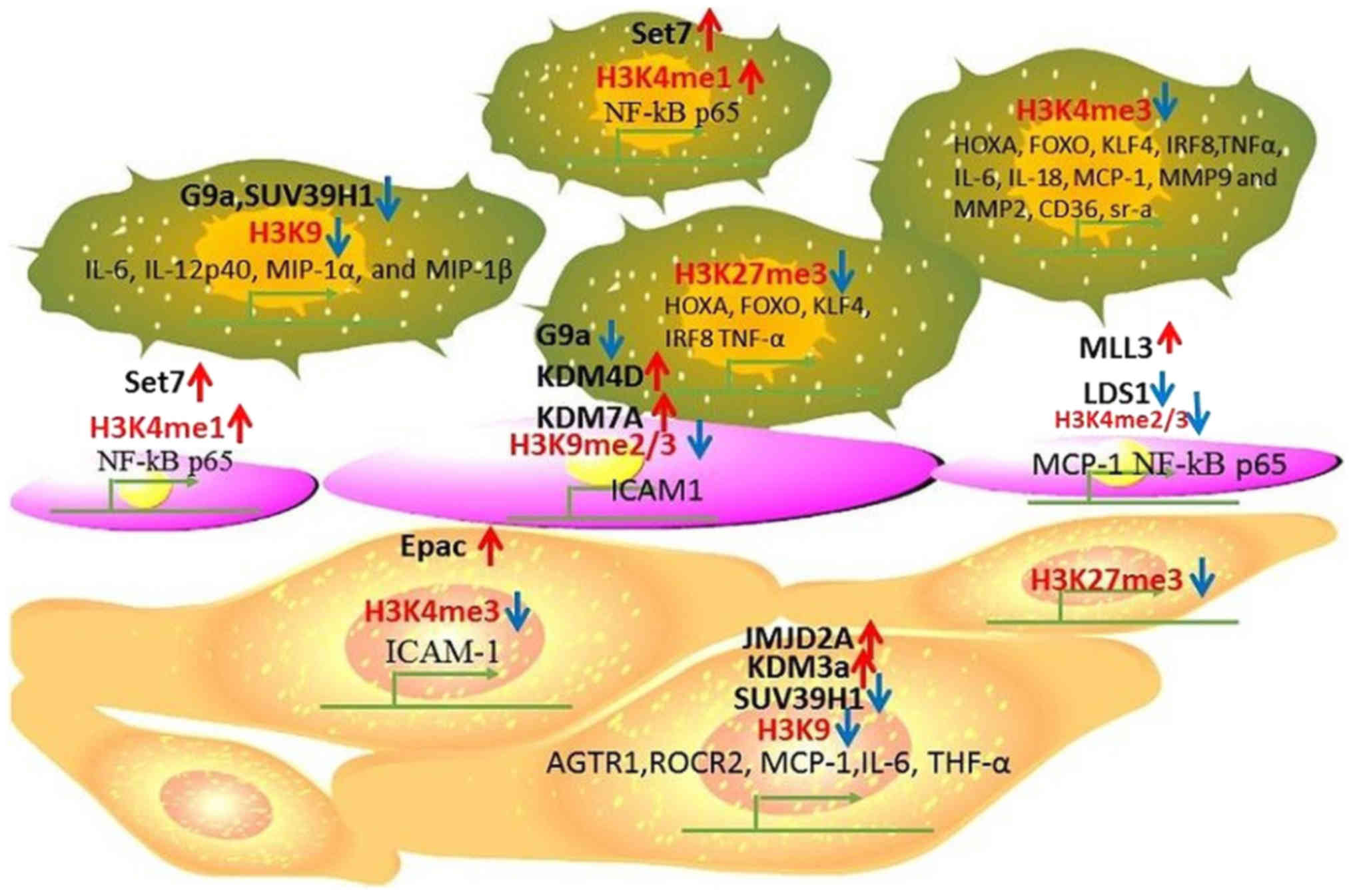
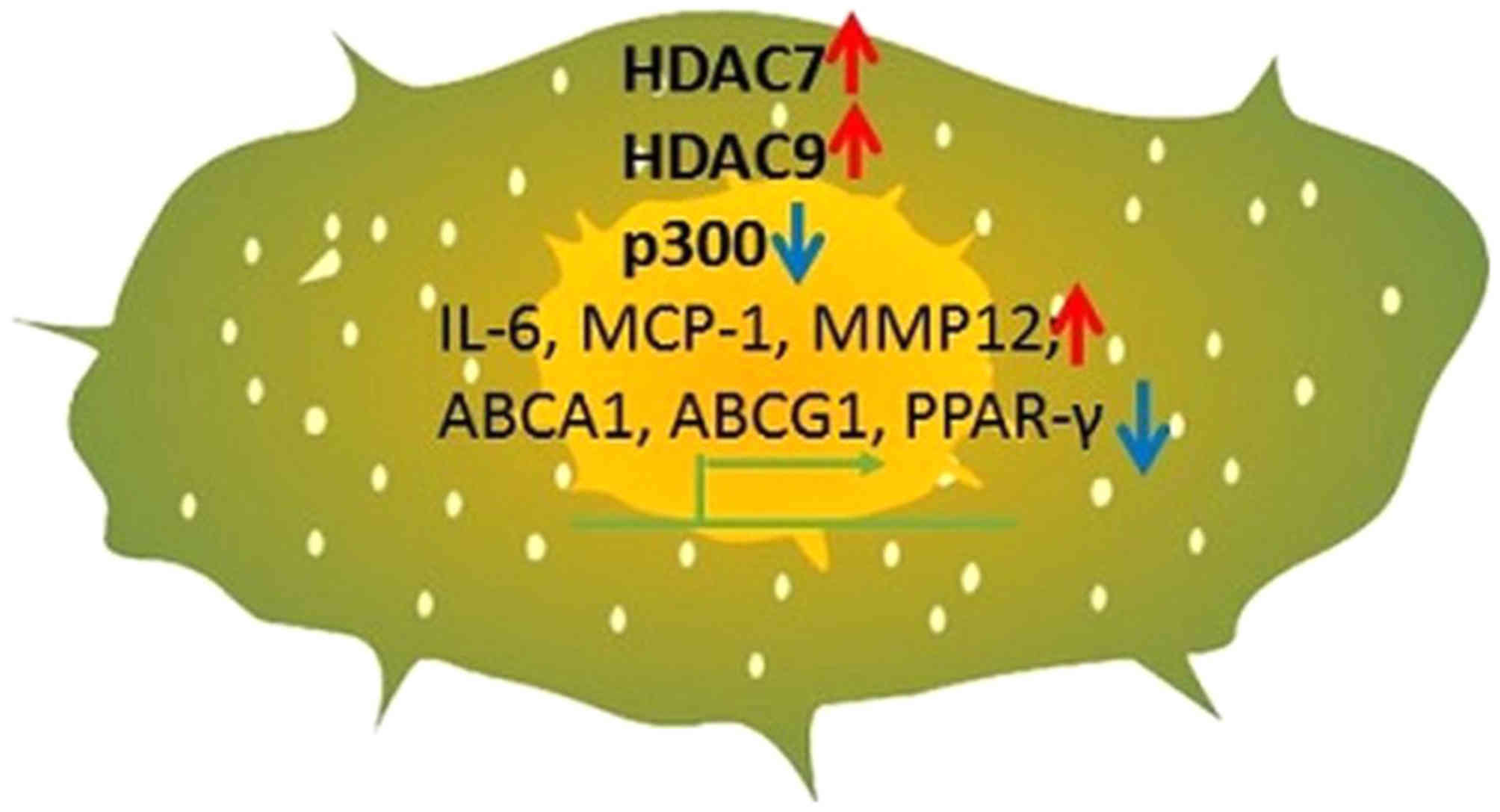
|
1
|
Seneviratne A, Hulsmans M, Holvoet P and
Monaco C: Biomechanical factors and macrophages in plaque
stability. Cardiovasc Res. 99:284–293. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Karlić R, Chung HR, Lasserre J, Vlahovicek
K and Vingron M: Histone modification levels are predictive for
gene expression. Proc Natl Acad Sci USA. 107:2926–2931. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dong X and Weng Z: The correlation between
histone modifications and gene expression. Epigenomics. 5:113–116.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Greer EL and Shi Y: Histone methylation: A
dynamic mark in health, disease and inheritance. Nat Rev Genet.
