|
1
|
GBD 2015 Disease and Injury Incidence and
Prevalence Collaborators: Global, regional, and national incidence,
prevalence, and years lived with disability for 310 diseases and
injuries, 1990–2015: A systematic analysis for the global burden of
disease study 2015. Lancet. 388:1545–1602. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ojha N, Roy S, Radtke J, Simonetti O,
Gnyawali S, Zweier JL, Kuppusamy P and Sen CK: Characterization of
the structural and functional changes in the myocardium following
focal ischemia-reperfusion injury. Am J Physiol Heart Circ Physiol.
294:H2435–H2443. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Aversano T, Aversano LT, Passamani E,
Knatterud GL, Terrin ML, Williams DO and Forman SA; Atlantic
Cardiovascular Patient Outcomes Research Team (C-PORT), :
Thrombolytic therapy vs primary percutaneous coronary intervention
for myocardial infarction in patients presenting to hospitals
without on-site cardiac surgery: A randomized controlled trial.
Jama. 287:1943–1951. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sen CK, Khanna S and Roy S: Perceived
hyperoxia: Oxygen-induced remodeling of the reoxygenated heart.
Cardiovasc Res. 71:280–288. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Roy S, Khanna S, Bickerstaff AA,
Subramanian SV, Atalay M, Bierl M, Pendyala S, Levy D, Sharma N,
Venojarvi M, et al: Oxygen sensing by primary cardiac fibroblasts:
A key role of p21(Waf1/Cip1/Sdi1). Circ Res. 92:264–271. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Roy S, Khanna S, Wallace WA, Lappalainen
J, Rink C, Cardounel AJ, Zweier JL and Sen CK: Characterization of
perceived hyperoxia in isolated primary cardiac fibroblasts and in
the reoxygenated heart. J Biol Chem. 278:47129–47135. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
McNamara MT and Higgins CB: Magnetic
resonance imaging of chronic myocardial infarcts in man. AJR Am J
Roentgenol. 146:315–320. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Orn S, Manhenke C, Anand IS, Squire I,
Nagel E, Edvardsen T and Dickstein K: Effect of left ventricular
