cervical cancer SiHa cells
via down-regulation of inhibitor of apoptosis proteins. Eur Rev Med
Pharmacol Sci. 22:336–342. 2018.PubMed/NCBI
|
|
20
|
Cao Y, Song J, Ge J, Song Z, Chen J and Wu
C: MicroRNA-100 suppresses human gastric cancer cell proliferation
by targeting CXCR7. Oncol Lett. 15:453–458. 2018.PubMed/NCBI
|
|
21
|
Wan L, Tian Y, Zhang R, Peng Z, Sun J and
Zhang W: MicroRNA-103 confers the resistance to long-treatment of
adriamycin to human leukemia cells by regulation of COP1. J Cell
Biochem. 119:3846–3852. 2018. View Article : Google Scholar
|
|
22
|
Liu L, Ren W and Chen K: MiR-34a promotes
apoptosis and inhibits autophagy by targeting HMGB1 in acute
myeloid leukemia cells. Cell Physiol Biochem. 41:1981–1992. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang Y, Tang P, Chen Y, Chen J, Ma R and
Sun L: Overexpression of microRNA-125b inhibits human acute myeloid
leukemia cells invasion, proliferation and promotes cells apoptosis
by targeting NF-kappaB signaling pathway. Biochem Biophys Res
Commun. 488:60–66. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ke S, Li RC, Lu J, Meng FK, Feng YK and
Fang MH: MicroRNA-192 regulates cell proliferation and cell cycle
transition in acute myeloid leukemia via interaction with CCNT2.
Int J Hematol. 106:258–265. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang X, Zuo D, Yuan Y, Yang X, Hong Z and
Zhang R: MicroRNA-183 promotes cell proliferation via regulating
programmed cell death 6 in pediatric acute myeloid leukemia. J
Cancer Res Clin Oncol. 143:169–180. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang T and Wang N: miR-135a confers
resistance to gefitinib in non-small cell lung cancer cells by
upregulation of RAC1. Oncol Res. Jan 31–2018.(Epub ahead of print).
View Article : Google Scholar
|
|
27
|
Zhao X, Sun Z, Li H, Jiang F, Zhou J and
Zhang L: MiR-135a-5p modulates biological functions of thyroid
carcinoma cells via targeting VCAN 3′-UTR. Cancer Biomark.
20:207–216. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zubieta Gomez DM, Hamood MA, Beydoun R,
Pall AE and Kondapalli KC: MicroRNA-135a regulates NHE9 to inhibit
proliferation and migration of glioblastoma cells. Cell Commun
Signal. 15:552017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang X, Gao F, Zhou L, Wang H, Shi G and
Tan X: UCA1 regulates the growth and metastasis of pancreatic
cancer by sponging miR-135a. Oncol Res. 25:1529–1541. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
von Felden J, Heim D, Schulze K, Krech T,
Ewald F, Nashan B, Lohse AW and Wege H: High expression of micro
RNA-135A in hepatocellular carcinoma is associated with recurrence
within 12 months after resection. BMC Cancer. 17:602017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yan LH, Chen ZN, Li L, Chen J, Wei WE, Mo
XW, Qin YZ, Lin Y and Chen JS: miR-135a promotes gastric cancer
progression and resistance to oxaliplatin. Oncotarget.
7:70699–70714. 2006.
|
|
32
|
Yoshida H, Broaddus R, Cheng W, Xie S and
Naora H: Deregulation of the HOXA10 homeobox gene in endometrial
carcinoma: role in epithelial-mesenchymal transition. Cancer Res.
66:889–897. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Bei L, Lu Y, Bellis SL, Zhou W, Horvath E
and Eklund EA: Identification of a HoxA10 activation domain
necessary for transcription of the gene encoding beta3 integrin
during myeloid differentiation. J Biol Chem. 282:16846–16859. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhang HY, Li JH, Li G and Wang SR:
Activation of ARK5/miR-1181/HOXA10 axis promotes
epithelial-mesenchymal transition in ovarian cancer. Oncol Rep.
34:1193–1202. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Tang W, Jiang Y, Mu X, Xu L, Cheng W and
Wang X: MiR-135a functions as a tumor suppressor in epithelial
ovarian cancer and regulates HOXA10 expression. Cell Signal.
26:1420–1426. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Sun S, Su C, Zhu Y, Li H, Liu N, Xu T, Sun
C and Lv Y: MicroRNA-544a regulates migration and invasion in
colorectal cancer cells via regulation of homeobox A10. Dig Dis
Sci. 61:2535–2544. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Carrera M, Bitu CC, de Oliveira CE, Graner
E, Manninen A, Salo T and Coletta RD: HOXA10 controls
proliferation, migration and invasion in oral squamous cell
carcinoma. Int J Clin Exp Pathol. 8:3613–3623. 2015.PubMed/NCBI
|
|
38
|
Xiao ZD, Jiao CY, Huang HT, He LJ, Zhao
JJ, Lu ZY and Liu LX: miR-218 modulate hepatocellular carcinoma
cell proliferation through PTEN/AKT/PI3K pathway and HoxA10. Int J
Clin Exp Pathol. 7:4039–4044. 2014.PubMed/NCBI
|
|
39
|
Li B, Cao X, Weng C, Wu Y, Fang X, Zhang X
and Liu G: HoxA10 induces proliferation in human prostate carcinoma
PC-3 cell line. Cell Biochem Biophys. 70:1363–1368. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Cui XP, Qin CK, Zhang ZH, Su ZX, Liu X,
Wang SK and Tian XS: HOXA10 promotes cell invasion and MMP-3
expression via TGFβ2-mediated activation of the p38 MAPK pathway in
pancreatic cancer cells. Dig Dis Sci. 59:1442–1451. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Shah CA, Bei L, Wang H, Platanias LC and
Eklund EA: HoxA10 protein regulates transcription of gene encoding
fibroblast growth factor 2 (FGF2) in myeloid cells. J Biol Chem.
287:18230–18248. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wang H, Lindsey S, Konieczna I, Bei L,
Horvath E, Huang W, Saberwal G and Eklund EA: Constitutively active
SHP2 cooperates with HoxA10 overexpression to induce acute myeloid
leukemia. J Biol Chem. 284:2549–2567. 2009. View Article : Google Scholar : PubMed/NCBI
|