13:343–357. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Xiaoling Y, Li Z, ShuQiang L, Shengchao M,
Anning Y, Ning D, Nan L, Yuexia J, Xiaoming Y, Guizhong L and
Yideng J: Hyperhomocysteinemia in ApoE-/-Mice leads to
overexpression of enhancer of zeste homolog 2 via miR-92a
regulation. PLoS One. 11:e01677442016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li Z, Cao R, Wang M, Myers MP, Zhang Y and
Xu R: Structure of a Bmi-1-ring1B polycomb group ubiquitin ligase
complex. J Biol Chem. 281:20643–20649. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wierda RJ, Rietveld IM, van Eggermond MC,
Belien JA, van Zwet EW, Lindeman JH and van den Elsen PJ: Global
histone H3 lysine 27 triple methylation levels are reduced in
vessels with advanced atherosclerotic plaques. Life Sci. 129:3–9.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kadakol A, Malek V, Goru SK, Pandey A and
Gaikwad AB: Esculetin reverses histone H2A/H2B ubiquitination, H3
dimethylation, acetylation and phosphorylation in preventing type 2
diabetic cardiomyopathy. J Funct Foods. 17:127–136. 2015.
View Article : Google Scholar
|
|
9
|
Xiao Y, Huang W, Zhang J, Peng C, Xia M
and Ling W: Increased plasma S-adenosylhomocysteine-accelerated
atherosclerosis is associated with epigenetic regulation of
endoplasmic reticulum stress in apoE-/-mice. Arterioscler Thromb
Vasc Biol. 35:60–70. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yuan Q, Xie X, Fu Z, Ma X, Yang Y, Huang
D, Liu F, Dai C and Ma Y: Association of the histone-lysine
N-methyltransferase MLL5 gene with coronary artery disease in
Chinese Han people. Meta Gene. 2:514–524. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen WL, Sun HP, Li DD, Wang ZH, You QD
and Guo XK: G9a-an appealing antineoplastic target. Curr Cancer
Drug Targets. 17:555–568. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cong G, Yan R, Huang H, Wang K, Yan N, Jin
P, Zhang N, Hou J, Chen D and Jia S: Involvement of histone
methylation in macrophage apoptosis and unstable plaque formation
in methionine-induced hyperhomocysteinemic ApoE-/-mice. Life Sci.
173:135–144. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zheng QF, Wang HM, Wang ZF, Liu JY, Zhang
Q, Zhang L, Lu YH, You H and Jin GH: Reprogramming of histone
methylation controls the differentiation of monocytes into
macrophages. FEBS J. 284:1309–1323. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Greißel A, Culmes M, Burgkart R,
Zimmermann A, Eckstein HH, Zernecke A and Pelisek J: Histone
acetylation and methylation significantly change with severity of
atherosclerosis in human carotid plaques. Cardiovasc Pathol.
25:79–86. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bekkering S, van den Munckhof I, Nielen T,
Lamfers E, Dinarello C, Rutten J, de Graaf J, Joosten LA, Netea MG,
Gomes ME and Riksen NP: Innate immune cell activation and
epigenetic remodeling in symptomatic and asymptomatic
atherosclerosis in humans in vivo. Atherosclerosis. 254:228–236.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Vaquero A, Scher M, Erdjument-Bromage H,
Tempst P, Serrano L and Reinberg D: SIRT1 regulates the histone
methyl-transferase SUV39H1 during heterochromatin formation.
