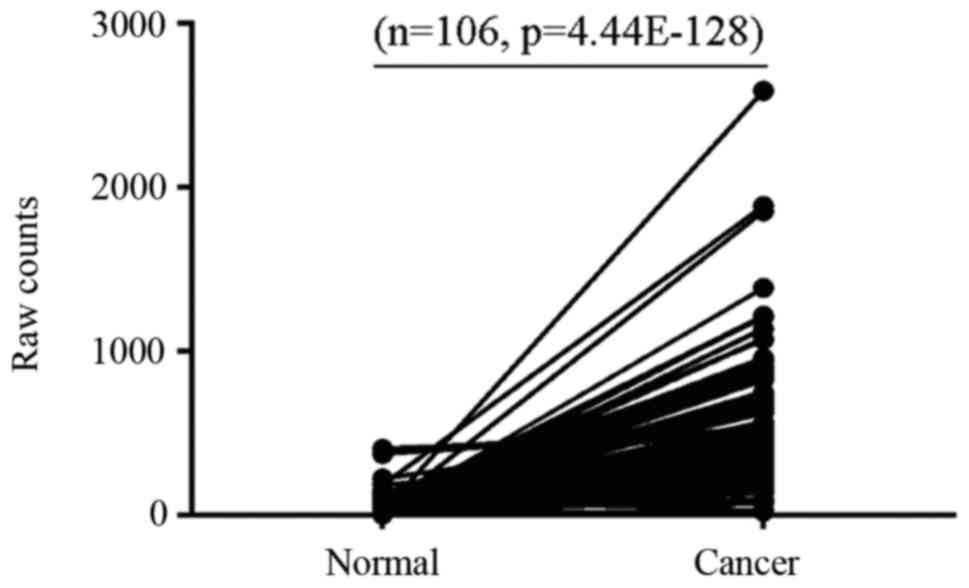
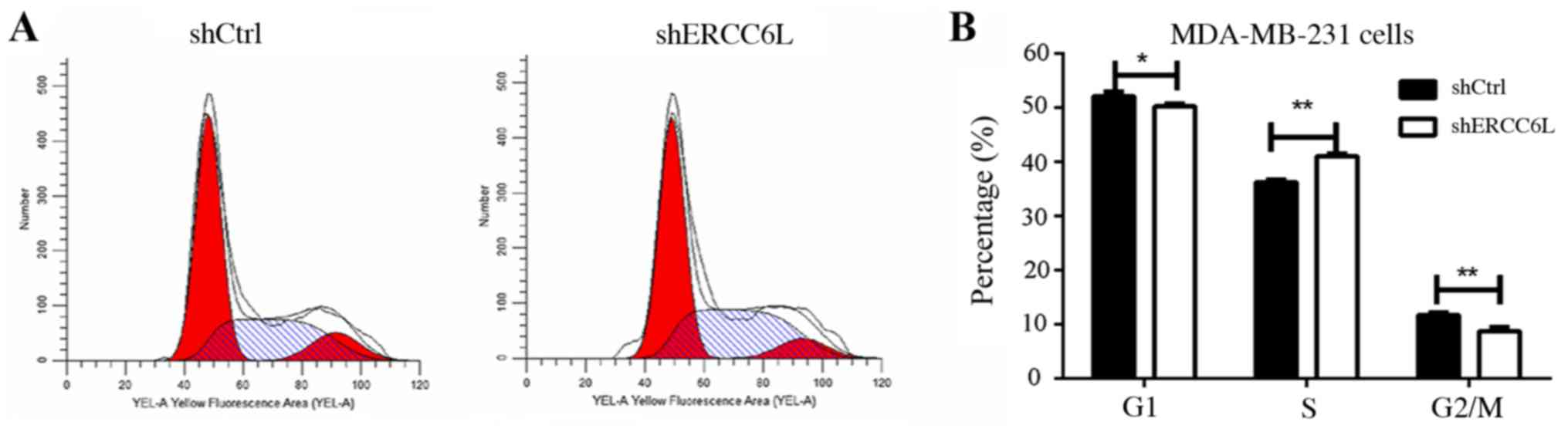
|
1
|
Torre LA, Siegel RL, Ward EM and Jemal A:
Global cancer incidence and mortality rates and trends-an update.
Cancer Epidemiol Biomarkers Prev. 25:16–27. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Johann DJ, Rodriguez-Canales J, Mukherjee
S, Prieto DA, Hanson JC, Emmert-Buck M and Blonder J: Approaching
solid tumor heterogeneity on a cellular basis by tissue proteomics
using laser capture microdissection and biological mass
spectrometry. J Proteome Res. 8:2310–2318. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Polyak K: Heterogeneity in breast cancer.