scar size, location, and transmurality on left ventricular
remodeling with healed myocardial infarction. Am J Cardiol.
99:1109–1114. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
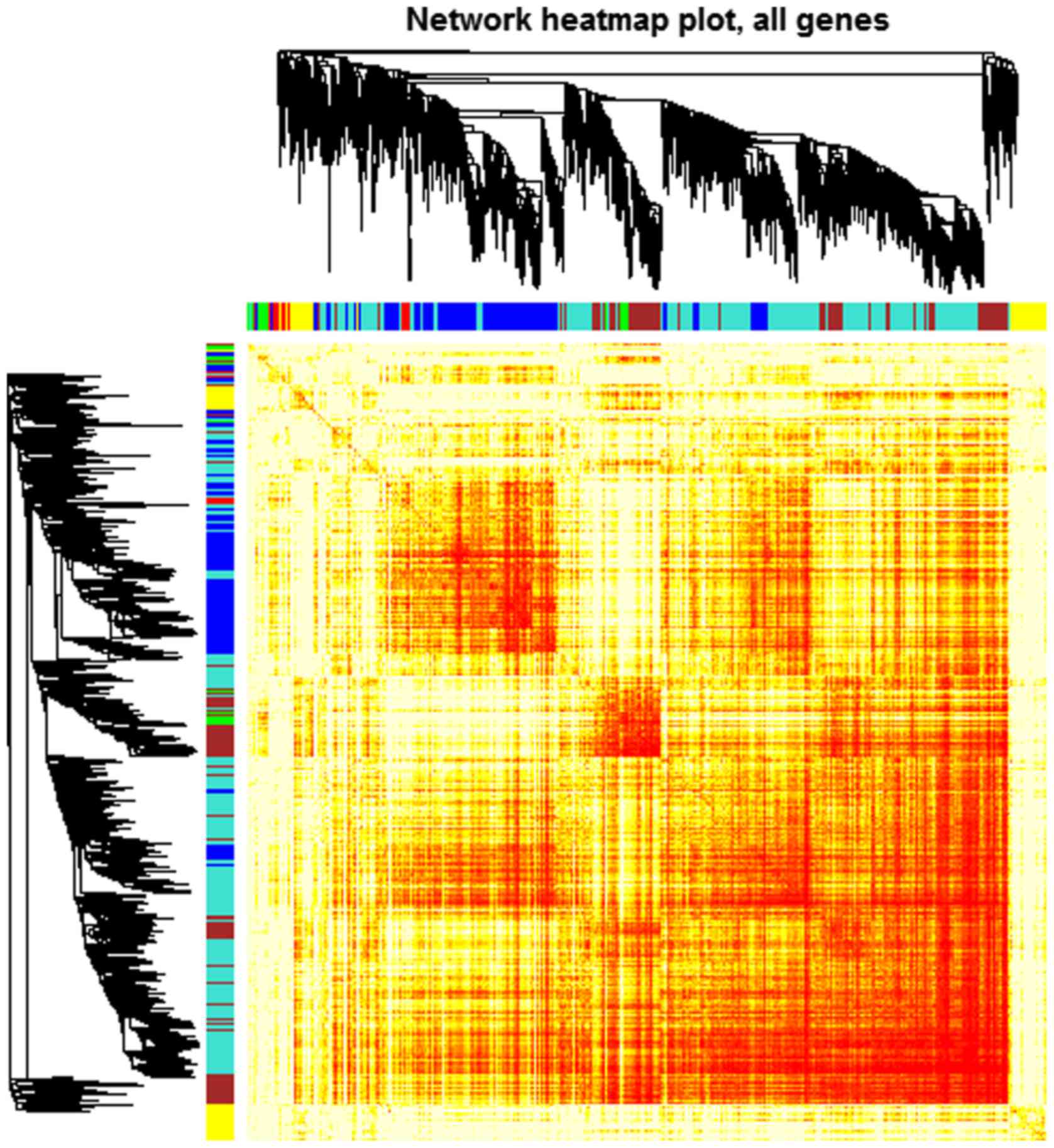
Zhang B and Horvath S: A general framework
for weighted gene co-expression network analysis. Stat Appl Genet
Mol Biol. 4:172005. View Article : Google Scholar
|
|
10
|
Maschietto M, Tahira AC, Puga R, Lima L,
Mariani D, Bda Paulsen S, Belmonte-de-Abreu P, Vieira H, Krepischi
AC, Carraro DM, et al: Co-expression network of
neural-differentiation genes shows specific pattern in
schizophrenia. BMC Med Genomics. 8:232015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Guo Y and Xing Y: Weighted gene
co-expression network analysis of pneumocytes under exposure to a
carcinogenic dose of chloroprene. Life Sci. 151:339–347. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tian F, Zhao J, Fan X and Kang Z: Weighted
gene co-expression network analysis in identification of
metastasis-related genes of lung squamous cell carcinoma based on
the cancer genome atlas database. J Thorac Dis. 9:42–53. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Roy S, Khanna S, Kuhn DE, Rink C, Williams
WT, Zweier JL and Sen CK: Transcriptome analysis of the
ischemia-reperfused remodeling myocardium: Temporal changes in
inflammation and extracellular matrix. Physiol Genomics.
25:364–374. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: Limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Langfelder P and Horvath S: WGCNA: An R
package for weighted correlation network analysis. BMC
Bioinformatics. 9:5592008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The gene
ontology consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
da Huang W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
da Huang W, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Szklarczyk D, Morris JH, Cook H, Kuhn M,
Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al:
The STRING database in 2017: Quality-controlled protein-protein
association networks, made broadly accessible. Nucleic Acids Res.