Nature. 450:4402007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li MF, Zhang R, Li TT, Chen MY, Li LX, Lu
JX and Jia WP: High glucose increases the expression of
inflammatory cytokine genes in macrophages through H3K9
methyltransferase mechanism. J Interferon Cytokine Res. 36:48–61.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Paneni F, Costantino S, Battista R,
Castello L, Capretti G, Chiandotto S, Scavone G, Villano A, Pitocco
D, Lanza G, et al: Adverse epigenetic signatures by histone
methyltransferase Set7 contribute to vascular dysfunction in
patients with type 2 diabetes. Circ Cardiovasc Genet. 8:150–158.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bekkering S, Quintin J, Joosten LA, van
der Meer JW, Netea MG and Riksen NP: Oxidized low-density
lipoprotein induces long-term proinflammatory cytokine production
and foam cell formation via epigenetic reprogramming of
monocytessignificance. Arterioscler Thromb Vasc Biol. 34:1731–1738.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Choi J, Yoon S, Kim S and Jo Ahn S: KDM4B
histone demethylase and G9a regulate expression of vascular
adhesion proteins in cerebral microvessels. Sci Rep. 7:450052017.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Barroso M, Kao D, Blom HJ, De Almeida
Tavares I, Castro R, Loscalzo J and Handy DE:
S-adenosylhomocysteine induces inflammation through NFκB: A
possible role for EZH2 in endothelial cell activation. Biochem
Biophys Acta. 1862:82–92. 2016.PubMed/NCBI
|
|
22
|
Choi J and Jo SA: KDM7A histone
demethylase mediates TNF-α-induced ICAM1 protein upregulation by
modulating lysosomal activity. Biochem Biophys Res Commun.
478:1355–1362. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Balcerczyk A, Rybaczek D, Wojtala M,
Pirola L, Okabe J and El-Osta A: Pharmacological inhibition of
arginine and lysine methyltransferases induces nuclear
abnormalities and suppresses angiogenesis in human endothelial
cells. Biochem Pharmacol. 121:18–32. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gu L, Hitzel J, Moll F, Kruse C, Malik RA,
Preussner J, Looso M, Leisegang MS, Steinhilber D, Brandes RP and
Fork C: The histone demethylase PHF8 is essential for endothelial
cell migration. PLoS One. 11:e1466452016. View Article : Google Scholar
|
|
25
|
Han P, Gao D, Zhang W, Liu S, Yang S and
Li X: Puerarin suppresses high glucose-induced MCP-1 expression via
modulating histone methylation in cultured endothelial cells. Life
Sci. 130:103–107. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zernecke A: Dendritic cells in
atherosclerosis. Arterioscler Thromb Vasc Biol. 35:763–770. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kumar A, Kumar S, Vikram A, Hoffman TA,
Naqvi A, Lewarchik CM, Kim YR and Irani K: Histone and DNA
methylation-mediated epigenetic downregulation of endothelial
kruppel-like factor 2 by low-density lipoprotein cholesterol.
Arterioscler Thromb Vasc Biol. 33:1936–1942. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Brasacchio D, Okabe J, Tikellis C,
Balcerczyk A, George P, Baker EK, Calkin AC, Brownlee M, Cooper ME
and El-Osta A: Hyperglycemia induces a dynamic cooperativity of
histone methylase and demethylase enzymes associated with
gene-activating epigenetic marks that coexist on the lysine tail.
Diabetes. 58:1229–1236. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
El-Osta A, Brasacchio D, Yao D, Pocai A,
Jones PL, Roeder RG, Cooper ME and Brownlee M: Transient high
glucose causes persistent epigenetic changes and altered gene
expression during subsequent normoglycemia. J Exp Med.
205:2409–2417. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chen J, Zhang J, Yang J, Xu L, Hu Q, Xu C,
Yang S and Jiang H: Histone demethylase KDM3a, a novel regulator of
vascular smooth muscle cells, controls vascular neointimal
hyperplasia in diabetic rats. Atherosclerosis. 257:152–163. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yousefipour Z, Newaz MA, Esmaeilli M and
Ranganna K: [PP.36.07] Modification of histone induced by acrolein
in rat vascular smooth muscle cells. J Hypertens. 34:e3372016.