J Clin Invest. 121:3786–3788. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Park SY, Gönen M, Kim HJ, Michor F and
Polyak K: Cellular and genetic diversity in the progression of in
situ human breast carcinomas to an invasive phenotype. J Clin
Invest. 120:636–644. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Beca F and Polyak K: Intratumor
heterogeneity in breast cancer. Adv Exp Med Biol. 882:169–189.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Karlsson E, Appelgren J, Solterbeck A,
Bergenheim M, Alvariza V and Bergh J: Breast cancer during
follow-up and progression - A population based cohort on new
cancers and changed biology. Eur J Cancer. 50:2916–2924. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Badve S and Gökmen-Polar Y: Tumor
heterogeneity in breast cancer. Adv Anat Pathol. 22:294–302. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bai X, Zhang E, Ye H, Nandakumar V, Wang
Z, Chen L, Tang C, Li J, Li H, Zhang W, et al: PIK3CA and TP53 gene
mutations in human breast cancer tumors frequently detected by ion
torrent DNA sequencing. PLoS One. 9:e993062014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Van Keymeulen A, Lee MY, Ousset M, Brohée
S, Rorive S, Giraddi RR, Wuidart A, Bouvencourt G, Dubois C, Salmon
I, et al: Reactivation of multipotency by oncogenic PIK3CA induces
breast tumour heterogeneity. Nature. 525:119–123. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Koren S, Reavie L, Couto JP, De Silva D,
Stadler MB, Roloff T, Britschgi A, Eichlisberger T, Kohler H, Aina
O, et al: PIK3CA(H1047R) induces multipotency and multi-lineage
mammary tumours. Nature. 525:114–118. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
King KL and Cidlowski JA: Cell cycle
regulation and apoptosis. Annu Rev Physiol. 60:601–617. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Alenzi FQ: Links between apoptosis,
proliferation and the cell cycle. Br J Biomed Sci. 61:99–102. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Foote FW Jr and Stewart FW: A histologic
classification of carcinoma of the breast. Surgery. 19:74–99.
1946.PubMed/NCBI
|
|
15
|
McGranahan N and Swanton C: Biological and
therapeutic impact of intratumor heterogeneity in cancer evolution.
Cancer Cell. 27:15–26. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Pu SY, Yu Q, Wu H, Jiang JJ, Chen XQ, He
YH and Kong QP: ERCC6L, a DNA helicase, is involved in cell
proliferation and associated with survival and progress in breast
and kidney cancers. Oncotarget. 8:42116–42124. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Baumann C, Körner R, Hofmann K and Nigg
EA: PICH, a centromere-associated SNF2 family ATPase, is regulated
by Plk1 and required for the spindle checkpoint. Cell. 128:101–114.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lin Z, Zhang X, Tuo J, Guo Y, Green B,
Chan CC, Tan W, Huang Y, Ling W, Kadlubar FF, et al: A variant of
the Cockayne syndrome B gene ERCC6 confers risk of lung cancer. Hum
Mutat. 29:113–122. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Abbasi R, Ramroth H, Becher H, Dietz A,
Schmezer P and Popanda O: Laryngeal cancer risk associated with
smoking and alcohol consumption is modified by genetic
polymorphisms in ERCC5, ERCC6 and RAD23B but not by polymorphisms
in five other nucleotide excision repair genes. Int J Cancer.
125:1431–1439. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu JW, He CY, Sun LP, Xu Q, Xing CZ and
Yuan Y: The DNA repair gene ERCC6 rs1917799 polymorphism is
associated with gastric cancer risk in Chinese. Asian Pac J Cancer
Prev. 14:6103–6108. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chang CH, Chiu CF, Wang HC, Wu HC, Tsai
RY, Tsai CW, Wang RF, Wang CH, Tsou YA and Bau DT: Significant
association of ERCC6 single nucleotide polymorphisms with bladder
cancer susceptibility in Taiwan. Anticancer Res. 29:5121–5124.
2009.PubMed/NCBI
|
|
22
|
Ma H, Hu Z, Wang H, Jin G, Wang Y, Sun W,
Chen D, Tian T, Jin L, Wei Q, et al: ERCC6/CSB gene polymorphisms
and lung cancer risk. Cancer Lett. 273:172–176. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ramaniuk VP, Nikitchenko NV, Savina NV,
Kuzhir TD, Rolevich AI, Krasny SA, Sushinsky VE and Goncharova RI:
Polymorphism of DNA repair genes OGG1, XRCC1, XPD and ERCC6 in
bladder cancer in Belarus. Biomarkers. 19:509–516. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yin Y, Tang L, Zhang J, Tang B and Li Z:
Molecular cloning and gene expression analysis of Ercc6l in Sika
Deer (Cervus nippon hortulorum). PLoS One. 6:e209292011. View Article : Google Scholar : PubMed/NCBI
|