45:D362–D368. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
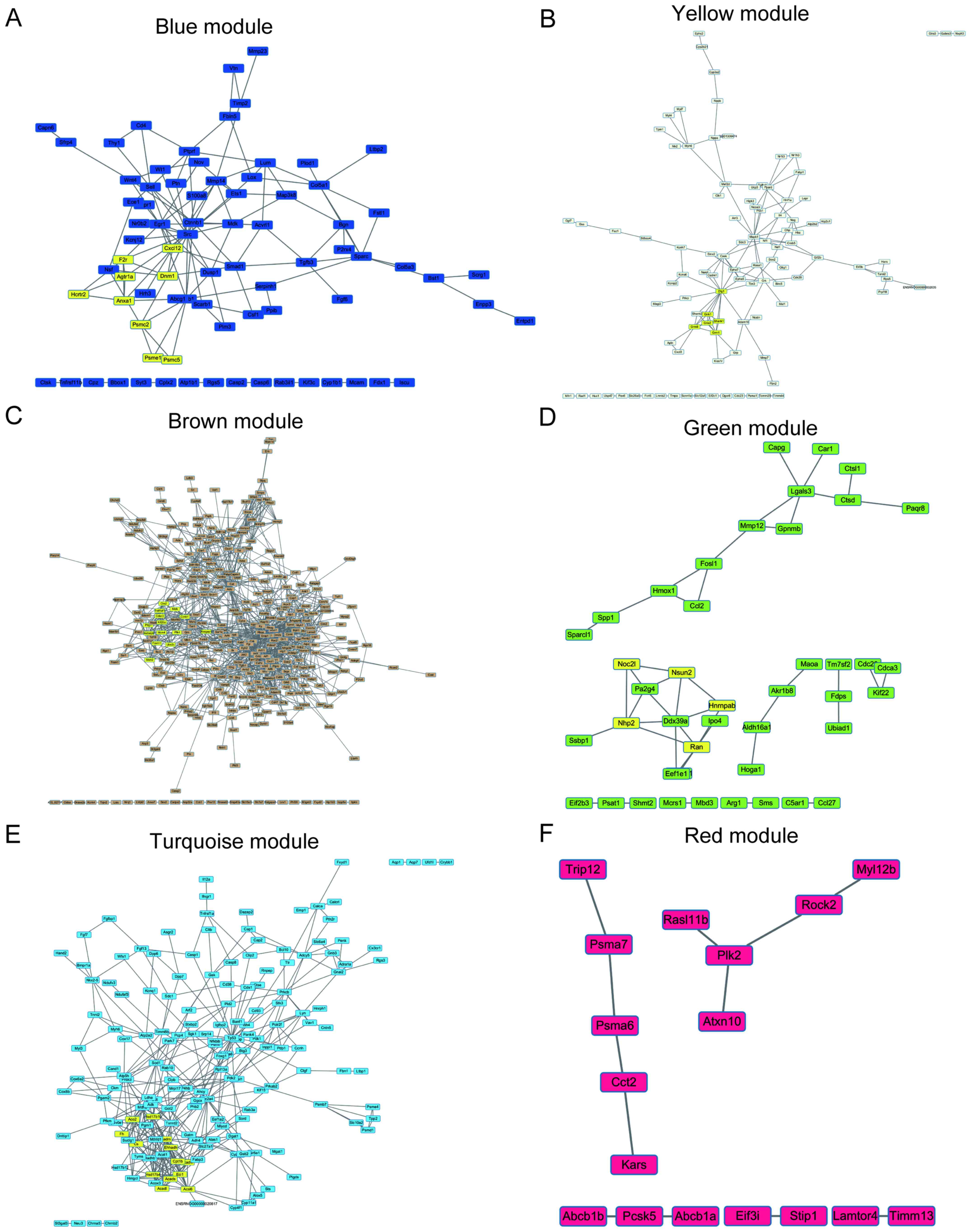
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bader GD and Hogue CW: An automated method
for finding molecular complexes in large protein interaction
networks. BMC Bioinformatics. 4:22003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Haendeler J and Berk BC: Angiotensin II
mediated signal transduction. Important role of tyrosine kinases.
Regul Pept. 95:1–7. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
McMullen JR, Shioi T, Zhang L, Tarnavski
O, Sherwood MC, Kang PM and Izumo S: Phosphoinositide
3-kinase(p110alpha) plays a critical role for the induction of
physiological, but not pathological, cardiac hypertrophy. Proc Natl
Acad Sci USA. 100:12355–12360. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Raaf L, Noll C, Mel Cherifi H, Samuel JL,
Delcayre C, Delabar JM, Benazzoug Y and Janel N: Myocardial
fibrosis and TGFB expression in hyperhomocysteinemic rats. Mol Cell
Biochem. 347:63–70. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shanaki M, Hossein-Nezhad A, Meshkani R,
Beigy M, Shirzad M, Pasalar P and Golmohammadi T: Effects of
resveratrol on crosstalk between canonical B-Catenin/Wnt and FOXO
pathways in coronary artery disease patients with metabolic
syndrome: A case control study. Iran J Pharm Res. 15:547–559.