View Article : Google Scholar
|
|
32
|
Lehrke M, Kahles F, Makowska A, Tilstam
PV, Diebold S, Marx J, Stöhr R, Hess K, Endorf EB, Bruemmer D, et
al: PDE4 inhibition reduces neointima formation and inhibits VCAM-1
expression and histone methylation in an Epac-dependent manner. J
Mol Cell Cardiol. 81:23–33. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Qi H, Jing Z, Xiaolin W, Changwu X,
Xiaorong H, Jian Y, Jing C and Hong J: Histone demethylase JMJD2A
inhibition attenuates neointimal hyperplasia in the carotid
arteries of balloon-injured diabetic rats via transcriptional
silencing: Inflammatory gene expression in vascular smooth muscle
cells. Cell Physiol Biochem. 37:719–734. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Villeneuve LM, Reddy MA, Lanting LL, Wang
M, Meng L and Natarajan R: Epigenetic histone H3 lysine 9
methylation in metabolic memory and inflammatory phenotype of
vascular smooth muscle cells in diabetes. Proc Natl Acad Sci USA.
105:9047–9052. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Xia J, Fang M, Wu X, Yang Y, Yu L, Xu H,
Kong H, Tan Q, Wang H, Xie W and Xu Y: A2b adenosine signaling
represses CIITA transcription via an epigenetic mechanism in
vascular smooth muscle cells. Biochim Biophys Acta. 1849:665–676.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Tarling EJ, Ryan KJ, Austin R, Kugler SJ,
Salter AM and Langley-Evans SC: Maternal high-fat feeding in
pregnancy programs atherosclerotic lesion size in the ApoE*3 Leiden
mouse. J Dev Orig Health Dis. Feb 2–2016.(Epub ahead of print).
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Alkemade FE, van Vliet P, Henneman P, van
Dijk KW, Hierck BP, van Munsteren JC, Scheerman JA, Goeman JJ,
Havekes LM, Gittenberger-de Groot AC, et al: Prenatal exposure to
apoe deficiency and postnatal hypercholesterolemia are associated
with altered cell-specific lysine methyltransferase and histone
methylation patterns in the vasculature. Am J Pathol. 176:542–548.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Bannister AJ and Kouzarides T: Regulation
of chromatin by histone modifications. Cell Res. 21:381–395. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Witt O, Deubzer HE, Milde T and Oehme I:
HDAC family: What are the cancer relevant targets? Cancer Lett.
277:8–21. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Tikoo K, Patel G, Kumar S, Karpe PA,
Sanghavi M, Malek V and Srinivasan K: Tissue specific up regulation
of ACE2 in rabbit model of atherosclerosis by atorvastatin: Role of
epigenetic histone modifications. Biochem Pharmacol. 93:343–351.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chaturvedi P, Kalani A, Givvimani S, Kamat
PK, Familtseva A and Tyagi SC: Differential regulation of DNA
methylation versus histone acetylation in cardiomyocytes during
HHcy in vitro and in vivo: An epigenetic mechanism. Physiol
Genomics. 46:245–255. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kumar A and Chauhan S: How much successful
are the medicinal chemists in modulation of SIRT1: A critical
review. Eur J Med Chem. 119:45–69. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Sathishkumar C, Prabu P, Balakumar M,
Lenin R, Prabhu D, Anjana RM, Mohan V and Balasubramanyam M:
Augmentation of histone deacetylase 3 (HDAC3) epigenetic signature
at the interface of proinflammation and insulin resistance in
patients with type 2 diabetes. Clin Epigenetics. 8:1252016.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Hoeksema MA, Gijbels MJ, Van den Bossche
J, van der Velden S, Sijm A, Neele AE, Seijkens T, Stöger JL,
Meiler S, Boshuizen MC, et al: Targeting macrophage Histone
deacetylase 3 stabilizes atherosclerotic lesions. EMBO Mol Med.
6:1124–1132. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Van den Bossche J, Neele AE, Hoeksema MA,
de Heij F, Boshuizen MC, van der Velden S, de Boer VC, Reedquist KA
and de Winther MPJ: Inhibiting epigenetic enzymes to improve
atherogenic macrophage functions. Biochem Biophys Res Commun.
455:396–402. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Xie X, Song X, Yuan S, Cai H, Chen Y,
Chang X, Liang B and Huang D: Histone acetylation regulates orphan
nuclear receptor NR4A1 expression in hypercholesterolaemia. Clin
Sci (Lond). 129:1151–1161. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Oksala NKJ, Seppälä I, Rahikainen R,
Mäkelä KM, Raitoharju E, Illig T, Klopp N, Kholova I, Laaksonen R,
Karhunen PJ, et al: Synergistic expression of histone deacetylase 9
and matrix metalloproteinase 12 in M4 macrophages in advanced
carotid plaques. Eur J Vasc Endovasc. 53:632–640. 2017. View Article : Google Scholar
|
|
48
|
Smith JD: New role for histone deacetylase
9 in atherosclerosis and inflammation. Arterioscler Thromb Vasc
Biol. 34:1798–1799. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Cao Q, Rong S, Repa JJ, Clair RS, Parks JS
and Mishra N: Histone deacetylase 9 represses cholesterol efflux
and generation of alternatively activated macrophages in
atherosclerosis development. Arterioscler Thrombosis Vasc Biol.
34:1871–1879. 2014. View Article : Google Scholar
|
|
50
|
Salah Ud-Din IA, Tikhomirova A and
Roujeinikova A: Structure and Functional diversity of GCN5-related
n-acetyltransferases (GNAT). Int J Mol Sci. 17:E10182016.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Chistiakov DA, Orekhov AN and Bobryshev
YV: Treatment of cardiovascular pathology with epigenetically
active agents: Focus on natural and synthetic inhibitors of DNA
methylation and histone deacetylation. Int J Cardiol. 227:66–82.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Vecellio M, Spallotta F, Nanni S, Colussi
C, Cencioni C, Derlet A, Bassetti B, Tilenni M, Carena MC, Farsetti
A, et al: The histone acetylase activator pentadecylidenemalonate
1b rescues proliferation and differentiation in the human cardiac
mesenchymal cells of type 2 diabetic patients. Diabetes.
63:2132–2147. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Zeidler R, de Freitas Soares BL, Bader A
and Giri S: Molecular epigenetic targets for liver diseases:
Current challenges and future prospects. Drug Discov Today.
22:1620–1636. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Miranda TB, Cortez CC, Yoo CB, Liang G,
Abe M, Kelly TK, Marquez VE and Jones PA: DZNep is a global histone
methylation inhibitor that reactivates developmental genes not
silenced by DNA methylation. Mol Cancer Ther. 8:1579–1588. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Wierda RJ, Goedhart M, van Eggermond MC,
Muggen AF, Miggelbrink XM, Geutskens SB, van Zwet E, Haasnoot GW
and van den Elsen PJ: A role for KMT1c in monocyte to dendritic
cell differentiation. Hum Immunol. 76:431–437. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Zhao Q, Li S, Li N, Yang X, Ma S, Yang A,
Zhang H, Yang S, Mao C, Xu L, et al: miR-34a targets
HDAC1-regulated H3K9 acetylation on lipid accumulation induced by
homocysteine in foam cells. J Cell Biochem. 118:4617–4627. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Makino J, Ogasawara R, Kamiya T, Hara H,
Mitsugi Y, Yamaguchi E, Itoh A and Adachi T: Royal jelly
constituents increase the expression of extracellular superoxide
dismutase through histone acetylation in monocytic thp-1 cells. J
Nat Prod. 79:1137–1143. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Svennerholm K, Haney M, Biber B, Ulfhammer
E, Saluveer O, Larsson P, Omerovic E, Jern S and Bergh N: Histone
deacetylase inhibition enhances tissue plasminogen activator
release capacity in atherosclerotic man. PLoS One. 10:e1211962015.