2016.PubMed/NCBI
|
|
27
|
Mueller-Hennessen M, Sigl J, Fuhrmann JC,
Witt H, Reszka R, Schmitz O, Kastler J, Fischer JJ, Müller OJ,
Giannitsis E, et al: Metabolic profiles in heart failure due to
non-ischemic cardiomyopathy at rest and under exercise. ESC Heart
Fail. 4:178–189. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen J, Wang D, Wang F, Shi S, Chen Y,
Yang B, Tang Y and Huang C: Exendin-4 inhibits structural
remodeling and improves Ca2+ homeostasis in rats with
heart failure via the GLP-1 receptor through the eNOS/cGMP/PKG
pathway. Peptides. 90:69–77. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ziehr J, Sheibani N and Sorenson CM:
Alterations in cell-adhesive and migratory properties of proximal
tubule and collecting duct cells from bcl-2 -/- mice. Am J Physiol
Renal Physiol. 287:F1154–F1163. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chetty S, Engquist EN, Mehanna E, Lui KO,
Tsankov AM and Melton DA: A Src inhibitor regulates the cell cycle
of human pluripotent stem cells and improves directed
differentiation. J Cell Biol. 210:1257–1268. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Pandey P, Hawkes W, Hu J, Megone WV,
Gautrot J, Anilkumar N, Zhang M, Hirvonen L, Cox S, Ehler E, et al:
Cardiomyocytes sense matrix rigidity through a combination of
muscle and non-muscle myosin contractions. Dev Cell. 44(326–336):
e3232018.
|
|
32
|
Laprise P, Viel A and Rivard N: Human
homolog of disc-large is required for adherens junction assembly
and differentiation of human intestinal epithelial cells. J Biol
Chem. 279:10157–10166. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Rivera C, Simonson SJ, Yamben IF, Shatadal
S, Nguyen MM, Beurg M, Lambert PF and Griep AE: Requirement for
Dlgh-1 in planar cell polarity and skeletogenesis during vertebrate
development. PLoS One. 8:e544102013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Elshourbagy NA, Near JC, Kmetz PJ, Wells
TN, Groot PH, Saxty BA, Hughes SA, Franklin M and Gloger IS:
Cloning and expression of a human ATP-citrate lyase cDNA. Eur J
Biochem. 204:491–499. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Mariño G, Pietrocola F, Kong Y, Eisenberg
T, Hill JA, Madeo F and Kroemer G: Dimethyl α-ketoglutarate
inhibits maladaptive autophagy in pressure overload-induced
cardiomyopathy. Autophagy. 10:930–932. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Dallol A, Hesson LB, Matallanas D, Cooper
WN, O'Neill E, Maher ER, Kolch W and Latif F: RAN GTPase is a
RASSF1A effector involved in controlling microtubule organization.
Curr Biol. 19:1227–1232. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Moss DK, Wilde A and Lane JD: Dynamic
release of nuclear RanGTP triggers TPX2-dependent microtubule
assembly during the apoptotic execution phase. J Cell Sci.
122:644–655. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Jiang X, Zhang D, Zhang H, Huang Y and
Teng M: Corrigendum: Role of Ran-regulated nuclear-cytoplasmic
trafficking of pVHL in the regulation of microtubular
stability-mediated HIF-1α in hypoxic cardiomyocytes. Sci Rep.
5:113072015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Fujita T and Ishikawa Y: Apoptosis in
heart failure. -The role of the β-adrenergic receptor-mediated
signaling pathway and p53-mediated signaling pathway in the
apoptosis of cardiomyocytes. Circ J. 75:1811–1818. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Oka T, Morita H and Komuro I: Novel
molecular mechanisms and regeneration therapy for heart failure. J
Mol Cell Cardiol. 92:46–51. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Teng H, Sui X, Zhou C, Shen C, Yang Y,
Zhang P, Guo X and Huo R: Fatty acid degradation plays an essential
role in proliferation of mouse female primordial germ cells via the
p53-dependent cell cycle regulation. Cell Cycle. 15:425–431. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Li J, Ma W, Wang PY, Hurley PJ, Bunz F and
Hwang PM: Polo-like kinase 2 activates an antioxidant pathway to
promote the survival of cells with mitochondrial dysfunction. Free
Radic Biol Med. 73:270–277. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Matsumoto T, Wang PY, Ma W, Sung HJ,
Matoba S and Hwang PM: Polo-like kinases mediate cell survival in
mitochondrial dysfunction. Proc Natl Acad Sci USA. 106:14542–14546.
2009. View Article : Google Scholar : PubMed/NCBI
|