View Article : Google Scholar
|
|
59
|
Anderson SJ, Feye KM, Schmidt-McCormack
GR, Malovic E, Mlynarczyk GSA, Izbicki P, Arnold LF, Jefferson MA,
de la Rosa BM, Wehrman RF, et al: Off-target drug effects resulting
in altered gene expression events with epigenetic and
Quasi-Epigenetic origins. Pharmacol Res. 107:229–233. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Cedar H and Bergman Y: Linking DNA
methylation and histone modification: Patterns and paradigms. Nat
Rev Gene. 10:295–304. 2009. View Article : Google Scholar
|
|
61
|
Yang F, Huo XS, Yuan SX, Zhang L, Zhou WP,
Wang F and Sun SH: Repression of the long noncoding RNA-LET by
histone deacetylase 3 contributes to hypoxia-mediated metastasis.
Mol Cell. 49:1083–1096. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Chen R, Kong P, Zhang F, Shu YN, Nie X,
Dong LH, Lin YL, Xie XL, Zhao LL, Zhang XJ and Han M: EZH2-mediated
α-actin methylation needs lncRNA TUG1 and promotes the cortex
cytoskeleton formation in VSMCs. Gene. 616:52–57. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Tee AE, Ling D, Nelson C, Atmadibrata B,
Dinger ME, Xu N, Mizukami T, Liu PY, Liu B, Cheung B, et al: The
histone demethylase JMJD1A induces cell migration and invasion by
up-regulating the expression of the long noncoding RNA MALAT1.
Oncotarget. 5:1793–1804. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Carrer A, Parris JLD, Trefely S, Henry RA,
Montgomery DC, Torres A, Viola JM, Kuo Y, Blair IA, Meier JL, et
al: Impact of a high-fat diet on tissue Acyl-CoA and histone
acetylation levels. J Biol Chem. 292:3312–3322. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Short JD, Tavakoli S, Nguyen HN, Carrera
A, Farnen C, Cox LA and Asmis R: Dyslipidemic diet-induced monocyte
‘priming’ and dysfunction in non-human primates is triggered by
elevated plasma cholesterol and accompanied by altered histone
acetylation. Front Immunol. 8:9582017. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Friso S, Carvajal CA, Fardella CE and
Olivieri O: Epigenetics and arterial hypertension: The challenge of
emerging evidence. Transl Res. 165:154–165. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Høgh Kølbæk Kjær AS, Brinkmann CR,
Dinarello CA, Olesen R, Østergaard L, Søgaard OS, Tolstrup M and
Rasmussen TA: The histone deacetylase inhibitor panobinostat lowers
biomarkers of cardiovascular risk and inflammation in HIV patients.
AIDS. 29:1195–1200. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Choi JH, Nam KH, Kim J, Baek MW, Park JE,
Park HY, Kwon HJ, Kwon OS, Kim DY and Oh GT: Trichostatin A
exacerbates atherosclerosis in low density lipoprotein
receptor-deficient mice. Arterioscler Thromb Vasc Biol.
25:2404–2409. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Okamoto H, Fujioka Y, Takahashi A,
Takahashi T, Taniguchi T, Ishikawa Y and Yokoyama M: Trichostatin
A, an inhibitor of histone deacetylase, inhibits smooth muscle cell
proliferation via induction of p21 (WAF1). J Atheroscler Thromb.
13:183–191. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Pandey D, Sikka G, Bergman Y, Kim JH, Ryoo
S, Romer L and Berkowitz D: Transcriptional regulation of
endothelial arginase 2 by histone deacetylase 2. Arterioscler
Thromb Vasc Biol. 34:1556–1566. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Liokatis S, Klingberg R, Tan S and
Schwarzer D: Differentially isotope-labeled nucleosomes to study
asymmetric histone modification crosstalk by time-resolved NMR
spectroscopy. Angew Chem Int Ed Engl. 55:8262–8265. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Clark SJ, Lee HJ, Smallwood SA, Kelsey G
and Reik W: Single-cell epigenomics: Powerful new methods for
understanding gene regulation and cell identity. Genome Biol.
17:722016. View Article : Google Scholar : PubMed/NCBI
|